Intracellular oxidative stress in cells transformed by the BCR-ABL oncogene is associated with increased DNA double-strand breaks. Imprecise repair of these breaks can result in the accumulation of mutations, leading to therapy-related drug resistance and disease progression. Using several BCR-ABL model systems, we found that BCR-ABL specifically promotes the repair of double-strand breaks through single-strand annealing (SSA), a mutagenic pathway that involves sequence repeats. Moreover, our results suggest that mutagenic SSA repair can be regulated through the interplay between BCR-ABL and extrinsic growth factors. Increased SSA activity required Y177 in BCR-ABL, as well as a functional PI3K and Ras pathway downstream of this site. Furthermore, our data hint at a common pathway for DSB repair whereby BCR-ABL, Tel-ABL, Tel-PDGFR, FLT3-ITD, and Jak2V617F all increase mutagenic repair. This increase in SSA may not be sufficiently suppressed by tyrosine kinase inhibitors in the stromal microenvironment. Therefore, drugs that target growth factor receptor signaling represent potential therapeutic agents to combat tyrosine kinase-induced genomic instability.
Introduction
Myeloproliferative diseases and myeloid leukemias are frequently associated with constitutively activated tyrosine kinases that enhance the proliferation and viability of hematopoietic cells. In chronic myelogenous leukemia (CML), a hematopoietic stem cell disorder, the BCR-ABL oncogene, generates a constitutively active cytoplasmic tyrosine kinase that enhances the proliferation and viability of myeloid lineage cells.1 Fusions between TEL (a member of the Ets family of transcription factors) and ABL resulting from a t(9;12) translocation are seen in atypical CML, acute lymphocytic leukemia, and acute myeloid leukemia. TEL fusions with PDGFR, characterized by a t(5;12) translocation, are present in a subset of patients with chronic myelomonocytic leukemia. In addition, a Jak2 point mutation (Jak2V617F) is frequently associated with human myeloproliferative disorders and results in constitutive Jak2 tyrosine kinase activity and transformation. Finally, the FMS-like tyrosine kinase 3 (FLT3) receptor tyrosine kinase is a frequent target of mutations, internal tandem duplications (ITDs), and other rearrangements in acute myeloid leukemia.2
The BCR-ABL oncogene is thought to be the only oncogene in patients in stable phase, but other genetic events may accumulate over time and the disease may progress to blast crisis. Targeted therapy with the ABL tyrosine kinase inhibitor imatinib results in complete hematologic remission in approximately 95% of patients with chronic-phase CML. However, most patients in blast crisis are either nonresponsive or relapse shortly after an initial response.3 Resistance to imatinib can result from point mutations within the adenosine triphosphate-binding pocket of BCR-ABL.1 Of particular importance, BCR-ABL increases levels of reactive oxygen species (ROS),4,5 causing oxidative DNA damage that, when imprecisely repaired or left unrepaired, could result in BCR-ABL mutations that promote imatinib resistance.6,7
ROS can result in a variety of DNA lesions. Of these, double-strand breaks (DSBs) are thought to be the most mutagenic, as neither strand remains intact to serve as a template for repair. Pathways for the faithful repair of DSBs either use a homologous template or involve nonhomologous end-joining (NHEJ).8,9 In NHEJ, DNA ends are rejoined without the use of significant sequence homology.10 Although NHEJ plays an important role in maintaining the overall integrity of chromosomes, it is potentially mutagenic because DNA ends may undergo modifications before ligation. NHEJ in mammalian cells is thought to primarily involve the classic NHEJ pathway, which includes the heterodimer Ku70/Ku80 (Ku), the serine/threonine kinase DNA-PKcs, the XRCC4/XLF/LIG4 complex, and the Artemis nuclease.10 This pathway is essential for normal V(D)J recombination, but cells deficient in classic NHEJ factors remain capable of efficiently orchestrating other forms of NHEJ.11,–13 DSB repair pathways that use a homologous template include homology-directed repair (HDR) and single-strand annealing (SSA). In both pathways, DSB ends are processed to single-strand 3′ tails. HDR involves the RAD51-dependent invasion of a single-strand tail into a donor DNA duplex, which is followed by template-dependent polymerization. HDR is precise if an identical sequence (eg, from the sister chromatid) is used to direct repair.14 In contrast, SSA proceeds through the annealing of complementary single-strand tails formed at repeated sequences and is inhibited by RAD51.15 SSA is always mutagenic because the sequence between repeats is deleted.
Previous studies described increased rates of HDR and error-prone NHEJ in myeloid cells expressing BCR-ABL.7,16 The enhanced efficiency of repair is thought to promote resistance to therapeutic clastogens, whereas the loss of repair precision contributes to mutagenesis and disease progression. Here, we have used quantitative techniques to examine the repair of DSBs. Specifically, we demonstrate that BCR-ABL and other oncogenic kinases specifically promote the mutagenic SSA pathway. We also show that stromal cell–conditioned medium is sufficient to increase SSA frequency in BCR-ABL-expressing cells in the presence of imatinib. Enhanced SSA activity is dependent on activated PI3K/Ras pathways, which occurs downstream of Y177, a major regulatory site for ROS induction.17,18 Together, these studies create a model of transformation, whereby altered SSA repair activity has the potential to contribute to disease progression, and mutagenesis in CML that involves both intrinsic kinase signaling and paracrine factors from the stromal microenvironment.
