Abstract
A total of 186 patients with primary myelofibrosis (PMF) were genotyped for JAK2V617F at diagnosis aimed at analyzing the correlation of mutational status and mutated allele burden with outcome variables, including time to anemia, leukocytosis, leukopenia, thrombocytopenia, massive splenomegaly, leukemia, and with overall survival. A total of 127 JAK2V617F-mutated patients (68% of whole series) were divided in quartiles of V617F allele burden. After a median follow-up of 17.2 months, 23 patients died, 15 because of leukemia. A JAK2V617F mutated status did not impact on the rate of leukemia transformation or overall survival. Patients in the lower quartile had shorter time to anemia and leukopenia and did not progress to large splenomegaly. Furthermore, survival was significantly reduced in the lower quartile compared with upper quartiles and JAK2 wild-type patients. In multivariate analysis, factors associated with reduced survival were age, a blast count more than 1%, and a JAK2V617F burden within first quartile. Causes of death in the lower quartile were represented mainly by systemic infections. We conclude that a low JAK2V617F allele burden at diagnosis is preferentially associated with a myelodepletive rather than myeloproliferative phenotype and represents an independent factor associated with shortened survival in patients with PMF.
Introduction
Sixty percent of patients with primary myelofibrosis (PMF) present an acquired recurrent mutation located in exon 14 of JAK2 (JAK2V617F)1-4 ; an additional 8% to 10% harbor a mutation at codon 515 in MPL, ie, W515L/K/A.5-7 These mutations represent a major criterion for diagnosis according to the 2008 World Health Organization classification of myeloid neoplasms8 ; however, they are not specific of this disorder and are shared by other Philadelphia chromosome-negative myeloproliferative neoplasms, which include polycythemia vera (PV), essential thrombocythemia (ET), and refractory anemia with ring sideroblasts and thrombocytosis.
A JAK2V617F mutated genotype as well as different V617F allele burden have been variably associated with unique hematologic and clinical characteristics in patients with PV or ET.9 In particular, it has been shown that increasing amounts of V617F allele corresponded to a more pronounced myeloproliferative phenotype favoring higher hemoglobin level and leukocyte count, delineating a “continuum” of the 2 disorders.10 Furthermore, the higher the mutational burden, the higher the risk of presenting aquagenic pruritus, developing splenomegaly, having thrombosis, requiring cytotoxic therapy, or transforming to post-PV/post-ET myelofibrosis.9
On the contrary, information in the setting of PMF is limited to a few studies that generally included patients genotyped at different times from diagnosis and used qualitative or semiquantitative assays11-13 ; therefore, biases resulting from disease duration and previous therapy on hematologic and clinical phenotype and on patients' outcome, as well as the effects of different mutated allele load, could not be adequately controlled. Only one recent study used a quantitative assay for measuring allele burden and evaluating its clinical correlates.14 The aim of current study was to analyze the prognostic relevance of JAK2V617F mutational status and of mutated allele burden in a population of newly diagnosed patients with PMF.
Methods
Patients
This study involved 186 patients with a diagnosis of PMF satisfying the 2008 World Health Organization criteria.15 Patients were eligible to enter the study if they had a blood sample collected for JAK2V617F genotyping at the time of diagnosis or within the following 6 months, before institution of any treatment. Patients who had MPL mutated were excluded from the analysis. Rules for collection, storage, and use of blood samples, and for data analysis, were approved by the local Institutional Review Board of the Azienda Ospedaliera Universitaria Careggi, and all subjects provided informed consent. This study was conducted in accordance with the Declaration of Helsinki. Conventional cytogenetic analysis was performed according to standard techniques using unstimulated bone marrow aspirates collected during diagnostic workup. Results were reported following the International System for Human Cytogenetic Nomenclature. According to recent literature,16,17 we considered as “favorable” cytogenetics the sole deletion of 13q, 20q, trisomy 9, as well as a normal diploid karyotype; “unfavorable” cytogenetics included the presence of abnormalities of chromosome 5 or 7 as well as complex karyotype with more than 3 abnormalities. No abnormality of chromosome 17, considered “very unfavorable,”16 was found in this series.
Purpose of the study
Our primary aim was to determine the correlations, if any, between a JAK2V617F mutated status or the burden of V617F allele and the rate of major outcome events, which included overall survival and transformation to acute leukemia. We also determined whether the mutated status or the burden of JAK2 mutated allele correlated with specific laboratory parameters or clinical features, which included red blood cell indexes, leukocyte or platelet count, serum lactate dehydrogenase levels, peripheral blood (PB) CD34+ cell count, percentage of PB blasts, splenomegaly, constitutional symptoms, and the ranking of patients according to the Dupriez et al18 or the Cervantes et al prognostic scores.19 Constitutional symptoms included loss of 10% or more of body weight in the last 6 months, night sweats, or unexplained fever. Splenomegaly was measured as centimeters from the left costal margin (LCM); we considered 2 groups of patients who presented a spleen enlargement smaller or greater than 15 cm from LCM, respectively.
Determination of JAK2V617F allele burden
Measurement of JAK2V617F allele burden was performed in genomic DNA purified from granulocytes. Granulocytes were separated from PB by differential centrifugation over a Ficoll-Paque gradient, and contaminating erythrocytes were removed by hypotonic lysis; DNA was purified using the QIAmp DNA blood kit (QIAGEN GmbH) and quantified with NanoDrop technology.
The burden of mutated allele was measured by a quantitative real-time polymerase chain reaction (PCR) assay, using 40 ng DNA.20 PCR amplification and detection were performed on an ABI Prism 7300 analyzer (Applied Biosystems) using the following cycling conditions: 10 minutes at 95°C followed by 50 cycles of 15 seconds at 95°C and 60 seconds at 60°C. Primer sequence was as follows: forward primer 5′-TTATGGACAACAGTCAAACAACAAT-3′; allele G reverse primer 5′-TTTACTTACTCTCGTCTCCACAGtC3′ and allele T reverse primer 5′-TTTACTTACTCTCGTCTCCACAGtA-3′ (lowercase nucleotides are locked nucleic acid–modified). 6-carboxyfluorescein–labeled minor groove binder probe sequence was: 5′-CTTGCTCATCATACTTGC-3′. All samples were analyzed in triplicate, and the amount of JAK2V617F allele was calculated by comparison with serial dilutions of mutated and wild-type DNA or with cloned JAK2 plasmids. The mean of triplicate ΔCT determinations (CTJAK2V617F-CTJAK2WT) was used to calculate the percentage of mutated allele. Positive and negative controls were included in each assay; inter- and intra-assay variation was 3% and 5%, respectively.
Statistical analysis
The JAK2 mutated allele burden was considered as an ordered categorical variable based on the following groups: 0% (JAK2 wild-type patients), 1% to 25%, greater than 25% to 50%, greater than 50% to 75%, and greater than 75%.
We used the χ2 or Fisher exact test (2 × 2 table) or χ2 test for trend (larger contingency table), where appropriate, to compare the variables among the different patient groups that had been categorized according to mutational status or mutated allele burden. The analysis of continuous variables among the groups was performed using the Mann-Whitney U test (2 groups) or Kruskal-Wallis test with the use of Dunn method for multiple comparison. Kaplan-Meier analysis and the log-rank test were used to estimate overall survival. Cox regression models were used to perform multivariate analysis. A P value of less than .05 was considered to indicate statistical significance; all tests were 2-tailed. Data were processed using the SPSS software.
Results
A total of 186 patients with a diagnosis of PMF were genotyped with a quantitative assay for JAK2V617F mutation in granulocyte DNA collected within 6 months from diagnosis, provided that no treatment, except for transfusional support, had been delivered in the meantime. A total of 127 patients were JAK2V617F-positive, giving an overall frequency of 68.3%; the median value of V617F allele burden was 54% (range, 5%-100%; Table 1). The distribution of mutated patients according to the V617F allele burden, shown in Table 2, corresponded to overall frequencies of 13.3% in the first quartile, 33.8% in the second, 24.4% in the third, and 28.3% in the fourth. Neither a JAK2V617F mutated status nor the categorized burden of mutated allele was correlated with age or sex (Tables 1–2). Results of cytogenetic analysis at diagnosis were available in 94 patients (50.5% of total population); 77 patients (81.9%) had favorable abnormalities (58 of whom presented a normal diploid karyotype), whereas 17 (18.1%) had unfavorable cytogenetic abnormalities (Table 3). Of the 26 informative patients who were JAK2 wild-type, 6 patients (23.0%) had unfavorable cytogenetic abnormalities compared with 11 of 68 (16.2%) of those who were JAK2V617F mutated (P = .021).
Hematologic and clinical features at presentation in patients with PMF according to their JAK2 V617F mutational status
| Feature . | JAK2 WT . | JAK2 V617F . | P . |
|---|---|---|---|
| No. | 59 | 127 | |
| Median age, y (range) | 57 (21-88) | 62 (21-90) | .47 |
| Median follow-up, mo (range) | 27 (1-185) | 29 (1-237) | .72 |
| Females, no. (%) | 18 (30.5) | 51 (40.1) | .23 |
| Median white blood cell count, ×109/L (range) | 7.70 (1.9-90.78) | 10.10 (3.01-16.15) | .009* |
| Median hemoglobin, g/L (range) | 106 (46-190) | 128.5 (54-180) | <.001* |
| Median platelets, ×109/L (range) | 251 (8.0-1488) | 386 (42-2011) | .02* |
| Blast >1, no. (%) | 16 (30.5) | 38 (29.9) | .89 |
| Median LDH, U/L (range) | 704 (197-2970) | 718 (120-2981) | .96 |
| Median CD34+ cell, % (range) | 0.25 (0.0-6.6) | 0.19 (0-34) | .65 |
| Median CD34+ cell, ×109/L (range) | 18.84 (0.0-3417) | 22 (0.3-800) | .77 |
| Median JAK2V617F allele burden (range) | — | 54 (5-100) | — |
| Splenomegaly >15 cm from LCM, no. (%) | 11 (18.6) | 26 (19.7) | .89 |
| Constitutional symptoms, no. (%) | 15 (25.4) | 38 (29.9) | .15 |
| Leukemia transformation, no. (%) | 6 (10.1%) | 9 (7.1) | .45 |
| Deaths, no. (%) | 8 (13.5) | 15 (11.8) | .64 |
| Lille score, no. (%) | |||
| 0 | 22 (37.2) | 80 (63.0) | .017* |
| 1 | 26 (44.0) | 38 (30.0) | |
| 2 | 11 (18.8) | 9 (7.0) | |
| IWG-MRT score, no. (%) | |||
| low | 18 (30.5) | 47 (37.0) | .60 |
| Intermediate-1 | 15 (25.5) | 32 (25.2) | |
| Intermediate-2 | 16 (27.1) | 20 (15.7) | |
| High | 10 (16.9) | 28 (22.1) |
| Feature . | JAK2 WT . | JAK2 V617F . | P . |
|---|---|---|---|
| No. | 59 | 127 | |
| Median age, y (range) | 57 (21-88) | 62 (21-90) | .47 |
| Median follow-up, mo (range) | 27 (1-185) | 29 (1-237) | .72 |
| Females, no. (%) | 18 (30.5) | 51 (40.1) | .23 |
| Median white blood cell count, ×109/L (range) | 7.70 (1.9-90.78) | 10.10 (3.01-16.15) | .009* |
| Median hemoglobin, g/L (range) | 106 (46-190) | 128.5 (54-180) | <.001* |
| Median platelets, ×109/L (range) | 251 (8.0-1488) | 386 (42-2011) | .02* |
| Blast >1, no. (%) | 16 (30.5) | 38 (29.9) | .89 |
| Median LDH, U/L (range) | 704 (197-2970) | 718 (120-2981) | .96 |
| Median CD34+ cell, % (range) | 0.25 (0.0-6.6) | 0.19 (0-34) | .65 |
| Median CD34+ cell, ×109/L (range) | 18.84 (0.0-3417) | 22 (0.3-800) | .77 |
| Median JAK2V617F allele burden (range) | — | 54 (5-100) | — |
| Splenomegaly >15 cm from LCM, no. (%) | 11 (18.6) | 26 (19.7) | .89 |
| Constitutional symptoms, no. (%) | 15 (25.4) | 38 (29.9) | .15 |
| Leukemia transformation, no. (%) | 6 (10.1%) | 9 (7.1) | .45 |
| Deaths, no. (%) | 8 (13.5) | 15 (11.8) | .64 |
| Lille score, no. (%) | |||
| 0 | 22 (37.2) | 80 (63.0) | .017* |
| 1 | 26 (44.0) | 38 (30.0) | |
| 2 | 11 (18.8) | 9 (7.0) | |
| IWG-MRT score, no. (%) | |||
| low | 18 (30.5) | 47 (37.0) | .60 |
| Intermediate-1 | 15 (25.5) | 32 (25.2) | |
| Intermediate-2 | 16 (27.1) | 20 (15.7) | |
| High | 10 (16.9) | 28 (22.1) |
— indicates not applicable; and IWG-MRT, International Working Group for Myelofibrosis Research and Treatment.
Statistically significant difference.
Hematologic and clinical features at presentation in patients with PMF according to their JAK2 V617F allele burden
| Feature . | JAK2V617F allele burden (%) . | P . | |||
|---|---|---|---|---|---|
| 1-25 . | 26-50 . | 51-75 . | 76-100 . | ||
| No. | 17 | 43 | 31 | 36 | |
| Median age, y (range) | 58 (38-90) | 63 (28-90) | 65 (28-77) | 60.31 (21-86) | .72 |
| Median follow-up, mo (range) | 16 (1-46) | 24 (1-84) | 26 (3-101) | 45 (1-237) | <.001* |
| Females, no. (%) | 4 (23.5) | 21 (48.8) | 12 (38.7) | 14 (38.9) | .40 |
| Median JAK2 V617F allele burden (range) | 15.5 (5-25) | 41 (27-50) | 66 (52-75) | 90.5 (77-100) | — |
| Median white blood cell count, ×109/L (range) | 10.26 (3.01-26.85) | 9.47 (3.73-106.15) | 9.40 (3.70-40.0) | 13.40 (3.40-40.60) | .03* |
| Median hemoglobin, g/L (range) | 128 (54-173) | 122 (61-175) | 134 (70-171) | 136 (56-180) | .05* |
| Median platelets, ×109/L (range) | 380.5 (45-1392) | 386 (49-1089) | 410 (42-1088) | 379 (75-2011) | .95 |
| Median LDH, U/L (range) | 712 (193-2981) | 745 (162-2497) | 600 (120-1958) | 700 (225-2166) | .83 |
| Blast ≥1, no. (%) | 10 (58.8) | 9 (33.3) | 7 (35) | 12 (50) | .21 |
| Median CD34+ cell, % (range) | 0.23 (0-3.1) | 0.19 (0.01-34) | 0.09 (0.02-3.43) | 0.25 (0.02-1.32) | .98 |
| Median CD34+ cell, ×109/L (range) | 22.8 (1.4-244) | 17.54 (0.26-800) | 7.06 (2.72-476) | 43.0 (3.0-130.0) | .93 |
| Splenomegaly >15 cm from LCM, no. (%) | 1 (5.8) | 7 (16.2) | 8 (25.8) | 9 (25.0) | .003* |
| Constitutional symptoms, % | 4 (23.5) | 11 (25.6) | 6 (19.3) | 17 (47.2) | .04* |
| Leukemia transformation, no. (%) | 0 | 4 (9.3) | 2 (6.4) | 3 (8.3) | .09 |
| Deaths, no. (%) | 6 (35.3) | 7 (16.3) | 1 (3.2) | 1 (2.8) | .007* |
| Feature . | JAK2V617F allele burden (%) . | P . | |||
|---|---|---|---|---|---|
| 1-25 . | 26-50 . | 51-75 . | 76-100 . | ||
| No. | 17 | 43 | 31 | 36 | |
| Median age, y (range) | 58 (38-90) | 63 (28-90) | 65 (28-77) | 60.31 (21-86) | .72 |
| Median follow-up, mo (range) | 16 (1-46) | 24 (1-84) | 26 (3-101) | 45 (1-237) | <.001* |
| Females, no. (%) | 4 (23.5) | 21 (48.8) | 12 (38.7) | 14 (38.9) | .40 |
| Median JAK2 V617F allele burden (range) | 15.5 (5-25) | 41 (27-50) | 66 (52-75) | 90.5 (77-100) | — |
| Median white blood cell count, ×109/L (range) | 10.26 (3.01-26.85) | 9.47 (3.73-106.15) | 9.40 (3.70-40.0) | 13.40 (3.40-40.60) | .03* |
| Median hemoglobin, g/L (range) | 128 (54-173) | 122 (61-175) | 134 (70-171) | 136 (56-180) | .05* |
| Median platelets, ×109/L (range) | 380.5 (45-1392) | 386 (49-1089) | 410 (42-1088) | 379 (75-2011) | .95 |
| Median LDH, U/L (range) | 712 (193-2981) | 745 (162-2497) | 600 (120-1958) | 700 (225-2166) | .83 |
| Blast ≥1, no. (%) | 10 (58.8) | 9 (33.3) | 7 (35) | 12 (50) | .21 |
| Median CD34+ cell, % (range) | 0.23 (0-3.1) | 0.19 (0.01-34) | 0.09 (0.02-3.43) | 0.25 (0.02-1.32) | .98 |
| Median CD34+ cell, ×109/L (range) | 22.8 (1.4-244) | 17.54 (0.26-800) | 7.06 (2.72-476) | 43.0 (3.0-130.0) | .93 |
| Splenomegaly >15 cm from LCM, no. (%) | 1 (5.8) | 7 (16.2) | 8 (25.8) | 9 (25.0) | .003* |
| Constitutional symptoms, % | 4 (23.5) | 11 (25.6) | 6 (19.3) | 17 (47.2) | .04* |
| Leukemia transformation, no. (%) | 0 | 4 (9.3) | 2 (6.4) | 3 (8.3) | .09 |
| Deaths, no. (%) | 6 (35.3) | 7 (16.3) | 1 (3.2) | 1 (2.8) | .007* |
— indicates not applicable; and LCM, left costal margin.
Statistically significant difference.
Distribution of cytogenetic abnormalities in patients with PMF according to their JAK2 V617F allele burden
| JAK2V617F allele burden, percentage . | Patients with cytogenetic information available . | Cytogenetic categories . | ||
|---|---|---|---|---|
| No. . | Percentage of total . | Favorable, no. (%) . | Unfavorable, no. (%) . | |
| 0 | 26 | 44.0 | 20 (76.9) | 6 (23.0) |
| 1-25 | 8 | 47.0 | 7 (87.5) | 1 (12.5) |
| 26-50 | 17 | 39.5 | 15 (88.2) | 2 (11.7) |
| 51-75 | 17 | 54.8 | 12 (70.5) | 5 (29.4) |
| 76-100 | 26 | 72.2 | 23 (88.4) | 3 (11.5) |
| Total | 94 | 50.5 | 77 (81.9) | 17 (18.1) |
| JAK2V617F allele burden, percentage . | Patients with cytogenetic information available . | Cytogenetic categories . | ||
|---|---|---|---|---|
| No. . | Percentage of total . | Favorable, no. (%) . | Unfavorable, no. (%) . | |
| 0 | 26 | 44.0 | 20 (76.9) | 6 (23.0) |
| 1-25 | 8 | 47.0 | 7 (87.5) | 1 (12.5) |
| 26-50 | 17 | 39.5 | 15 (88.2) | 2 (11.7) |
| 51-75 | 17 | 54.8 | 12 (70.5) | 5 (29.4) |
| 76-100 | 26 | 72.2 | 23 (88.4) | 3 (11.5) |
| Total | 94 | 50.5 | 77 (81.9) | 17 (18.1) |
Favorable cytogenetic category included sole deletion of 13q or 20q or trisomy 9, and a normal karyotype; unfavorable cytogenetic category included abnormalities of chromosome 5 and/or 7 and complex cytogenetics.
Association of V617F mutated genotype or mutated allele burden with hematologic and clinical characteristics
At presentation, patients harboring a JAK2V617F mutation had significantly higher hemoglobin level (P < .001), leukocyte count (P = .009), and platelet count (P = .02) compared with those without the mutation (Table 1). Accordingly, the proportion of anemic patients (hemoglobin < 100 g/L) at presentation was significantly lower among JAK2V617 mutated patients compared with wild-types (22% vs 44%; P = .03). By considering the different categories of patients in dependence of the V617F allele burden, we observed a statistically significant correlation with leukocytosis (P = .03) and an almost significant trend for higher hemoglobin level (P = .05; Table 2). On the contrary, we found no impact of the mutated genotype or the mutated allele burden on other hematologic parameters, including peripheral blasts more than 1%, lactate dehydrogenase level, and circulating CD34+ cells. JAK2V617F mutated and wild-type patients did not differ from each other regarding the presence of a palpable splenomegaly greater than 15 cm from LCM or of constitutional symptoms; however, considering the group of mutated patients only, the frequency of those with large splenomegaly or constitutional symptoms increased significantly in relation to V617F allele burden (P = .003 and P = .04, respectively; Table 2).
To analyze whether the JAK2V617F mutational status correlated with prognostic scoring systems, we evaluated the distribution of patients in the different risk categories of the Dupriez et al18 or Cervantes et al19 score; however, because of the relatively low number of patients in the quartiles of V617F allele burden, we abstained from performing subgroup analysis. We observed that the number of JAK2V617F mutated patients in the low-risk category of the Dupriez et al18 scoring system was significantly greater compared with unmutated patients (P = .017), possibly the result of the higher hemoglobin level found in mutated patients; on the other hand, the distribution of patients in the 4 risk categories of the Cervantes et al19 score was independent of JAK2V617F mutational status.
Association of V617F mutated genotype or mutated allele burden with disease progression
To investigate the impact of JAK2V617F mutational status or categorized V617F allele burden on parameters associated with disease progression, we calculated the time elapsing from diagnosis to hematologic and clinical outcome events, which included: changes in hematologic parameters not the result of cytoreductive therapy (anemia, defined as an hemoglobin steadily < 100 g/L; leukopenia, as a leukocyte count < 4 × 109/L; leukocytosis, as a leukocyte count > 25 × 109/L; thrombocytopenia, as a platelet count < 100 × 109/L); the development of massive splenomegaly (considered as a spleen enlarged > 15 cm from LCM in patients who presented smaller spleen size at diagnosis); and the evolution to acute leukemia. In the comparison of JAK2V617F mutated vs unmutated patients, we found that time to anemia or time to leukopenia was significantly longer (P = .017 and P = .001, respectively) in those harboring the mutation, whereas the statistical significance was not attained for other outcome events (Figure 1). The relative risk (RR) associated with a JAK2 wild-type status was 1.64 (95% confidence interval [CI], 1.04-2.58) in case of anemia and 3.43 (95% CI, 1.59-7.41) in case of leukopenia compared with JAK2V617F mutated patients. In particular, we observed that a JAK2V617F mutated status did not represent a risk factor for transformation to acute leukemia (Figure 1); the RR was 1.57 (95% CI, 0.55-4.41) in JAK2V617F mutated compared with wild-type patients.
Kaplan-Meier analysis of the time to developing hematologic and clinical outcome events in PMF patients according to their JAK2V617F mutational status. The plots show time to anemia (hemoglobin < 100 g/L), leukopenia (leukocytes < 4 × 109/L), leukocytosis (> 25 × 109/L), thrombocytopenia (< 100 × 109/L), large splenomegaly (> 15 cm from left costal margin in patients presenting smaller spleen at diagnosis), and time to leukemic transformation. P values are shown inside each box. WT indicates JAK2 wild-type patients; V617F, JAK2V617F mutated patients.
Kaplan-Meier analysis of the time to developing hematologic and clinical outcome events in PMF patients according to their JAK2V617F mutational status. The plots show time to anemia (hemoglobin < 100 g/L), leukopenia (leukocytes < 4 × 109/L), leukocytosis (> 25 × 109/L), thrombocytopenia (< 100 × 109/L), large splenomegaly (> 15 cm from left costal margin in patients presenting smaller spleen at diagnosis), and time to leukemic transformation. P values are shown inside each box. WT indicates JAK2 wild-type patients; V617F, JAK2V617F mutated patients.
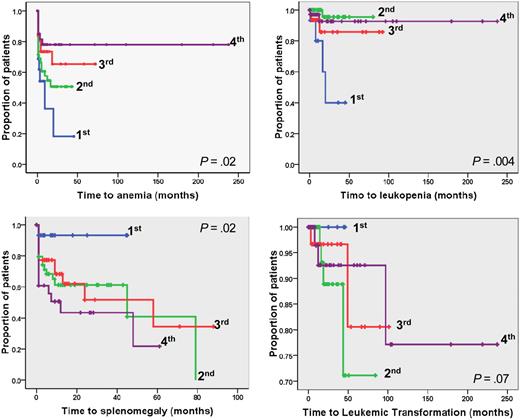
The relevance of different V617F allele burden on those outcomes was also evaluated. We found that patients in the lower quartile had significantly shorter time to anemia (P = .02) and leukopenia (P = .004) compared with upper quartiles (Figure 2); the corresponding RR values were 2.11 (95% CI, 1.1-4.43) for anemia and 6.84 (95% CI, 1.9-24.59) for leukopenia. We also found that time to large splenomegaly was significantly shorter in patients of the upper quartiles compared with the lower quartile (P = .02); only one of 17 patients in the lower quartile (5.8%) developed large splenomegaly during the follow-up compared with 16.2%, 25.8%, and 25.0% in the other quartiles (Figure 3; Table 2). On the other hand, there was no significant difference in the time to leukemia progression among the 4 quartiles (P = .07); however, it must be noted that none of the patients in the lower burden group evolved to leukemia (Figure 3; Table 2).
Kaplan-Meier analysis of the time to developing hematologic and clinical outcome events in JAK2V617F mutated PMF patients categorized according to quartiles of mutated allele burden. The plots show time to anemia, leukopenia, large splenomegaly, and leukemic transformation in the 4 quartiles of JAK2V617F allele burden. P values are shown inside each box. Quartiles of V617F allele burden were as follows: first, 1% to 25%; second, greater than 25% to 50%; third, greater than 50% to 75%; fourth, greater than 75% to 100%.
Kaplan-Meier analysis of the time to developing hematologic and clinical outcome events in JAK2V617F mutated PMF patients categorized according to quartiles of mutated allele burden. The plots show time to anemia, leukopenia, large splenomegaly, and leukemic transformation in the 4 quartiles of JAK2V617F allele burden. P values are shown inside each box. Quartiles of V617F allele burden were as follows: first, 1% to 25%; second, greater than 25% to 50%; third, greater than 50% to 75%; fourth, greater than 75% to 100%.
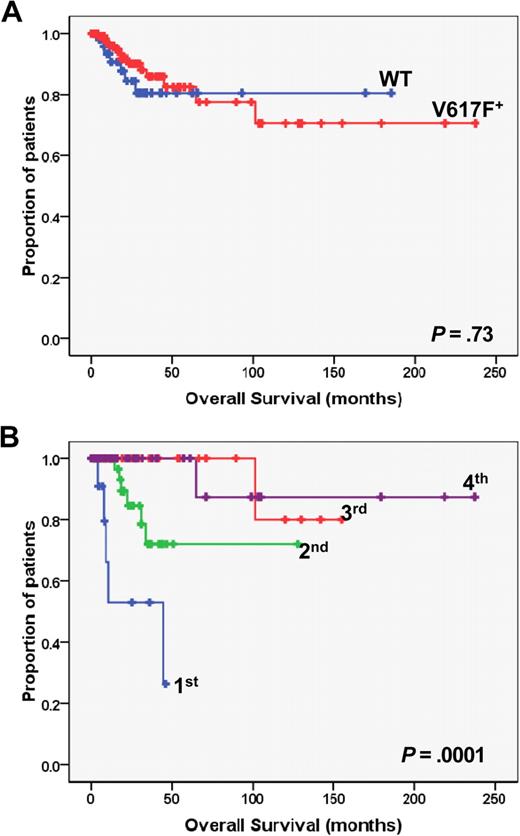
Kaplan-Meier analysis of overall survival in PMF patients. (A) The plot shows the overall survival in JAK2 wild-type (WT; n = 59) vs JAK2V617F mutated (V617F; n = 127) patients. (B) Overall survival of JAK2V617F mutated patients who had been categorized in quartiles according to the burden of mutated allele. P values are shown inside each box. Quartiles of V617F allele burden were as follows: first, 1% to 25%; second, greater than 25% to 50%; third, greater than 50% to 75%; fourth, greater than 75% to 100%.
Kaplan-Meier analysis of overall survival in PMF patients. (A) The plot shows the overall survival in JAK2 wild-type (WT; n = 59) vs JAK2V617F mutated (V617F; n = 127) patients. (B) Overall survival of JAK2V617F mutated patients who had been categorized in quartiles according to the burden of mutated allele. P values are shown inside each box. Quartiles of V617F allele burden were as follows: first, 1% to 25%; second, greater than 25% to 50%; third, greater than 50% to 75%; fourth, greater than 75% to 100%.
Association of V617F mutated genotype or mutated allele burden with disease outcome
After a median follow-up of 17.2 months, there were 23 patients (12.3%) who died, accounting for 13.5% and 11.8% of the JAK2 wild-type and JAK2V617F mutated, respectively (P = .64; Table 1). Transformation to acute leukemia was the reason of death in 15 of them (65% of all deaths), accounting for 10.1% and 7.1% of JAK2 wild-type and JAK2V617F mutated patients, respectively (P = .078). Duration of survival was similar in JAK2 wild-type and JAK2V617F mutated patients (P = .73; Figure 3A). However, when we analyzed JAK2V671F mutated patients after stratification according to the burden of V617F allele, we found that survival was significantly shorter in the lower quartile compared with upper quartiles (P = .001, Figure 3B) as well as to JAK2 wild-type patients (P = .006; Figure 3A). The percentage of patients in the lower quartile who died was as high as 35.3% compared with 16.3%, 3.2%, and 2.8% in upper quartiles, respectively (P = .007, Table 2). Causes of death in the lower quartile were represented by systemic infections or sepsis (n = 5) and multiorgan failure (n = 1), whereas no case of leukemia transformation was observed. The distribution of patients with favorable or unfavorable cytogenetic abnormalities according to their V617F allele burden is presented in Table 3; there was no statistically significant preferential clustering of unfavorable cytogenetic abnormalities in any of the different allele burden categories (P = .073, χ2 test for trend).
Variables that were associated with overall survival in univariate and multivariate analysis are listed in Table 4. Age, PB blast count, and a lower V617F allele burden maintained their significant association with survival in multivariate analysis; of note, there was an almost significant association of hemoglobin level (hazard ratio = 0.60; 95% CI, 0.36-1.01; P = .051) with survival.
Cox proportional hazard models of factors independently predictive of overall survival of PMF patients
| Variable . | HR (95% CI) . | P . |
|---|---|---|
| Univariate | ||
| Age | 1.12 (1.07-1.18) | < .001 |
| Female sex | 7.58 (1.76-32.63) | .007 |
| Hemoglobin level | 0.77 (0.66-0.89) | .001 |
| Leukocyte count | 1.04 (1.02-1.05) | < .001 |
| PB blasts > 1% | 1.30 (1.16-1.45) | < .001 |
| Splenomegaly (> 15 cm LCM) | 2.41 (1.09-5.29) | .028 |
| V617F burden < 25% | 0.09 (0.03-0.32) | < .001 |
| Multivariate | ||
| Age | 1.21 (1.08-1.34) | .001 |
| PB blasts > 1% | 1.57 (1.13-2.16) | .022 |
| V617F burden < 25% | 0.023 (0.002-0.24) | .002 |
| Variable . | HR (95% CI) . | P . |
|---|---|---|
| Univariate | ||
| Age | 1.12 (1.07-1.18) | < .001 |
| Female sex | 7.58 (1.76-32.63) | .007 |
| Hemoglobin level | 0.77 (0.66-0.89) | .001 |
| Leukocyte count | 1.04 (1.02-1.05) | < .001 |
| PB blasts > 1% | 1.30 (1.16-1.45) | < .001 |
| Splenomegaly (> 15 cm LCM) | 2.41 (1.09-5.29) | .028 |
| V617F burden < 25% | 0.09 (0.03-0.32) | < .001 |
| Multivariate | ||
| Age | 1.21 (1.08-1.34) | .001 |
| PB blasts > 1% | 1.57 (1.13-2.16) | .022 |
| V617F burden < 25% | 0.023 (0.002-0.24) | .002 |
Discussion
PMF is associated with poorer prognosis compared with other classic BCR/ABL-negative chronic myeloproliferative neoplasms because of a substantial reduction of life expectancy21 in the order of 31% compared with sex- and age-adjusted control population.22 However, survival may range from a few months to an excess of a decade, especially in younger patients.23-25 Therefore, identification of variables associated with prognosis is of considerable importance for driving therapeutic decisions, particularly in the case of hematopoietic stem cell transplantation, which is the only potentially curative therapeutic approach, yet still burdened by considerable morbidity and mortality.26 Three main prognostic score systems have been developed. One is the so-called Lille score, which includes anemia and leukopenia or leukocytosis (leukocytes < 4 × 109/L or > 30 × 109/L, respectively) as covariates to identify 3 distinct prognostic groups.18 The second one, developed at the Mayo Clinic, introduced thrombocytopenia (< 100 × 109/L) and monocytosis (> 109/L) as additional variables resulting in an improved discriminatory power of the Lille score.27 More recently, the International Working Group for Myelofibrosis Research and Treatment developed a new prognostic score system that included age (> 65 years), presence of constitutional symptoms, anemia (hemoglobin < 100 g/L), leukocytosis (> 25 × 109/L), and blood blasts more than 1% and discriminated 4 categories of patients with significantly different median survival in the range of 27 to 135 months.19
The discovery of the JAK2V617F mutation and its association with distinct hematologic and clinical characteristics prompted further research to assessing the predictive value of mutational status for the most common complications of PV or ET, ie, vascular events, and for prognosis and survival more in general. The latter aspect is particularly relevant for PMF because survival in PV or ET may be only slightly reduced compared with the normal population. The purpose of present study was to evaluate the consequence of JAK2V617F mutational status and V617F mutated allele burden on disease outcome in a population of newly diagnosed patients with PMF. Similar to observations made in PV or ET, also in this series of PMF patients, we found statistically significant association of the JAK2V617F mutated status with a more pronounced myeloproliferative phenotype favoring raised hemoglobin level and leukocyte count; accordingly, the time to developing anemia or leukopenia during the follow-up was significantly longer, and conversely the time to develop leukocytosis significantly shorter, in mutated patients. These findings were generally in agreement with results from the English11 and GIMEMA13 groups and at variance with the Mayo Clinic series12,14 in which hemoglobin levels were not significantly modified by the presence of mutation.14 We also found that patients presenting anemia at diagnosis were significantly less frequent among JAK2V617F mutated than wild-type, and this probably accounted for more mutated patients to be included within the low-risk category according to Lille score system. However, the main outcome events of the study, namely, overall survival and leukemia transformation, were not appreciably different in the 2 categories of patients defined by their JAK2V617F genotype. These results are in line with the study from the Mayo Clinic14 but in disagreement with the results of the English study,11 which reported a hazard ratio of shortened survival of 3.3 (95% CI, 1.26-8.68) in patients harboring the V617F allele.11 In addition, in the GIMEMA study,13 overall survival was not influenced by a JAK2V617F mutated status, although a higher rate of leukemic transformation was observed; the reason for these discrepancies is not obvious and might be generically ascribed to series variability. Therefore, a conclusion of the present study is that a JAK2V617F mutated status per se does not reflect in a poorer prognosis in PMF.
However, the fact that overall survival in JAK2V617F mutated patients was similar to wild-type patients notwithstanding their higher hemoglobin level, which is considered as one major factor influencing survival in PMF, suggested possible heterogeneity within the category of mutated patients. Similarly, a correlation of JAK2V617F mutated status with outcome events, including thrombosis, in patients with PV resulted evident only after stratification according to their burden of mutated allele at diagnosis.28,29 Therefore, in this study, we used a sensitive real-time PCR approach to stratifying PMF patients in quartiles of burden of mutated allele at presentation. We found that patients included in the lower quartile showed significantly shorter progression time to anemia, leukopenia, and conversely longer time to large splenomegaly, compared with upper quartiles. More importantly, we found a striking difference in overall survival that was significantly reduced in patients of the lower quartile compared not only with upper quartiles but also with JAK2 wild-type patients. Reduced survival was not the result of leukemic transformation (no patient in the lower quartile evolved to leukemia) but mainly to systemic infections. Thus, patients in the lower quartiles apparently died because of the consequences of bone marrow failure, as indirectly suggested by the shorter time to developing leukopenia. The poorer survival of patients harboring low JAK2V617F allele burden was first described in a study from the Mayo Clinic,14 although, at variance with present results, also leukemia-free survival was reportedly shorter in patients included in the lower quartile. Because the follow-up time of the 2 series is comparable, differences in patient characteristics and/or of treatments used could unpredictably underlie these discrepancies. Furthermore, an additional variable might have been represented by the different source of genomic DNA used for quantification of JAK2V617F burden, that is, unfractionated whole bone marrow cells in the Mayo Clinic study compared with purified granulocytes in the current study.
In conclusion, results of this study validate and extend a recent report14 and point to a low JAK2V617F allele burden at diagnosis as a strong surrogate marker associated with shortened survival in PMF; at present, the biologic mechanisms underlying this correlation remain fully to be established. One additional implication of our findings is that, although JAK2 genotyping is recommended in the diagnostic workup of suspected PMF according to the WHO classification, a quantitative rather than a qualitative assay would be more appropriate. In this regard, an initiative of the European Leukemia Net working group is undergoing with the aim to harmonize different JAK2V617F quantitation methods; indeed, a recent multicenter study found that several of the quantitative assays described in the literature produced discrepant results.30 Indeed, the finding of a low JAK2V617F allele burden at diagnosis would point to a poorer prognosis that cannot be appropriately recognized by current score systems. This information might drive early therapeutic decisions, involving the use of stem cell transplantation in younger patients and/or patient inclusion in trials with investigational drugs. However, we acknowledge that a preferential clustering of low-allele burden subjects in the high-risk category of the International Working Group for Myelofibrosis Research and Treatment scoring system might have been missed because of the relatively small size of our patient population. Therefore, we suggest the opportunity of a large prospective study on a well-characterized population of patients and with a standardized JAK2V617F quantitation method, which might ideally find place within international consortia and potentially provide definite information within a relatively short time.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by Istituto Toscano Tumori, Ente Cassa di Risparmio di Firenze, Ministero della Università e Ricerca (PRIN projects), and Università di Firenze (institutional funds, ex-60%). A.P. was supported by Associazione Italiana per le Leucemie, Firenze.
Authorship
Contribution: P.G. designed and performed research and contributed to manuscript writing; G.B. designed research and contributed to manuscript writing; G.S., A.R., F.L.C., E. Antonioli, L.P., F.D., and E. Ammatuna contributed patients and revised the manuscript; A.P. and V.P. performed research; G.L. analyzed data and revised the manuscript; V.L. and A.B. contributed patients and revised the manuscript; T.B. designed research and contributed to manuscript writing; and A.M.V. designed and performed research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alessandro M. Vannucchi, UF di Ematologia, Università degli Studi, Viale Morgagni, 85, 50134 Florence, Italy; e-mail: amvannucchi@unifi.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal