Abstract
Total plasma homocysteine concentration (tHcy) is a biomarker for atherothrombotic disease, but causality remains uncertain. Polymorphisms in the genes involved in methionine metabolism explain only a small fraction of the heritability of tHcy levels. In a genome-wide association study, we examined the genetic determinants of tHcy using a 2-stage design. First, 283 437 single nucleotide polymorphisms (SNPs) were tested for association with tHcy in 387 persons recruited from 21 large Spanish families. Of those, 17 SNPs showed equal or stronger association with tHcy level compared with the MTHFR 677C>T SNP (β = 0.10, P = .0001). Second, a replication analysis of these 17 SNPs was performed in patients with premature myocardial infarction (n = 1238). Novel associations were found for SNPs near the ZNF366 gene (lead SNP rs7445013; discovery stage: adjusted β = −0.12, P = 5.30 × 10−6, replication stage: adjusted β = −0.13, P = .004) and the PTPRD gene (lead SNP rs973117; discovery stage: adjusted β = 0.11, P = 5.5 × 10−6, replication stage: adjusted β = 0.10, P = .005). These associations were independent of known confounders, including creatinine clearance and plasma fibrinogen concentration. Our findings implicate novel pathways in homocysteine metabolism, and highlight the need for investigation of the associated genes in the etiology of vascular diseases.
Introduction
Homocysteine is formed during the metabolism of the essential amino acid methionine. Extreme elevations of total plasma homocysteine (tHcy) (free and protein-bound) are observed among children with homocystinuria, a condition closely associated with the occurrence of vascular disease and venous thrombosis.1 In the general population, moderate elevation of tHcy levels is associated with increased risk of coronary artery disease (CAD),2,3 stroke,3 and venous thromboembolism (VTE)4 but causality has not been established.5,6
The issue of whether tHcy is causally related to atherothrombotic events has been addressed by genetic association studies of the 677C>T single nucleotide polymorphism (SNP) in the methylenetetrahydrofolate reductase (MTHFR) gene (rs1801133). The 677C>T substitution leads to a nonsynonymous amino acid change resulting in reduced enzymatic activity of MTHFR.7 Recent meta-analyses of this genetic variant show weak support for homocysteine as an etiologically important factor for CAD, whereas the support for a causal relationship with stroke and VTE is stronger.4,5,8 In contrast with the monogeneic homocystinuria,8 moderate elevations in tHcy levels are believed to reflect the effects of several genetic variants encoding enzymes involved in methionine metabolism,9,10 or their B-vitamin cofactors (folate and vitamin B12)11 or the effects of renal impairment.12
Studies in related persons have indicated that the tHcy concentration has a strong genetic component, with an estimated total heritability of tHcy ranging from 25% in family studies to 57% in twin studies.13,14 Linkage study signals typically reflect the effects of rare genetic variants and several such studies have reported linkage at different chromosomal regions: 11q23, 12q14, 13q31, and 16q12.13,15 In addition, common SNPs in the MTHFR, methionine synthase (MTR),16 methionine synthase reductase (MTRR),14 and CBS16 genes have been associated with plasma tHcy, of which the most well studied is MTHFR 677C>T.8,14,17,18 However, the high heritability of tHcy levels remains largely unexplained by common variants in the genes involved in methionine metabolism detected to date. Accordingly, we searched for novel genetic determinants of plasma tHcy using a genome-wide set of SNPs in the GAIT (Genetic Analysis of Idiopathic Thrombophilia) study and sought to replicate any associations detected in the PROCARDIS study of patients with myocardial infarction (MI).
Methods
The GAIT discovery cohort included 387 persons belonging to 21 Spanish families; 12 were selected through a proband with idiopathic thrombophilia, whereas the remaining 9 families were randomly selected (Table 1).19 The plasma tHcy concentration was measured by high-performance liquid chromatography (Millipore) and fluorescence detection (Kontron Instruments).13 Genotyping of a genome-wide set of 307 984 SNPs was performed using the Illumina Infinium 317k Beadchip. SNPs were filtered out if they had a genotype call rate less than 95%, a minor allele frequency (MAF) less than 2.5%, or failed the Hardy-Weinberg (HWE) test of expected genotype distribution (P < 5 × 10−7). After quality control, 283 437 SNPs remained for analysis.
Baseline characteristics of the GAIT and PROCARDIS study cohorts
| Characteristic . | GAIT study (n = 399) . | PROCARDIS study (n = 1238) . |
|---|---|---|
| Median age, y (range) | 37.7 (1-87) | 62 (57-66) |
| Women, no. (%) | 217 (54.4) | 236 (18.7) |
| Current smoker, no. (%) | 151 (37.8) | 708 (56.0) |
| Lipid-lowering treatment, no. (%) | 28 (7.0) | 910 (71.9) |
| Antihypertensive treatment, no. (%) | 30 (7.5) | 1010 (79.8) |
| Mean body mass index, kg/m2 (SD) | 24.8 (4.9) | 28.1 (4.12) |
| Mean HDL cholesterol (SD) | 1.42 (0.37) | 1.14 (0.29) |
| Mean LDL cholesterol (SD) | 3.27 (1.0) | 2.82 (0.81) |
| Mean triglycerides (SD) | 1.12 (0.88) | 1.87 (1.14) |
| Median fibrinogen, g/L (interquartile range) | 3.2 (1.6-5.8) | 3.7 (3.3-4.3) |
| Median homocysteine, total, mg/L (interquartile range) | 7.74 (2.7-23.8) | 12.2 (9.9-14.9) |
| Median estimated glomerular filtration rate, mL/min/kg2 (interquartile range) | NA | 74.8 (62-89) |
| Characteristic . | GAIT study (n = 399) . | PROCARDIS study (n = 1238) . |
|---|---|---|
| Median age, y (range) | 37.7 (1-87) | 62 (57-66) |
| Women, no. (%) | 217 (54.4) | 236 (18.7) |
| Current smoker, no. (%) | 151 (37.8) | 708 (56.0) |
| Lipid-lowering treatment, no. (%) | 28 (7.0) | 910 (71.9) |
| Antihypertensive treatment, no. (%) | 30 (7.5) | 1010 (79.8) |
| Mean body mass index, kg/m2 (SD) | 24.8 (4.9) | 28.1 (4.12) |
| Mean HDL cholesterol (SD) | 1.42 (0.37) | 1.14 (0.29) |
| Mean LDL cholesterol (SD) | 3.27 (1.0) | 2.82 (0.81) |
| Mean triglycerides (SD) | 1.12 (0.88) | 1.87 (1.14) |
| Median fibrinogen, g/L (interquartile range) | 3.2 (1.6-5.8) | 3.7 (3.3-4.3) |
| Median homocysteine, total, mg/L (interquartile range) | 7.74 (2.7-23.8) | 12.2 (9.9-14.9) |
| Median estimated glomerular filtration rate, mL/min/kg2 (interquartile range) | NA | 74.8 (62-89) |
The replication cohort consisted of 1238 patients enrolled in the PROCARDIS study, all of whom had suffered MI before or at the age of 65 years (Table 1).20,21 Recruitment was carried out in Germany, Italy, Sweden, and the United Kingdom. Total homocysteine was determined by fluorescence high-performance liquid chromatography.21 Genotyping was performed using the Illumina Infinium II Human 1M Beadchip. All SNPs selected for in silico replication analysis in PROCARDIS conformed to HWE (P > .05), with the exception of rs10488697, for which the P value was .01. The average genotype call rate for the SNPs prioritized for replication was 98.7%.
GAIT protocols were approved by the Institutional Review Board of the Hospital de la Santa Creu i Sant Pau, and the PROCARDIS protocol was approved by the Ethics Committees of the participating institutions. All subjects gave informed consent in accordance with the Declaration of Helsinki.
The statistical analyses in GAIT were carried out using variance components-based methodology and SOLAR Version 4.0 software package.22 Variables associated with plasma tHcy including age, sex, body mass index, smoking, and MTHFR 677C>T were included in the variance components framework as linear predictors of log-transformed plasma tHcy level. Measured genotype analysis was used for association testing, assuming an additive model of allelic effect.23 A P value less than .0001 was required for a SNP to be included in the replication study. The choice of significance level was based on the strength of the MTHFR 677C>T SNP association with plasma tHcy, that is, SNPs in the GAIT GWAS with a similar effect size and frequency as MTHFR 677C>T were prioritized for replication (Table 2).
Top SNP associations with plasma tHcy concentration in GAIT and replication results in PROCARDIS
| CHR . | SNP . | Position, kbp . | Nearest gene* . | A1 . | GAIT, n = 387 . | PROCARDIS, n = 1238 . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| MAF, % . | β . | P . | MAF, % . | β . | P† . | |||||
| 1 | rs1801133 | 11 779 | MTHFR | G | 39 | 0.11 | 9.30 × 10−5 | 36 | 0.12 | .003 |
| 3 | rs10934885 | 130 782 | PLXND1 | T | 27 | −0.13 | 2.10 × 10−5 | 22 | −0.01 | .905 |
| 5 | rs7445013 | 71 734 | ZNF366 | A | 45 | −0.12 | 5.30 × 10−6 | 45 | −0.11 | .009 |
| 5 | rs2338216 | 71 738 | ZNF366 | C | 45 | −0.13 | 3.30 × 10−6 | 45 | −0.10 | .011 |
| 5 | rs4703872 | 71 759 | ZNF366 | C | 45 | −0.12 | 5.30 × 10−6 | 45 | −0.10 | .010 |
| 6 | rs2071481 | 32 928 | TAP1 | T | 12 | −0.16 | 8.20 × 10−5 | 10 | −0.06 | .401 |
| 6 | rs154986 | 32 987 | HLA-DMB | G | 10 | 0.17 | 7.60 × 10−5 | 7 | −0.08 | .328 |
| 7 | rs2701039 | 107 411 | LAMB1 | G | 34 | −0.12 | 2.20 × 10−5 | 39 | −0.07 | .112 |
| 7 | rs1559971 | 124 749 | T | 25 | 0.13 | 1.00 × 10−5 | 33 | 0.01 | .765 | |
| 7 | rs7795867 | 124 765 | T | 19 | 0.13 | 5.40 × 10−5 | 25 | 0 | .995 | |
| 9 | rs973117 | 8 990 | PTPRD | C | 33 | 0.11 | 5.50 × 10−6 | 36 | 0.09 | .042 |
| 11 | rs10488697 | 113 475 | ZBTB16 | A | 11 | 0.17 | 2.20 × 10−5 | 8 | 0.07 | .311 |
| 12 | rs2190679 | 124 886 | G | 33 | 0.11 | 6.60 × 10−5 | 36 | −0.02 | .564 | |
| 12 | rs1859943 | 124 893 | G | 36 | 0.11 | 6.40 × 10−5 | 41 | −0.01 | .892 | |
| 15 | rs10162946 | 21 689 | G | 10 | −0.19 | 2.00 × 10−5 | 13 | 0.02 | .721 | |
| 18 | rs9962181 | 73 441 | GALR1 | T | 13 | 0.14 | 9.20 × 10−5 | 10 | −0.04 | .547 |
| 19 | rs2250066 | 56 221 | KLK11 | C | 12 | 0.14 | 8.90 × 10−5 | 15 | 0 | .945 |
| 21 | rs2826520 | 21 085 | C | 33 | −0.11 | 3.20 × 10−5 | 32 | 0.02 | .725 | |
| CHR . | SNP . | Position, kbp . | Nearest gene* . | A1 . | GAIT, n = 387 . | PROCARDIS, n = 1238 . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| MAF, % . | β . | P . | MAF, % . | β . | P† . | |||||
| 1 | rs1801133 | 11 779 | MTHFR | G | 39 | 0.11 | 9.30 × 10−5 | 36 | 0.12 | .003 |
| 3 | rs10934885 | 130 782 | PLXND1 | T | 27 | −0.13 | 2.10 × 10−5 | 22 | −0.01 | .905 |
| 5 | rs7445013 | 71 734 | ZNF366 | A | 45 | −0.12 | 5.30 × 10−6 | 45 | −0.11 | .009 |
| 5 | rs2338216 | 71 738 | ZNF366 | C | 45 | −0.13 | 3.30 × 10−6 | 45 | −0.10 | .011 |
| 5 | rs4703872 | 71 759 | ZNF366 | C | 45 | −0.12 | 5.30 × 10−6 | 45 | −0.10 | .010 |
| 6 | rs2071481 | 32 928 | TAP1 | T | 12 | −0.16 | 8.20 × 10−5 | 10 | −0.06 | .401 |
| 6 | rs154986 | 32 987 | HLA-DMB | G | 10 | 0.17 | 7.60 × 10−5 | 7 | −0.08 | .328 |
| 7 | rs2701039 | 107 411 | LAMB1 | G | 34 | −0.12 | 2.20 × 10−5 | 39 | −0.07 | .112 |
| 7 | rs1559971 | 124 749 | T | 25 | 0.13 | 1.00 × 10−5 | 33 | 0.01 | .765 | |
| 7 | rs7795867 | 124 765 | T | 19 | 0.13 | 5.40 × 10−5 | 25 | 0 | .995 | |
| 9 | rs973117 | 8 990 | PTPRD | C | 33 | 0.11 | 5.50 × 10−6 | 36 | 0.09 | .042 |
| 11 | rs10488697 | 113 475 | ZBTB16 | A | 11 | 0.17 | 2.20 × 10−5 | 8 | 0.07 | .311 |
| 12 | rs2190679 | 124 886 | G | 33 | 0.11 | 6.60 × 10−5 | 36 | −0.02 | .564 | |
| 12 | rs1859943 | 124 893 | G | 36 | 0.11 | 6.40 × 10−5 | 41 | −0.01 | .892 | |
| 15 | rs10162946 | 21 689 | G | 10 | −0.19 | 2.00 × 10−5 | 13 | 0.02 | .721 | |
| 18 | rs9962181 | 73 441 | GALR1 | T | 13 | 0.14 | 9.20 × 10−5 | 10 | −0.04 | .547 |
| 19 | rs2250066 | 56 221 | KLK11 | C | 12 | 0.14 | 8.90 × 10−5 | 15 | 0 | .945 |
| 21 | rs2826520 | 21 085 | C | 33 | −0.11 | 3.20 × 10−5 | 32 | 0.02 | .725 | |
P values were adjusted for age, sex, smoking, BMI, and the MTHFR rs1801133 SNP.
CHR indicates chromosome; MAF, minor allele frequency; A1, major allele; and β, standardized β coefficient, obtained in the linear regression analyses.
No nearest gene given for SNPs more than 100 kbp from the closest protein-coding gene.
Adjusted for age, sex, smoking, BMI, country, and the MTHFR rs1801133 SNP. For the rs1801133 SNP, the above adjustments were applied, with the obvious exception of SNP rs1801133 itself.
In PROCARDIS, the log-transformed plasma tHcy concentration was tested for association with prioritized SNPs using linear regression, assuming an additive genetic model, with adjustments for age, sex, body mass index, smoking status, country of origin, and MTHFR 677C>T genotype. The MTR SNP rs1805087was tagged using SNP rs16879418. The threshold for statistical significance was set at .05 in the replication study. Statistical analyses were carried out in PLINK 1.0524 and SPSS 16.0 for Windows, and calculations of linkage disequilibrium were performed in Haploview 4.0.25
Results
Table 1 shows selected characteristics of participants in the GAIT study and the PROCARDIS study with data available on genotypes and tHcy levels. Of a total of 283 437 SNPs tested in GAIT, 18 SNPs, including MTHFR 677C>T, were associated with plasma tHcy at P < .0001 (Table 2). The strongest associations were observed for 3 SNPs positioned between 20- and 30-kb upstream of the ZNF366 gene (P = 5 × 10−6; Table 2). The SNPs identified as associated with plasma tHcy levels in GAIT were tested for replication in patients with MI.
We replicated the associations with tHcy for SNP rs1801133 in the MTHFR gene (P = .003), for SNP rs973117 located 18-kb upstream of the PTPRD gene (P = .04), and for the SNPs adjacent to the ZNF366 gene (lead SNP rs7445013, P = .009), with a consistent direction of effect (Table 2). Plasma tHcy concentration (log transformed) correlated with both plasma fibrinogen (Pearson R2 = 0.11) and eGFR (Pearson R2 = −0.24) in PROCARDIS. However, the association between genetic variants and tHcy concentration persisted after additional adjustment for plasma fibrinogen and eGFR (MTHFR rs1801133, β = 0.12, P = .001; ZNF366 rs7445013, β = −0.13, P = .0004; PTPRD rs973117, β = 0.10, P = .005). The geometric mean tHcy concentration was 12.1, 12.4, and 13.8 μM, respectively, for CC, CT, and TT genotypes of MTHFR rs1801133; 11.9, 12.5, and 12.8 μM, respectively, for AA, GA, and GG genotypes of ZNF366 rs7445013; and 12.1, 12.6, and 12.9 μM, respectively, for CC, AC and AA genotypes of PTPRD rs973117.
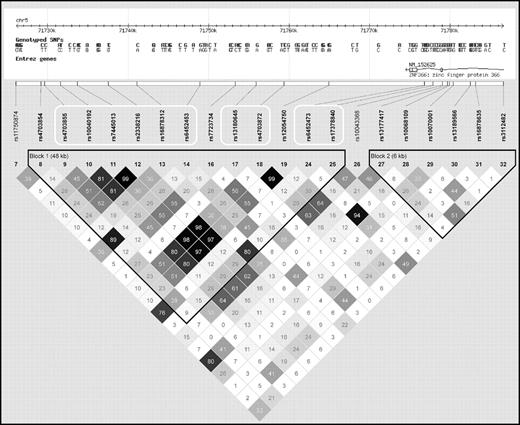
Analyses of SNPs positioned near ZNF366 rs7445013 (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article) and PTPRD rs973117 (supplemental Table 2) revealed additional significant SNP associations with plasma tHcy. Importantly, not all of the additional tHcy-associated SNPs in the ZNF366 locus were strongly linked to the lead rs7445013 SNP (Figure 1). In contrast, the nominally associated SNPs near PTPRD rs973117 were all in near complete allelic association (data not shown).
Linkage disequilibrium between all genotyped SNPs positioned in or near the ZNF366 gene in the PROCARDIS study. The white frames denote SNPs associated with plasma tHcy concentration at a P < .05.
Linkage disequilibrium between all genotyped SNPs positioned in or near the ZNF366 gene in the PROCARDIS study. The white frames denote SNPs associated with plasma tHcy concentration at a P < .05.
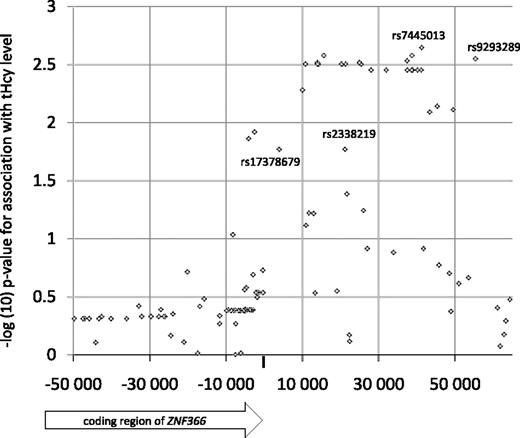
The ZNF366 locus was subsequently saturated with SNPs inferred by imputation, using the CEU HapMap phase II samples as reference and the PROCARDIS MI patient sample as target. Using PLINK, only SNPs with an average maximal posterior probability greater than 90% were kept, which resulted in 52 usable SNPs that were tested for association with the plasma tHcy concentration together with the experimental SNPs. Overall, most of the imputed SNPs showed similar association test-statistics compared with rs7445013 (Figure 2). However, SNPs rs9293289, rs2338219, and rs17378679 were significantly associated with plasma tHcy levels with effect size larger than that for rs7445013. Specifically, SNP rs17378679 is less common than rs7445013 (MAF = 14%), is positioned close to the 3′ UTR region of ZNF366, and shows markedly stronger association test statistics with the plasma tHcy concentration (β = −0.18, P = .019).
Association of plasma tHcy concentration with experimental and imputed SNPs in the PROCARDIS study. SNPs rs9293289, rs2338219, and rs17378679 were significantly associated with plasma tHcy (P < .05) and had an effect size larger than that of rs7445013 (β = −0.10).
Association of plasma tHcy concentration with experimental and imputed SNPs in the PROCARDIS study. SNPs rs9293289, rs2338219, and rs17378679 were significantly associated with plasma tHcy (P < .05) and had an effect size larger than that of rs7445013 (β = −0.10).
To investigate the relative contribution to the variation in plasma tHcy made by SNPs ZNF366 rs7445013 and PTPRD rs973117, models for each of the 2 studies were generated that also took other suggested loci for tHcy as well as nongenetic factors into account (Table 3). The result obtained for ZNF366 rs7445013 suggested that this SNP contributes to 3.8% of the variation in GAIT, whereas the corresponding figure in PROCARDIS MI patients was 0.9%. The PTPRD rs973117 SNP contributed to 1.3% of the variation in GAIT and to 0.5% of the variation in PROCARDIS.
Partition of plasma tHcy variance according to clinical and genetic variables obtained by multiple stepwise linear regression analysis in the GAIT and PROCARDIS samples
| Category . | Category R2 . | |
|---|---|---|
| GAIT . | PROCARDIS . | |
| Age, sex, smoking, BMI, country | 0.188 | 0.063 |
| Plasma fibrinogen | 0.008 | 0.013 |
| eGFR | NA | 0.035 |
| MTHFR (rs1801133), MTR (rs1805087), MTRR (rs16879418), CBS (rs6586282) | 0.050 | 0.015 |
| CPS1 (rs7422339), MUT (rs4267943), NOX4 (rs11018628), DPEP1 (rs460879) | 0.002 | 0.011 |
| ZNF366 (rs7445013) | 0.038 | 0.009 |
| PTPRD (rs973117) | 0.013 | 0.005 |
| Total | 0.299 | 0.151 |
| Category . | Category R2 . | |
|---|---|---|
| GAIT . | PROCARDIS . | |
| Age, sex, smoking, BMI, country | 0.188 | 0.063 |
| Plasma fibrinogen | 0.008 | 0.013 |
| eGFR | NA | 0.035 |
| MTHFR (rs1801133), MTR (rs1805087), MTRR (rs16879418), CBS (rs6586282) | 0.050 | 0.015 |
| CPS1 (rs7422339), MUT (rs4267943), NOX4 (rs11018628), DPEP1 (rs460879) | 0.002 | 0.011 |
| ZNF366 (rs7445013) | 0.038 | 0.009 |
| PTPRD (rs973117) | 0.013 | 0.005 |
| Total | 0.299 | 0.151 |
BMI indicates body mass index; and NA, not available.
Discussion
In the present study, we performed a genome-wide SNP association study of plasma tHcy concentration and identified SNPs adjacent to and in the ZNF366 and PTPRD genes as novel determinants of plasma tHcy. In our replication sample, the lead SNP rs7445013 at the ZNF366 locus was an equally strong predictor of plasma tHcy concentration as MTHFR 677C>T. An advantage of using a sample of MI patients for replication is that it allows for testing the SNP-tHcy associations in the context of impaired renal function, low-grade inflammation, and hypercoagulability. Therefore it is important that the genotype-phenotype associations were not attenuated by additional adjustment for eGFR and plasma fibrinogen concentration. In contrast, the predictive values of the linear regression models testing lead SNPs in the 2 loci were improved. With the exception of MTHFR, none of the genes positioned close to the associated SNPs have previously been implicated in the regulation of plasma tHcy levels, nor do they have any known function in methionine metabolism.
To date, common sequence variants in a total of 8 genes have been suggested to be associated with plasma tHcy. These include the MTHFR, MTR, MTRR, and CBS genes in the methionine synthesis pathway and 4 additional loci that were recently identified in the Women's Genome Health Study.26 The newly identified loci were positioned in or adjacent to the CPS1, MUT, NOX4, and DPEP1 genes. Taken together, the associated SNPs in these 8 genes accounted for a total of 2.6% of the variation in tHcy in the PROCARDIS sample, whereas the corresponding figure was 5.2% in GAIT (Table 3). In addition, we have previously reported a linkage signal on chromosome 11 in the GAIT project.14 Fine-mapping of this genomic region identified a haplotype built with 10 SNPs within the NNMT gene that was strongly associated with higher tHcy levels.14 This is a rare haplotype (only 18 carriers among 398 persons) that explained 5% of the tHcy variation in GAIT, whereas the whole QTL on chromosome 11 accounted for as much as 19%. However, SNPs within or nearby the NNMT gene included in the GAIT GWAS did not show significant associations with the plasma tHcy concentration (data not shown). This latter observation indicates that the power of the GAIT GWAS was too low to detect any rare variants in this locus. Taken together, the current and previous studies on GAIT using SNP-based genotype-phenotype association analysis and microsatellite marker-based linkage analysis, respectively, demonstrate that common and rare variants at different loci contribute to the genetic architecture of an intermediate phenotype such as plasma tHcy concentration. Collectively, common and rare homocysteine-associated genetic variants identified to date seem to account for at least 15% of the variation in plasma tHcy in GAIT. Accordingly, they account for a relatively limited proportion of the estimated heritability for plasma tHcy.
One limitation of the present study was the inability to adjust for folate and B-vitamin status, which is known to influence the plasma tHcy concentration. A recent tHcy-lowering trial in patients with CAD demonstrated a decrease by 30% after 1 year of intervention in the groups receiving folic acid and vitamin B12.27 Thus, intake of folic acid and B-vitamins through food fortification or oral capsules is an important confounder when addressing genetic influences on tHcy level, and the inability to adjust for this confounder may result in an underestimation of the genetic effects on plasma tHcy concentration. However, we do not expect the folate and B-vitamin status to differ significantly between the study cohorts, as both the GAIT and PROCARDIS studies were conducted in populations without mandatory folic acid fortification. In this context, it is interesting that a recent genome-wide association study demonstrated novel associations of vitamin B6 and B12 concentration with SNPs in the FUT2 and ALPL genes.28 These findings suggest that vitamin-B metabolism is genetically determined and imply the existence of interindividual variation in the effectiveness of tHcy-lowering treatment with B-vitamins or food fortification, which could influence the plasma tHcy concentration.
It is noteworthy that SNPs with low MAF or structural variants were not included in the models, mainly because of low statistical power. Thus, other types of genetic variation at the loci we identified and tested may account for a significant proportion of the heritability. Consequently, larger studies including the novel ZNF366 and PTPRD loci are required to obtain final estimations of the genetic contribution to the variation in plasma tHcy. These restrictions notwithstanding, the associations between common SNPs in the ZNF366 and PTPRD genes and plasma tHcy concentration are robust and firmly implicate ZNF366 and PTPRD in homocysteine metabolism.
The ZNF366 gene on chromosome 5 encodes a Kruppel-type zinc finger protein. Zinc-finger proteins constitute a large family of transcriptional regulators, many of which have been strongly conserved during evolution. A recent study investigated the transcriptional regulation of estrogen receptor-α (ERalpha) and concluded that ZNF366 acts as a strong corepressor of ERalpha activity and thus may play an important role in the regulation of expression of genes in response to estrogen.29 A link between ERalpha activity and homocysteine is the decrease of plasma tHcy, observed after treatment with selective estrogen receptor modulators in postmenopausal women.30 The PTPRD gene on chromosome 9 encodes the protein tyrosine phosphatase receptor type delta. Two recent GWAS reports demonstrated that SNPs in this gene were associated with the restless legs syndrome, a condition known to be more prevalent among patients with cardiovascular disease and end-stage renal disease.31
We conclude that common genetic variants at the ZNF366 and PTPRD loci were associated with the plasma tHcy concentration. The proteins encoded at these loci may be part of unexplored molecular pathways influencing homocysteine metabolism and accordingly warrant further investigation in stroke, VTE, and other thrombotic diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Genotyping of the GAIT samples was performed at the Center National de Génotypage. PROCARDIS samples were genotyped at the Center National de Génotypage and the SNP Technology Platform. The GAIT study was supported in part by grants from the National Institutes of Health (NIH) (2 R01 HL070751-05), Ministerio de Ciencia y Tecnología and FEDER (PI-05/1361, PI-05/1382, SAF2005-04738) and La Marató TV3 (060410). J.M.S. was supported by “Programa d'Estabilització d'Investigadors de la Direcció d'Estrategia i Coordinació del Departament de Salut” (Generalitat de Catalunya). PROCARDIS was funded by the European Commission (LSHM-CT-2007-037273), the Knut and Alice Wallenberg Foundation, the British Heart Foundation, the Swedish Heart-Lung Foundation, the Swedish Research Council (8691), the Stockholm County Council (560183), and AstraZeneca AB.
National Institutes of Health
Authorship
Contribution: J.M.S. and J.C.S. (GAIT), R.C., H.W., and A.H. (PROCARDIS) designed research; U.S. contributed analytical tools; J.C.S., J.F., J.P., M.A., and A.S. collected data; A.M., J.M.S., F.B.-V., R.C., S.B., and A.H. analyzed and interpreted data; A.M., A.B., and L.A. performed statistical analysis; and A.M., J.M.S., and A.H. wrote the paper. All authors participated in the revision of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
See the supplemental Appendix for a complete list of GAIT and PROCARDIS consortia participants.
Correspondence: Anders Mälarstig, Center for Molecular Medicine, Building L8:03, Karolinska University Hospital Solna, S-171 76 Stockholm, Sweden; e-mail: anders.malarstig@ki.se.
References
Author notes
*A.H. and J.M.S. contributed equally to this work.