Methods
Cells
The parental murine pro–B cell line BaF3 and cells expressing the tyrosine kinase oncogenes Tel-ABL, Tel-PDGFR, and FLT3-ITD, as well as BaF3.EpoR cells expressing Jak2V617F and Jak2V617F.Y114A, were used for DNA repair studies. BCR-ABL was expressed under a doxycycline-inducible promoter in BaF3.TonB cells. Transformed cells were maintained in RPMI 1640 (Mediatech/Cellgro) containing 10% fetal bovine serum (FBS; Harlan Bioproducts), and the medium for factor-dependent cells was supplemented with 10% WEHI-3B–conditioned medium as a source of murine interleukin-3 (IL-3). BaF3.FLT3-ITD cells were cultured in media supplemented with G418 (1.5 mg/mL). BaF3.BCR-ABL and BaF3.BCR-ABL.Y177F with similar expression and autophosphorylation levels of the oncoprotein were maintained in the presence of imatinib (1 μM) with 10% WEHI–3B-conditioned medium, as described.18 Under experimental conditions, parental BaF3 and BaF3 cells expressing oncogenic kinases were treated with low amounts of IL-3 to support viability but not growth (10 pg/mL). The human BCR-ABL-expressing cell lines Ku812, K562, BV-173, and Meg-01 were grown in RPMI 1640 containing 10% FBS. The human HEL cell line expressing the Jak2V617F mutation was grown in RPMI 1640 containing 10% FBS and supplemented with 1 mM sodium pyruvate. Bone marrow stromal cell lines KM104, KM105, HS5, and HS27A were used for cell proliferation and ROS studies and maintained in RPMI 1640 containing 20% FBS.
Analysis of DNA repair fidelity by PCR
BaF3.BCR-ABL cells (5 × 106) were stably transfected with either the hprtDRGFP or the hprtSAGFP reporter (kindly provided by Maria Jasin, Memorial Sloan-Kettering Cancer Center8,15 ) and selected for puromycin resistance (3 μg/mL). DSBs were generated by transfecting cells with an I-SceI expression vector, pCBASce,19 and repair was assayed by flow cytometry 2 days later. Genomic DNA was amplified for the repaired GFP gene by polymerase chain reaction (PCR) using DRGFP primers, as previously described.20 Confirmational studies included cloning of the PCR products with the TA cloning system (Invitrogen) according to the manufacturer's instructions, followed by evaluation of single derived clones by nested PCR using the following primers: DRGFP forward, 5′-TTGGCAAAGAATTCAGATCCGCCG-3′, and DRGFP reverse,5′-AGAAGTCGTGCTGCTTCATGTGGT-3′. The PCR product using the DRGFP primers covers a 548-bp region, adjacent to the DSB site and homologous to the repair template. The mutation frequency within a portion of the remaining region with homology to the repair template was not determined. A total of 20 clones were analyzed. For SAGFP-transfected cells, mutation analysis was performed by amplifying the GFP gene using the following primers: SAGFP-1 forward: 5′-GCAACGTGCTGGTTATTGTGCTGT-3′ and SAGFP-1 reverse: 5′-TTACTTGTACAGCTCGTCCATGCC-3′. Confirmational studies included nested PCR with the following primers: SAGFP-2 forward: 5′-TGACCCTGAAGTTCATCTGCACCA-3′, SAGFP-2 reverse, 5′-TTGATGCCGTTCTTCTGCTTGTCG-3′, and a total of 20 clones were analyzed. PCR products were sequenced by the Dana-Farber Cancer Institute core-sequencing facility.
Analysis of SSA repair activity
BaF3 cells expressing different oncogenes were cotransfected with 5 μg each of the pSAGFP reporter and pCBASce in 100 μL Nucleofector solution V (Amaxa) according to the manufacturer's instructions using the Nucleofector device. The pmaxGFP vector (Amaxa) was transfected to evaluate transfection efficiency. Transfected cells were either left untreated or treated within 3 hours of electroporation with imatinib (1 μM; Novartis), midostaurin (50 nM; Novartis), Jak inhibitor (1 μM; Calbiochem), LY294002 (Calbiochem), Akt inhibitor X (Calbiochem), PD98059 (Promega), or U0126 (Promega) in the presence of IL-3 (1 ng/mL; Invitrogen). For knockdown studies, BaF3.BCR-ABL cells were cotransfected with 2.5 μg of pCBASce and 5 μg of nontargeting (control) siRNA, siRNA targeting Akt1/2, and MEK1 (Santa Cruz Biotechnology). SSA frequency was determined 2 days after transfection by flow cytometric analysis on a BD Biosciences FACScan. Percentage SSA repair was calculated by comparing percentage GFP expression in kinase inhibitor–treated cells and untreated cells. All error bars represent SEM.
Immunoblotting
Immunoblotting was performed as described using a standard chemiluminescence technique.21 Rabbit polyclonal antibodies against Akt1/2/3 (H-136) and MEK1 (C-18; Santa Cruz Biotechnology) and mouse monoclonal antibodies against β-actin (12H8; Sigma-Aldrich) or the hemagglutinin epitope (HA tag; 12CA5; Roche Diagnostics) were used to detect protein expression. Levels of HA-tagged I-SceI were determined 24 hours after electroporation of the expression plasmid.
Results
BCR-ABL increases the frequency of SSA
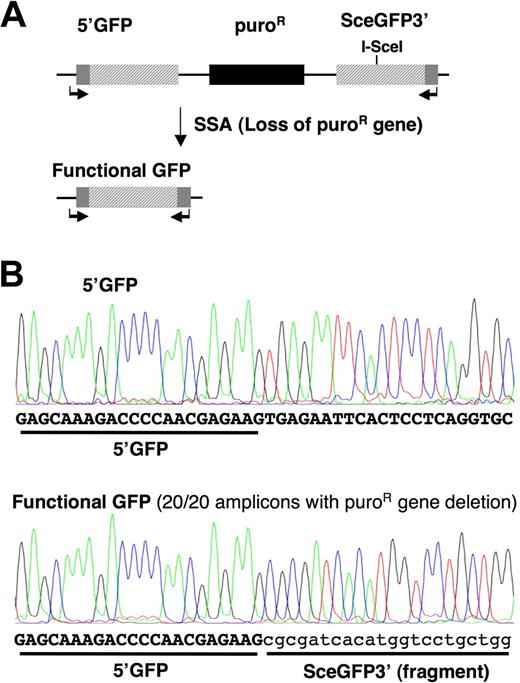
DSBs that result from ROS induced by BCR-ABL can be repaired by multiple repair pathways.7,16 Of these, both SSA and HDR require processing of DSB ends to 3′ single-strand tails and orchestrate repair using a homologous template. In contrast with HDR, which is precise, SSA is always mutagenic and is therefore suppressed by sequence divergence in mammalian cells.22 To evaluate the effects of BCR-ABL on different homologous DSB repair pathways, we generated unique BaF3 and BaF3.BCR-ABL–expressing cell lines containing the SSA reporter SAGFP (Figure 1A). We selected the BaF3 system, as the BaF3.BCR-ABL cell lines readily develop clinically relevant mutations within BCR-ABL,23,24 thus recapitulating the development of imatinib resistance in primary CML cells. SAGFP consists of 2 overlapping GFP fragments. Cleavage of the downstream fragment by the rare-cutting endonuclease I-SceI, followed by SSA between the fragments, establishes a functional GFP by deleting a 2.7-kb sequence that contains a puromycin resistance gene. To be certain that GFP+ cells recovered after transient expression of I-SceI had undergone SSA, we confirmed that GFP+ cells were sensitive to puromycin (data not shown). We also sequenced 20 GFP+ BaF3.BCR-ABL subclones and confirmed the expected GFP sequence from an SSA event. No mutations were identified in the GFP sequence (Figure 1B).
BCR-ABL initiates mutagenic SSA repair in SAGFP reporter containing BaF3 cells. (A) The SAGFP reporter for SSA consists of the GFP gene fragments 5′GFP and SceGFP3′ separated by a puromycin resistance gene. Repair of the I-SceI–generated DSB in SceGFP3′ by SSA results in a functional GFP gene and excision of the puromycin resistance gene. (B) BaF3.BCR-ABL cells stably expressing the SSA reporter were used to measure repair fidelity in response to I-SceI–induced DSBs. Genomic DNA from clonal populations of GFP+ BaF3.BCR-ABL cells containing the reporter was isolated to assess the repaired GFP region by PCR (SAGFP-1 forward and reverse primers indicated by arrows) and sequencing (partial DNA sequence shown).
BCR-ABL initiates mutagenic SSA repair in SAGFP reporter containing BaF3 cells. (A) The SAGFP reporter for SSA consists of the GFP gene fragments 5′GFP and SceGFP3′ separated by a puromycin resistance gene. Repair of the I-SceI–generated DSB in SceGFP3′ by SSA results in a functional GFP gene and excision of the puromycin resistance gene. (B) BaF3.BCR-ABL cells stably expressing the SSA reporter were used to measure repair fidelity in response to I-SceI–induced DSBs. Genomic DNA from clonal populations of GFP+ BaF3.BCR-ABL cells containing the reporter was isolated to assess the repaired GFP region by PCR (SAGFP-1 forward and reverse primers indicated by arrows) and sequencing (partial DNA sequence shown).
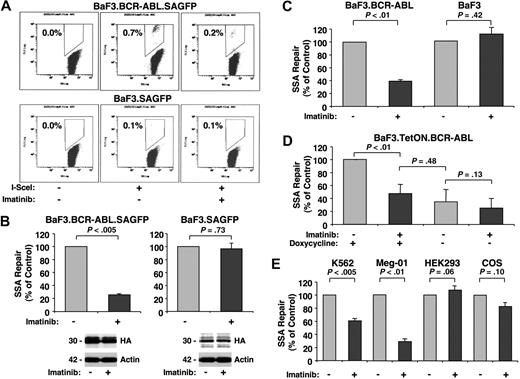
To assess carefully the effects of BCR-ABL kinase activity on SSA, we used 4 different model systems. Initially, we assayed BaF3 and BaF3.BCR-ABL cells with stably integrated SAGFP. DSBs were introduced by transfection of an I-SceI expression vector, and SSA frequency was determined after 2 days by flow cytometry. For a typical experiment, I-SceI expression in BaF3.BCR-ABL.SAGFP cells resulted in 0.7% GFP+ cells and BaF3.SAGFP cells resulted in 0.1% GFP+ cells (Figure 2A). We compared the effect of imatinib on SSA frequency between BaF3.BCR-ABL cells and parental BaF3 cells. Whereas imatinib (1 μM) reduced SSA frequency in BaF3.BCR-ABL cells approximately 4-fold, it had no effect on SSA frequency in the parental line (Figure 2B top panels). Imatinib treatment did not alter expression of I-SceI in either line (Figure 2B bottom panels), suggesting that BCR-ABL kinase activity specifically promotes SSA.
BCR-ABL increases SSA repair frequency. Changes in SSA repair frequency were determined by flow cytometry in response to I-SceI–induced DSBs in cells left untreated or treated with imatinib (1 μM) for 48 hours using the SAGFP reporter. BaF3 and BaF3.BCR-ABL cells stably (A-B) or transiently (C) expressing the SAGFP reporter were used. GFP+ cells were counted by flow cytometry (A dot plot of typical experiments) and compared with untreated samples (B-C, n = 3). Cellular expression of HA-tagged I-SceI (HA) and actin was detected by immunoblotting, as indicated (B bottom panels). (D) BaF3 cells with inducible BCR-ABL were either left untreated or treated with doxycycline to induce BCR-ABL expression (n = 4). (E) K562 (n = 3) and Meg-01 (n = 4) cells (expressing the BCR-ABL oncogene) as well as HEK293 (n = 4) and COS (n = 3) cells (BCR-ABL–negative) were used to measure SSA frequency, as indicated.
BCR-ABL increases SSA repair frequency. Changes in SSA repair frequency were determined by flow cytometry in response to I-SceI–induced DSBs in cells left untreated or treated with imatinib (1 μM) for 48 hours using the SAGFP reporter. BaF3 and BaF3.BCR-ABL cells stably (A-B) or transiently (C) expressing the SAGFP reporter were used. GFP+ cells were counted by flow cytometry (A dot plot of typical experiments) and compared with untreated samples (B-C, n = 3). Cellular expression of HA-tagged I-SceI (HA) and actin was detected by immunoblotting, as indicated (B bottom panels). (D) BaF3 cells with inducible BCR-ABL were either left untreated or treated with doxycycline to induce BCR-ABL expression (n = 4). (E) K562 (n = 3) and Meg-01 (n = 4) cells (expressing the BCR-ABL oncogene) as well as HEK293 (n = 4) and COS (n = 3) cells (BCR-ABL–negative) were used to measure SSA frequency, as indicated.
Next, to confirm these findings and exclude artifacts because of the chromosomal integration of the reporter, we transiently cotransfected BaF3 and BaF3.BCR-ABL cells with SAGFP and the I-SceI expression plasmid. After 2 days, SSA frequency was found to be reduced in imatinib-treated BaF3.BCR-ABL cells (39.07% ± 2.3%) compared with untreated cells, but not in parental cells treated with imatinib (111% ± 11.1%) compared with untreated cells (Figure 2C). In control experiments, we found that growth-stimulatory concentrations of IL-3 (1 ng/mL) did not alter SSA frequency in BaF3 cells (105.0% ± 7.6% of control, P = .58, n = 3). In addition, we used a BaF3 cell line with doxycycline-inducible BCR-ABL expression to assay SSA. This approach avoids confounding differences between cell lines. Cells cotransfected with SAGFP and the I-SceI expression vector were evaluated for SSA in both the presence and absence of doxycycline and imatinib (1 μM; Figure 2D). No significant changes were seen in SSA frequency in the absence of doxycycline between imatinib-treated and untreated cells. However, when BCR-ABL expression was induced with doxycycline, SSA frequency was increased nearly 3-fold. In line with previous experiments, this increase in SSA frequency could be reduced almost to control levels by imatinib treatment. SSA repair depends in part on expression of the DNA repair proteins RAD52 and ERCC1.15 In our experiments, we did not observe altered levels of these proteins in response to imatinib (1 μM, 27 hours; not shown), suggesting that the BCR-ABL-dependent decrease in SSA frequency is not regulated through modulation of RAD52 and ERCC1 expression.
Finally, although the murine BaF3 cell line has been used for studying BCR-ABL-dependent genomic instability, differences in repair between murine and human cell lines cannot be excluded. We therefore measured SSA frequency in the human BCR-ABL–expressing cell lines K562 and Meg-01. After cotransfection of SAGFP and the I-SceI expression vector, significant decreases in the frequency of SSA were observed in K562 (60.0% ± 2.3%) and Meg-01 (29.51% ± 3.4%) in response to imatinib (Figure 2E left panels). This is in contrast to the BCR-ABL-negative cell lines HEK293 (119.1% ± 6.5%) and COS (82.9% ± 6.0%), which are not imatinib-sensitive and did not show a significant change in frequency of SSA (Figure 2E right panels).
Bone marrow stromal cell–conditioned media increases mutagenic repair and oxidative stress in imatinib-treated K562 cells
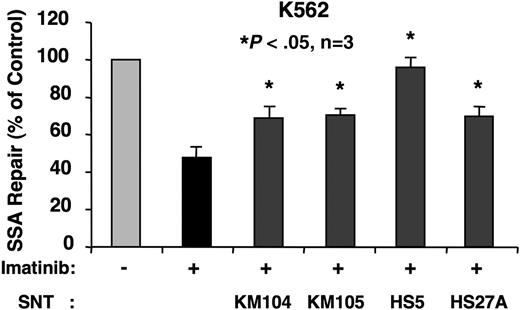
Increasing evidence suggests that imatinib-resistant CML cells develop from populations within the stromal cell microenvironment.25 The exact contribution of stromal cells to therapy-related drug resistance is not known. We sought to determine whether the stromal cell microenvironment provides receptor-mediated signals that could alter mutagenic DSB repair in BCR-ABL–transformed cells treated with imatinib. To begin to address this possibility, we determined the effect of stromal cell–conditioned media on SSA frequency in K562 cells. Culture media from 4 different human bone marrow stromal cell lines (KM104, KM105, HS5, and HS27A) were harvested after a 3-day culture. SSA frequency was measured in K562 cells in the presence or absence of imatinib (1 μM) and treatment with different conditioned media. Strikingly, conditioned media derived from all 4 stromal cell lines blocked the suppressive effect of imatinib on SSA, although to various degrees (Figure 3). It is of interest that none of the conditioned media supported cell growth under these experimental conditions (not shown). These data are consistent with a model supporting a role for the stromal cell microenvironment in promoting mutagenesis, resulting in disease progression and the development of drug resistance, even in the presence of BCR-ABL kinase inhibitors.
Stromal cell–conditioned media promotes SSA in imatinib-treated K562 cells. K562 cells were cotransfected with SAGFP and the I-SceI expression vector. SSA frequency was determined by flow cytometry after 48 hours in the presence of imatinib (1 μM) and stromal cell–conditioned media derived from KM104, KM105, HS5, or HS27A, as indicated (n = 3).
Stromal cell–conditioned media promotes SSA in imatinib-treated K562 cells. K562 cells were cotransfected with SAGFP and the I-SceI expression vector. SSA frequency was determined by flow cytometry after 48 hours in the presence of imatinib (1 μM) and stromal cell–conditioned media derived from KM104, KM105, HS5, or HS27A, as indicated (n = 3).
Various oncogenic tyrosine kinases promote SSA
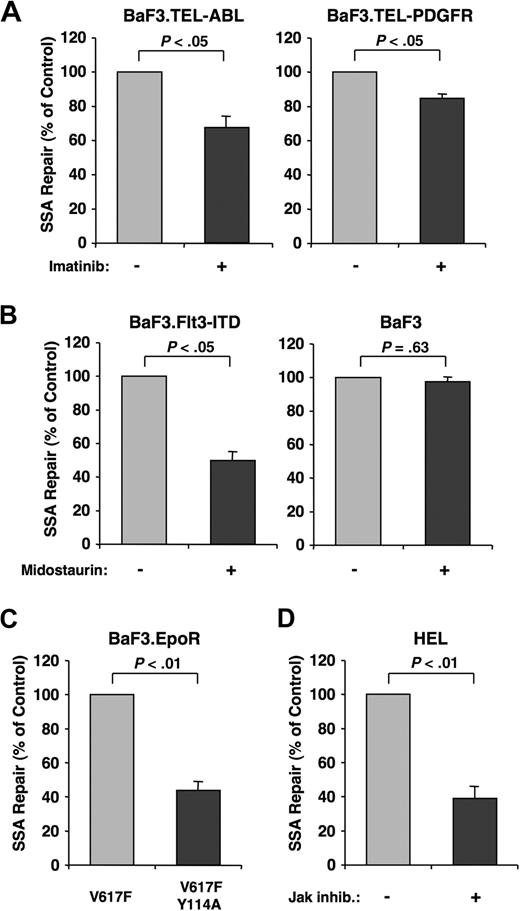
In addition to the BCR-ABL oncogene, other oncogenic tyrosine kinases have also been associated with genomic instability.26 We therefore hypothesized that heightened SSA activity was not unique to BCR-ABL. To test this, we used BaF3 cell lines transformed by Tel-ABL, Tel-PDGFR, and FLT3-ITD. In addition to BCR-ABL, imatinib also effectively inhibits Tel-ABL and Tel-PDGFR27 kinase activity. The FLT3 inhibitor midostaurin (PKC412)28 was used in cells transformed with FLT3-ITD. Transformed cells were treated with their respective inhibitors after cotransfection of SAGFP and the I-SceI expression plasmid. Similar to imatinib-treated BaF3.BCR-ABL cells, imatinib reduced the frequency of SSA in BaF3.Tel-ABL cells (67.0% ± 6.7%) and BaF3.Tel-PDGFR cells (84.64% ± 2.54%) compared with untreated cells (Figure 4A). In addition, midostaurin reduced the frequency of SSA in BaF3.FLT3-ITD cells (49.75% ± 4.8%), but not in the parental BaF3 cells (97.49% ± 4.4%; Figure 4B).
Oncogenic tyrosine kinases increase SSA repair. SSA frequency was determined in BaF3 cells transformed with different oncogenic tyrosine kinases. Cells cotransfected with SAGFP and I-SceI expression vector were evaluated by flow cytometry after 48 hours. (A) BaF3 cells transformed with the Tel-ABL (n = 4) and Tel-PDGFR (n = 5) oncogenes were used to measure SSA frequency in response to imatinib (1 μM) treatment, as indicated. (B) BaF3 cells transformed with FLT3-ITD and parental BaF3 cells were used to measure SSA frequency in response to midostaurin (50 nM) treatment, as indicated (n = 3). (C) Parental BaF3.EpoR cells containing the Jak2.V617F mutation or the double mutation, V617F.Y114A, were used to measure SSA frequency (n = 3). (D) HEL cells expressing mutant Jak2.V617F were used to measure SSA frequency in response to a Jak inhibitor (1 μM) (n = 3).
Oncogenic tyrosine kinases increase SSA repair. SSA frequency was determined in BaF3 cells transformed with different oncogenic tyrosine kinases. Cells cotransfected with SAGFP and I-SceI expression vector were evaluated by flow cytometry after 48 hours. (A) BaF3 cells transformed with the Tel-ABL (n = 4) and Tel-PDGFR (n = 5) oncogenes were used to measure SSA frequency in response to imatinib (1 μM) treatment, as indicated. (B) BaF3 cells transformed with FLT3-ITD and parental BaF3 cells were used to measure SSA frequency in response to midostaurin (50 nM) treatment, as indicated (n = 3). (C) Parental BaF3.EpoR cells containing the Jak2.V617F mutation or the double mutation, V617F.Y114A, were used to measure SSA frequency (n = 3). (D) HEL cells expressing mutant Jak2.V617F were used to measure SSA frequency in response to a Jak inhibitor (1 μM) (n = 3).
To measure the effects on SSA frequency resulting from Jak2V617F, we used BaF3.EpoR cells that express either Jak2V617F or mutant Jak2V617F.Y114A. The erythropoietin receptor (EpoR) in this BaF3 cell model is required for transformation by Jak2V617F. The additional Y114A mutation in the FERM domain renders Jak2 nontransforming,29 allowing for a comparison with the Jak2V617F mutant. The frequency of SSA in BaF3.EpoR cells containing the Jak2V617F.Y114A double mutation was significantly reduced (43.88% ± 5.3%) compared with Jak2V617F-expressing cells (Figure 4C). To exclude differences between murine and human cells, we assayed SSA frequency in the human HEL cell line, which contains a Jak2V617F mutation. Confirming the role of Jak2 signaling in promoting SSA, the frequency of SSA was reduced (39.17% ± 6.1%) in cells treated with a Jak inhibitor (Figure 4D). These data suggest that increased SSA activity is shared between the Tel-ABL, Tel-PDGFR, FLT3-ITD, and Jak2V617F proteins and can be targeted by small-molecule drugs that inhibit oncogenic tyrosine kinases.
SSA is regulated through the PI3K and Ras pathways
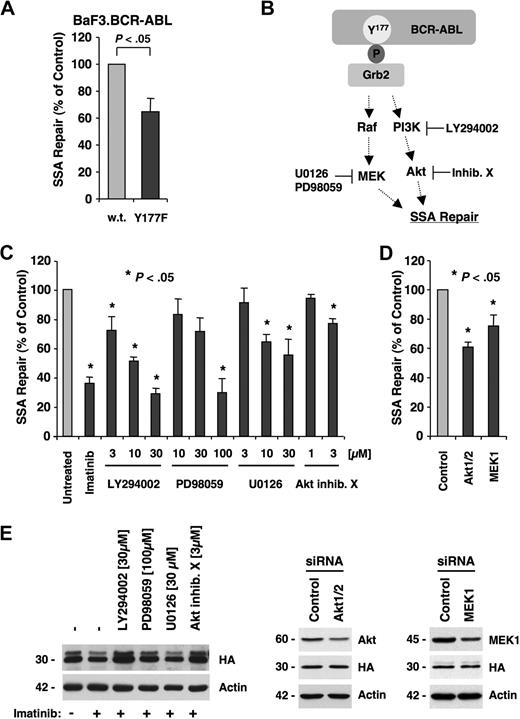
In BCR-ABL transformation, mutagenesis and the resulting induction of imatinib resistance are regulated in part through the Grb2-binding site (Tyr177/Y177).6 BCR-ABL requires an intact Grb2-binding site for optimal activation of the PI3K and potentially Ras pathways.18,30,,–33 This site is also required for the induction of a transforming phenotype in hematopoietic progenitor cells.34 To determine the role of Grb2 signaling pathways in promoting SSA, we compared the frequency of SSA in BaF3.BCR-ABL cells and BaF3 cells containing BCR-ABL.Y177F. After cotransfection of SAGFP and the I-SceI expression plasmid, SSA frequency in cells expressing BCR-ABL.Y177F (65.2% ± 9.8%) was reduced compared with BaF3.BCR-ABL cells (Figure 5A).
SSA is regulated by BCR-ABL through the PI3K and Ras/MEK pathways. (A) BaF3 cells containing BCR-ABL or BCR-ABL.Y177F were used. Cells were cotransfected with SAGFP and I-SceI expression vector and assayed for SSA frequency by flow cytometry (n = 5). (B) Phosphorylated Y177 in the active BCR-ABL oncoprotein provides a docking site for Grb2. Signaling downstream of Y177 includes activation of PI3K, Akt, and Ras/MEK pathways. Pharmacologic inhibition of PI3K with LY294002, Akt with Akt inhibitor X, MEK1 with PD98059, or MEK1/2 with U0126 blocks these pathways. (C) I-SceI was expressed in BaF3.BCR-ABL cells with integrated SAGFP and treated for 48 hours with LY294002 (n = 4), PD98059 (n = 4), U0126 (n = 4), or Akt inhibitor X (n = 3). (D) I-SceI was expressed in BaF3.BCR-ABL cells with integrated SAGFP and transfected with control siRNA, siRNA targeting Akt1/2, or MEK1 (n = 3). (E) Cellular expression of HA-tagged I-SceI (HA), actin, Akt1/2/3, or MEK1 was detected by immunoblotting in response to inhibitors or siRNA, as indicated.
SSA is regulated by BCR-ABL through the PI3K and Ras/MEK pathways. (A) BaF3 cells containing BCR-ABL or BCR-ABL.Y177F were used. Cells were cotransfected with SAGFP and I-SceI expression vector and assayed for SSA frequency by flow cytometry (n = 5). (B) Phosphorylated Y177 in the active BCR-ABL oncoprotein provides a docking site for Grb2. Signaling downstream of Y177 includes activation of PI3K, Akt, and Ras/MEK pathways. Pharmacologic inhibition of PI3K with LY294002, Akt with Akt inhibitor X, MEK1 with PD98059, or MEK1/2 with U0126 blocks these pathways. (C) I-SceI was expressed in BaF3.BCR-ABL cells with integrated SAGFP and treated for 48 hours with LY294002 (n = 4), PD98059 (n = 4), U0126 (n = 4), or Akt inhibitor X (n = 3). (D) I-SceI was expressed in BaF3.BCR-ABL cells with integrated SAGFP and transfected with control siRNA, siRNA targeting Akt1/2, or MEK1 (n = 3). (E) Cellular expression of HA-tagged I-SceI (HA), actin, Akt1/2/3, or MEK1 was detected by immunoblotting in response to inhibitors or siRNA, as indicated.
Because Y177 regulates multiple pathways implicated in transformation, including the induction of ROS and point mutations, we investigated the role of signaling pathways downstream of Y177 in the regulation of SSA. We targeted the PI3K and Ras pathways using small-molecule inhibitors to block PI3K (LY294002), Akt (Akt inhibitor X), and MEK1 (U0126), as well as MEK1/2 (PD98059) downstream of Ras (Figure 5B). We found that inhibition of these pathways in BaF3.BCR-ABL cells containing an integrated copy of SAGFP resulted in a dose-dependent reduction in SSA frequency. LY294002 decreased SSA at a concentration range of 3 to 30 μM (72.4% ± 9.2% to 29.0% ± 3.7%). The Akt inhibitor X decreased SSA at a concentration of 3 μM (77.5% ± 2.2%). Blocking the MEK pathway with PD98059 decreased SSA significantly at a concentration of 100 μM (29.8% ± 9.5%), and a significant dose-dependent decrease in SSA was observed with U0126 at 10 μM (71.5% ± 9.3%) and 30 μM (55.3% ± 10.8%; Figure 5C). Further, SSA was significantly decreased using specific siRNAs against Akt1/2 (61.0% ± 3.2%) and MEK1 (75.4% ± 7.3%; Figure 5D). The knockdown of Akt1/2 and MEK1 was determined by immunoblotting 24 hours after electroporation. Akt1/2 and MEK1 expression was found to be reduced at the time of I-SceI expression. We also confirmed that small-molecule inhibitors and targeted RNA interference did not reduce the expression of I-SceI (Figure 5E). Overall, these data indicate that SSA is regulated through the PI3K and Ras pathways downstream of Y177 and can be targeted using small-molecule drugs. Whether this regulation is specific to BCR-ABL or is a more generalized mechanism in eukaryotic cells remains to be determined.
BCR-ABL does not affect the precision of HDR in BaF3 cells
In general, HDR is considered to be a high-fidelity event; however, recent evidence in a myeloid cell line suggests that this process may be uniquely altered in the presence of BCR-ABL.7 To assay HDR, we used the DRGFP reporter, which consists of a full-length GFP gene disrupted by an I-SceI recognition site. After cleavage by I-SceI, repair by HDR using a downstream internal repeat can result in a functional GFP gene (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). To determine whether BCR-ABL affects the fidelity of HDR in our model system, we analyzed BaF3.BCR-ABL cells with an integrated copy of DRGFP by sequencing the GFP gene from 20 different GFP+ subclones recovered after expression of I-SceI. Surprisingly, we found no point mutations in any of the repaired GFP sequences (supplemental Figure 1). Further studies using nested PCR confirmed these data (data not shown), suggesting that, in BaF3.BCR-ABL cells, HDR is a high-fidelity repair mechanism. It is thus possible that the fidelity of HDR varies between different cell types. In contrast with the increased SSA in BCR-ABL transformed cells, the frequency of GFP+ cells recovered after I-SceI expression in BaF3.BCR-ABL cells containing DRGFP was decreased 1.7-fold relative to imatinib-treated cells (data not shown). Because HDR and SSA are known to compete for repair of DSBs,15 these data argue that BCR-ABL shifts the balance away from the precise HDR pathway and toward the mutagenic SSA pathway.
Discussion
Transformation by BCR-ABL and other oncogenic tyrosine kinases is associated with genomic instability in vivo. This instability may promote the acquisition of additional genomic alterations that result in therapy-related drug resistance and disease progression.1 The molecular mechanisms that regulate these processes seem, at least in part, to require the kinase activity of the transforming oncogene. BCR-ABL kinase activity is essential for promoting increased intracellular ROS levels that result in single-strand breaks and DSBs. Of note, BCR-ABL directly affects the efficiency of nucleotide excision repair35 and mismatch repair.36 Thus, single-strand breaks induced by ROS may be improperly or inefficiently repaired and ultimately result in additional DSBs.
One of the initial branchpoints in the response to a DSB is whether the ends undergo processing to 3′ single-strand tails or are protected from processing and repair through classic NHEJ.37,38 Cells deficient for classic NHEJ factors have increased rates of repair by other pathways, including HDR and alternate, error-prone NHEJ.8,12,39 The latter pathway is a probable cause for point mutations and short sequence deletions, such as those observed in BCR-ABL and associated with imatinib resistance. Importantly, expression of the classic NHEJ factors Artemis, DNA ligase IV, and possibly DNA-PKcs is reduced in BCR-ABL–expressing cells.16,40 Thus, BCR-ABL may suppress classic NHEJ and favor processing of ends to substrates that can participate in HDR, SSA, or error-prone NHEJ.37 Our data hint at potential overlap in the regulation of SSA by various oncoproteins.
The factors that determine whether a processed DSB is repaired by HDR, SSA, or error-prone NHEJ are partially understood. SSA is highly efficient between precise repeats but is suppressed by sequence divergence.22 RAD51 and BRCA2 promote HDR, whereas SSA is promoted by RAD52 or ERCC1 and suppressed by RAD51.15 BCR-ABL–associated changes in the expression of RAD52 or ERCC1 were not observed in our analysis (not shown). Previous data by others suggested that RAD51 expression is increased in BCR-ABL–transformed cells.41 However, the regulation of RAD51 by BCR-ABL remains controversial.42
Our preliminary findings suggest that BCR-ABL decreases the frequency of HDR (not shown) concomitant with an increase in mutagenic SSA. BCR-ABL–dependent changes in SSA were also observed by Cramer et al, published during revision of this manuscript.43 Although we did not directly assay NHEJ, previous studies also reported increased rates of error-prone NHEJ in BCR-ABL–expressing cells.7 We also found that BCR-ABL does not alter the sequence fidelity of either SSA or HDR in our model system. However, this does not exclude the possibility that BCR-ABL promotes error-prone HDR in some cellular contexts, as has been previously demonstrated.7 It is probable that yet to be identified factors lead to altered repair frequency and fidelity, which may vary in cell line models or, more importantly, among CML patients. We are currently attempting to develop methodologies to determine the fidelity and frequency of DSB repair in CML patient samples to answer this question.
Our data suggest that increased SSA activity depends on oncogenic BCR-ABL kinase activity and is regulated in part through signaling pathways downstream of Y177 in BCR-ABL. The Y177 autophosphorylation site is thought to regulate both Ras and PI3K signaling through recruitment of Grb2 complexes.18,30,,–33 Consistent with this mechanism, we found that pharmacologic inhibition of PI3K or MEK1/2 as well as targeted knockdown of Akt1/2 and MEK1 lead to a significant reduction in SSA. Y177 is already known to be required for optimal induction of ROS,17 and oxidative stress regulated through Y177 is thought to be sufficient to promote DNA mutations.7 Our findings add to the current model, whereby signaling pathways downstream of Y177 contribute to genomic instability. Thus, targeting Y177-dependent signaling probably affects genomic stability in CML. The same downstream pathways may also be important in related diseases with activated tyrosine kinases. It will now be interesting to further identify the exact repair mechanisms that are regulated through Y177. Methodologies that can overcome the inherent difficulties associated with the short-term culture of transfected primary patient samples will be helpful to validate targeted approaches in a fashion similar to our in vitro cell lines.
Normal hematopoiesis is supported by cytokines in the bone marrow microenvironment that help to maintain viability and self-renewal of stem cells. It is thought that this interaction provides a venue for a small population of cancer stem cells to escape targeted therapies.44 Nevertheless, the role of receptor-mediated signals from the tumor microenvironment in progression of the disease and development of genomic instability is not known. We have shown that stromal cell-conditioned media in K562 cells are sufficient to increase SSA activity and can increase intracellular ROS as well (not shown) in the presence of imatinib. These data suggest that the bone marrow microenvironment can overcome some effects of imatinib, by both promoting mutagenic repair and increasing oxidative stress. Secondary mutations caused by SSA may complement BCR-ABL during transformation. It will thus be important to determine factors involved in this process, as they are expected to play a role in the interactions between CML cells and bone marrow stroma. Targeting these interactions and SSA probably reduces genomic instability in CML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the National Institutes of Health (grants CA134660-01, M.S.; CA66996, CA36167, and DK50654, J.D.G.), the Leukemia & Lymphoma Society (SCOR grant, J.D.G.), the American Cancer Society, the US Department of Defense, and the Adams Barr Program in Innovative Cancer Research (M.S.).
National Institutes of Health
Authorship
Contribution: M.S.F. designed research, performed research, and wrote the manuscript; M.M.R., J.R.G., and S.C.D. performed research; J.D.G. provided vital reagents and revised the manuscript; K.P. and D.M.W. designed research and revised the manuscript; and M.S. designed research, performed research, and wrote the manuscript.
Conflict-of-interest disclosure: J.D.G. has a financial interest with Novartis Pharma, Basel, Switzerland. The remaining authors declare no competing financial interests.
Correspondence: Martin Sattler, Department of Medical Oncology, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: martin_sattler@dfci.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal