The naive phenotype of cord blood (CB) T cells may reduce graft-versus-host disease after umbilical cord blood transplantation, but this naivety and their low absolute numbers also delays immune reconstitution, producing higher infection-related mortality that is predominantly related to CMV, adenovirus (Adv), and EBV. Adoptive immunotherapy with peripheral blood-derived virus-specific cytotoxic T lymphocytes (CTLs) can effectively prevent viral disease after conventional stem cell transplantation, and we now describe the generation of single cultures of CTLs from CB that are specific for multiple viruses. Using EBV-infected B cells transduced with a clinical-grade Ad5f35CMVpp65 adenoviral vector as sources of EBV, Adv, and CMV antigens, we expanded virus-specific T cells even from CB T cells with a naive phenotype. After expansion, each CTL culture contained both CD8+ and CD4+ T-cell subsets, predominantly of effector memory phenotype. Each CTL culture also had HLA-restricted virus-specific cytotoxic effector function against EBV, CMV, and Adv targets. The CB CTLs recognized multiple viral epitopes, including CD4-restricted Adv-hexon epitopes and immunosubdominant CD4- and CD8-restricted CMVpp65 epitopes. Notwithstanding their naive phenotype, it is therefore possible to generate trivirus-specific CTLs in a single culture of CB, which may be of value to prevent or treat viral disease in CB transplant recipients. This study is registered at www.clinicaltrials.gov as NCT00078533.
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) is the treatment of choice for selected patients with high-risk hematologic malignancies. However, a significant proportion of patients—especially nonwhites—lack a marrow or peripheral blood stem cell donor. Umbilical cord blood (CB) has therefore emerged as an important alternative source of stem cells for patients who receive an allotransplant.1,2
The major advantages of CB transplants compared with unrelated adult donor stem cells include more rapid procurement of the graft, the requirement for less-stringent HLA matching, the higher likelihood of finding a match for ethnic minorities, and a decreased incidence of graft-versus-host disease. Although a major disadvantage of CB is the low stem cell dose, which results in delayed engraftment, there is also delayed immune recovery, particularly in mismatched unrelated cord blood transplantations with the use of antithymocyte globulin.3 This is attributed to the small numbers of T cells transferred, the absence of memory T cells within the CB grafts, and the apparent hyporesponsiveness of CB antigen-presenting cells. Consequently, CB recipients are susceptible to an array of viral and other infections that are the leading cause of death in these patients.3,,,–7
Cytomegalovirus (CMV), Epstein-Barr virus (EBV), and adenovirus (Adv) are particularly problematic in the immunocompromised recipient and are associated with significant rates of morbidity and mortality.8,–10 Although antiviral drugs can be beneficial for CMV and EBV, effective agents are unavailable for Adv; all have substantial toxicities and drug resistance may occur. These deficiencies in conventional therapeutics have increased interest in an immunotherapeutic approach to viral disorders, and adoptive transfer of monoculture-derived multivirus-specific cytotoxic T-lymphocytes (CTLs) from CMV-seropositive donors can prevent and treat CMV, EBV, and adenoviral disease after HSCT without significant toxicities.11
There are significant conceptual obstacles to extending this approach to the recipients of CB transplants. Only limited numbers of CB T cells are available for manipulation, and these cells have a naive phenotype, making them incapable of conferring protection against CMV, EBV, and Adv during engraftment.3,12 Hence, the development of virus-protective T-cell therapy for CB transplant recipients requires the priming and extensive expansion of naive T cells rather than the more limited and simple direct expansion of a preexisting virus-specific memory T-cell population. This task is made even more challenging because CB T cells have lower cytotoxic activity and higher activation-induced cell death than do peripheral blood–derived T cells.13,14 These limitations have prevented the successful generation of virus-specific CTLs in sufficient numbers for clinical use as prophylactic therapy of CMV, EBV, and adenoviral infections after CB transplantation (CBT).
It has become evident that, in addition to antigen presentation and intercellular costimulation, the ex vivo priming of antigen-specific T cells requires the presence of appropriate soluble cytokines. Hence, IL-7, IL-15, and IL-12, respectively, decrease the antigen concentration threshold, direct T cells toward a central memory phenotype, and influence the polarization of T cytotoxic type 1 and T helper type 1 cells. In combination these cytokines augment the generation of antigen-specific CTLs from naive T cells.15
We now describe how Ad5f35pp65-transduced CB-derived antigen-presenting cells can be used to generate large numbers of T cells that are specific for both CMV and Adv even from the naive T-cell population in human CB. Incorporation of EBV-transformed B-lymphoblastoid cell lines in the antigen-presenting cells further allows the Adv/CMV specificity of the CB T cells to be extended to EBV. The cells we describe have broad epitope specificity and are cytolytic to CMV, Adv, and EBV target cells. Hence, they may be protective in vivo.
Methods
CB units
Virus-specific CTLs were generated from CB units obtained frozen from the M. D. Anderson Cancer Center Cord Blood Bank or fresh from mothers who had consented to the protocol approved by the Institutional Review Board (IRB) of Baylor College of Medicine. To ensure the future clinical feasibility of this approach, we also froze the fresh CB units in DMSO containing 50% human serum before use for the generation of CB-derived CTLs, thereby mimicking the likely clinical setting. All CB samples were typed by the HLA laboratory of The Methodist Hospital, and we used CB from donors with multiple HLA types (Table 1). To further ensure that the approach would be feasible clinically, a total of only 40 million CB mononuclear cells (which can be obtained from the 20% fraction of frozen CB units) were thawed and used in the manufacturing process. According to our IRB approval, clinical protocol patients must have a single CB unit matched with the patient at 4, 5, or 6 of 6 HLA class I (serologic) and II (molecular) antigens. The unit must be cryopreserved in 2 fractions, with a minimum of 2.5 × 107 total nucleated cells/kg prethaw in the fraction which will be used for the primary transplant. The remaining fraction will be used to generate the CTLs to give at day 30 or beyond as described in section entitled “Generation of multivirus-specific cultures derived from umbilical CB.” This cell dose has been found to support acceptable engraftment in both pediatric and adult patients and is a commonly used minimal cell dose target in the CB transplantation community.16
Cord blood and maternal HLA typing and CMV serostatus
| Cord ID . | HLA typing cord . | HLA typing mother . | CMV serostatus of mother . |
|---|---|---|---|
| CB1 | A2;3/B44;51 | Unknown | Negative |
| CB2 | A2;24/B7 | Unknown | Unknown |
| CB3 | A2;29/B35;44 | Unknown | Positive |
| CB4 | A3/B7;44 | A2;3/B7;44 | Positive |
| CB5 | A3;29/B44;45 | A2;3/B7;44 | Positive |
| CB6 | A2;33/B40(60);44 | A2/B40(60);40(61) | Negative |
| CB7 | A11;30/B18;52 | Unknown | Negative |
| CB8 | A2;3/B8;18 | Unknown | Negative |
| CB9 | A29;31/B7;51 | Unknown | Unknown |
| Cord ID . | HLA typing cord . | HLA typing mother . | CMV serostatus of mother . |
|---|---|---|---|
| CB1 | A2;3/B44;51 | Unknown | Negative |
| CB2 | A2;24/B7 | Unknown | Unknown |
| CB3 | A2;29/B35;44 | Unknown | Positive |
| CB4 | A3/B7;44 | A2;3/B7;44 | Positive |
| CB5 | A3;29/B44;45 | A2;3/B7;44 | Positive |
| CB6 | A2;33/B40(60);44 | A2/B40(60);40(61) | Negative |
| CB7 | A11;30/B18;52 | Unknown | Negative |
| CB8 | A2;3/B8;18 | Unknown | Negative |
| CB9 | A29;31/B7;51 | Unknown | Unknown |
Generation of dendritic cells from cord blood
Umbilical CB was thawed and then purified by Ficoll (Lymphoprep; Nycomed) gradient separation. CB mononuclear cells (CBMCs; 30 × 106) were washed twice and resuspended in CellGenix media (CellGenix USA) and plated at 5 × 106 cells/well in dendritic cell (DC) medium (CellGenix media plus 2 mM l-glutamine; GlutaMAX; Invitrogen) in a 6-well plate (Costar) for 2 hours at 37°C in a humidified CO2 incubator. Nonadherent cells were removed by rinsing with 1× PBS (GibcoBRL) and refrozen. Loosely adherent cells were cultured in DC media with 800 U/mL GM-CSF (Sargramostim Leukine; Immunex) and 500 U/mL IL-4 (R&D Systems) for 7 days. IL-4 and GM-CSF were again added on day 3. On day 5, cells were harvested for transduction.
Transduction of DCs
As our source of CMV and Adv antigens, we used a clinical-grade recombinant Adv type 5 vector pseudotyped with an Adv type 35 fiber, that encoded CMVpp65.11
On day 5, the CB-derived DCs were transduced with the clinical-grade Ad5f35CMVpp65 vector11 at a multiplicity of infection of 10 IU/cell for 2 hours and matured in a cytokine cocktail of GM-CSF, IL-4, IL-1β, TNF-α, IL-6, (R&D Systems) and PGE2 (Sigma-Aldrich) for 2 days. On day 7, the DCs were harvested, irradiated (30 Gy) and then used to stimulate virus-specific CTLs.
Generation and transduction of EBV-transformed B-cell lines from umbilical CB
As the source of EBV antigens, 5 × 106 CBMCs were used for the establishment of an EBV-transformed lymphoblastoid cell line (EBV-LCL), by infection of CBMCs with concentrated clinical-grade supernatants from a B95-8 working cell bank.17
Two days before CTL stimulation, LCLs were harvested, pelleted, and incubated with Ad5f35CMVpp65 at a multiplicity of infection of 100 IU/cell for 90 minutes at 37°C. The cells were then resuspended at 5 × 105 cells/mL of complete media (RPMI [Hyclone] plus human serum and GlutaMAX) and transferred to a 24-well plate at 2 mL/well and cultured for 2 days before use as stimulator cells.
Generation of multivirus-specific cultures derived from umbilical CB
Frozen CBMCs were thawed, washed, and resuspended in 45% RPMI (Hyclone) and 45% CLICKS (Irvine Scientific) with 10% human serum plus GlutaMAX-I (CTL medium). Cells were resuspended at 2 × 106/mL and cocultured with autologous, irradiated transduced DCs at a ratio of 20 CBMCs to 1 DC in the presence of the cytokines IL-7, IL-12 (R&D Systems), and IL-15 (CellGenix), all at 10 ng/mL. Cultures were restimulated on day 10 with irradiated Ad5f35CMVpp65-transduced LCL at a responder-to-stimulator ratio of 4:1 plus IL-15 (10 ng/mL) and 1 week later with irradiated autologous Ad5f35CMVpp65-transduced LCL at a responder-to-stimulator ratio of 4:1. IL-2 (50-100 U/mL; Proleukin; Chiron) was added 3 days after the second stimulation and then twice weekly.
To define the origin of the virus-specific T-cell population we sorted CD45RA/CCR7 double-positive (DP) and double-negative (DN) CD3+ T-cell populations with the use of flow cytometry and stimulated the DP and DN populations with transduced DCs followed by transduced LCLs.
Peripheral blood–derived CTL lines
CTL lines from peripheral blood were prepared from stem cell donors who gave informed consent, in accordance with the Declaration of Helsinki, on enrollment in our clinical trial of virus-specific T cells for the treatment of viral infection after transplantation.11 All protocols were approved by the Baylor College of Medicine IRBs and the National Marrow Donor Program. For the purposes of this analysis, we have characterized 11 of these CTL lines.
Immunophenotyping
CTL lines were analyzed with monoclonal antibodies to CD19, CD4, CD8, CD56/16, TCRαβ, TCRγδ, CD45RA, CD3/28, CD62L, CD4/25, CCR7 (Becton Dickinson). Samples were acquired on a FACScan flow cytometer (Becton Dickinson), and the data were analyzed with the use of CellQuest software (Becton Dickinson).
Enzyme-linked immunospot assay
Enzyme-linked immunospot (ELISPOT) analysis was used to determine the frequency and function of T cells secreting IFN-γ in response to PepMixes (Jerini) for CMVpp65, AdvHexon, AdvPenton, and CMVIE1, which contain all 15mer peptides of each viral antigen in one pool.11 ELISPOT assays were performed on the CTL lines. In addition, to define the CD4- and CD8-restricted CMVpp65-specific activity we pulsed CD8+ or CD4+ T cells (sorted from CTL lines with flow cytometry) with CMV peptides.18 CBMCs stimulated with CMVIE1 PepMix and staphylococcal enterotoxin B (1 μg/mL; Sigma-Aldrich) served as controls. Spot-forming cells were enumerated by Zellnet Consulting and compared with input cell numbers to obtain the frequency of virus-reactive T cells.
Multimers and peptides
To detect CMVpp65-specific T cells in the CTL lines, we used the soluble HLA-peptide HLA-A*0201-MLN tetramer prepared by the Baylor College of Medicine Tetramer Core Facility. Peptides were synthesized by the Baylor College of Medicine Protein Core Facility or by Proimmune Inc. Tetramer staining of CTLs (5 × 105) was previously described.19 For each sample, 100 000 cells were analyzed using CellQuest software.
Panels of 20mer peptides (overlapping by 15 amino acids) covering the entire amino acid sequence of CMVpp65 from the AD169 strain and Adv-hexon from serotype 5 were synthesized.18,20 For CMVpp65,22 peptide pools comprising 2 to 12 15mer peptides were prepared, so that each 20mer peptide was represented in 2 pools.20 For Adv-hexon,11 peptide pools contained 17 or 18 peptides, so that each 20mer peptide was represented in one pool, and we then screened the positive pools as previously described.18 These CMVpp65 and Adv-hexon peptide libraries were designed to identify all possible HLA class I–restricted epitopes, which have a length of 9 to 11 amino acids, and to also identify HLA class II–restricted epitopes, which have lengths of 13 to 17 amino acids.21
Cytotoxicity assay
CTLs were tested for specific cytotoxicity against autologous LCLs,22 PHA blasts pulsed with CMVpp65 PepMix (Jerini), or Adv-hexon PepMix. As control target cells we used unpulsed PHA blasts, PHA blasts pulsed with irrelevant peptides, and HLA class I–mismatched LCLs. 51Cr-labeled target cells were mixed with effector cells at doubling dilutions to produce the effector-to-target (E/T) ratios specified. Target cells incubated in complete medium or 5% Triton X-100 (Sigma-Aldrich) were used to determine spontaneous and maximal 51Cr release, respectively. After 4 hours (LCLs and PHA blasts) or 6 hours (fibroblasts), supernatants were collected, and radioactivity was measured on a gamma counter. The mean percentage of specific lysis of triplicate wells was calculated as 100 × (experimental release − spontaneous release)/(maximal release − spontaneous release).
Statistical analysis
The Student t test was used to test for significance in each set of values, assuming equal variance. Mean values plus or minus SDs are given unless otherwise stated.
Results
Expanded CB-derived CTLs are virus specific and polyclonal
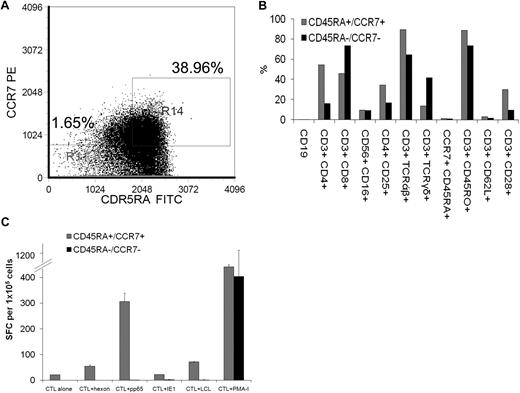
We prepared T-cell lines from 9 different CB units. After growth on Ad5f35CMVpp65-transduced DCs and LCLs in the presence of IL-2, IL-7, and IL-15, we harvested a median of 85.5 × 106 T cells (range, 60 × 106 to 2.65 × 108 T cells) by day 21 of culture. All of these cell numbers would be sufficient for current T-cell adoptive transfer protocols, in which 1 × 107/m2 effectively reconstitute immunity to CMV, EBV, and Adv11 and for product safety, function, and identity testing (Figure 1A). These cells were 78.7% CD8+ (range, 46%-94%) and 31% CD4+ (range, 12%-54%; Figure 1B). Flow cytometric analysis of memory markers showed a predominance of CD45RO+ CD62L− T cells (mean, 87.9% ± 5%) with a smaller population of CD45RA− CD62L+ T cells (mean, 14.6% ± 13%; Figure 1B).
Expansion, immunophenotype, and function of virus-specific CB-derived cytotoxic T lymphocytes. (A) For the generation of virus-specific CTLs from CB (virus-CTLs), a total of 2 × 106 CB mononuclear cells (CBMCs) were activated with autologous Ad5f35CMVpp65-transduced dendritic cells (DCs) in the presence of IL-12, IL-7, and IL-15 followed by weekly stimulations with Ad5f35CMVpp65-transduced LCL (LMP2-CTL) and twice weekly IL-2 feeds. CTL cell numbers (×106) are recorded from weekly cell counts comparing the expansion rates of 9 CB-derived CTL lines. (B) Reactivity of CTL lines (n = 9) with antibodies against the T-cell and natural killer cell surface antigens and memory markers CD3, CD4, CD8, CD16, CD56, TCRαβ, TCRγδ, CD45RA, CD28, CD62L, CD25, and the B-cell surface antigen CD19. Error bars indicate mean + SD. (C) 51Cr release at 4 hours after coincubation of a representative CTL line with PHA blasts pulsed with CMVpp65 PepMix (CMVpp65 target), or PHA blasts pulsed with CMVIE1 PepMix (CMVIE1 target), or PHA blasts pulsed with Adv-hexon PepMix (Adv-hexon target), nontransduced autologous EBV-LCL (EBV target), or allogeneic EBV-LCL (allogeneic target). The data are the percentage of lysis of targets by a representative CTL line at E/T of 40:1, 20:1, 10:1, and 5:1. (D) Virus-specific activity of all 9 CTL lines after 3 stimulations as determined with the IFN-γ ELISPOT assay in response to direct stimulation with CMVpp65 PepMix (CMVpp65), CMVIE1 PepMix (CMVIE1), Adv-hexon PepMix (Adv hexon), or irradiated autologous EBV-LCL at an E/T of 4:1 (EBV). For each CTL line, mean values of triplicate experiments are reported. SFCs indicate spot-forming cells.
Expansion, immunophenotype, and function of virus-specific CB-derived cytotoxic T lymphocytes. (A) For the generation of virus-specific CTLs from CB (virus-CTLs), a total of 2 × 106 CB mononuclear cells (CBMCs) were activated with autologous Ad5f35CMVpp65-transduced dendritic cells (DCs) in the presence of IL-12, IL-7, and IL-15 followed by weekly stimulations with Ad5f35CMVpp65-transduced LCL (LMP2-CTL) and twice weekly IL-2 feeds. CTL cell numbers (×106) are recorded from weekly cell counts comparing the expansion rates of 9 CB-derived CTL lines. (B) Reactivity of CTL lines (n = 9) with antibodies against the T-cell and natural killer cell surface antigens and memory markers CD3, CD4, CD8, CD16, CD56, TCRαβ, TCRγδ, CD45RA, CD28, CD62L, CD25, and the B-cell surface antigen CD19. Error bars indicate mean + SD. (C) 51Cr release at 4 hours after coincubation of a representative CTL line with PHA blasts pulsed with CMVpp65 PepMix (CMVpp65 target), or PHA blasts pulsed with CMVIE1 PepMix (CMVIE1 target), or PHA blasts pulsed with Adv-hexon PepMix (Adv-hexon target), nontransduced autologous EBV-LCL (EBV target), or allogeneic EBV-LCL (allogeneic target). The data are the percentage of lysis of targets by a representative CTL line at E/T of 40:1, 20:1, 10:1, and 5:1. (D) Virus-specific activity of all 9 CTL lines after 3 stimulations as determined with the IFN-γ ELISPOT assay in response to direct stimulation with CMVpp65 PepMix (CMVpp65), CMVIE1 PepMix (CMVIE1), Adv-hexon PepMix (Adv hexon), or irradiated autologous EBV-LCL at an E/T of 4:1 (EBV). For each CTL line, mean values of triplicate experiments are reported. SFCs indicate spot-forming cells.
We next used cytotoxicity and IFNγ ELISPOT assays to discover whether the T-cell lines were indeed trivirus-specific CTLs. Cytotoxic activity was tested against a panel of 51Cr-labeled autologous and allogeneic target cells and showed that the CTL lines killed PHA blasts only if they were pulsed with CMVpp65 PepMix or Adv-hexon PepMix (40% and 13%, respectively, at an E/T of 20:1; Figure 1C). Furthermore, autologous EBV-LCLs were also killed in this assay, showing an EBV-specific T-cell component. There was no cytotoxic activity against unpulsed PHA blasts, PHA blasts pulsed with CMVIE1 PepMix (Figure 1C),or HLA-mismatched EBV-LCLs (data not shown).
Similarly, Figure 1D shows that after 3 stimulations, virus-specific T-cell lines (n = 9) can secrete IFNγ in response to EBV, CMVpp65-, and Adv-hexon–expressing target cells with a mean (± SD) of 157 (± 134), 209 (± 210), and 74 (± 65) spot-forming cells per 105 T cells, after incubation with EBV-LCL, CMV-pp65, and Adv-hexon/penton peptides, respectively. In contrast, T cells did not respond to CMVIE1, the irrelevant peptide control.
Virus-specific CTLs are expanded from the naive CCR7/CD45RA+ T-cell population
We derived the virus-specific CTLs from CB, which should lack an EBV-, CMV-, or Adv-specific memory T-cell compartment. To determine whether the virus-specific T cells were indeed expanded from T cells with a naive phenotype (ie, CCR7+/CD45RA+ T-cell populations23 ), the mononuclear cells from 3 cord units (2 from CMV-seronegative mothers and 1 from a CMV-seropositive mother) were incubated with antibodies for CD45RA and CCR7. We used flow cytometry to sort T cells that were either DP for both CD45RA and CCR7 or DN for CD45RA and CCR7 (Figure 2A). Each of these populations was stimulated with transduced autologous antigen-presenting cells, and the virus-specific activity was evaluated with the use of the IFNγ ELISPOT. After 3 stimulations, the phenotypes of the resultant CTL lines showed a similar paucity of CD45RA+/CCR7+ T cells (Figure 2B). As shown in Figure 2C, T-cell lines derived from both CD45RA+/CCR7+ and CD45RA−/CCR7− populations secreted IFN-γ in response to PMA-ionomycin. However, only T cells derived from the CD45RA and CCR7 DP population secreted IFN-γ in response to EBV, CMV, and Adv antigen-expressing target cells.
Virus-specific cytotoxic T lymphocytes from CB are derived from the CD45RA+/CCR7+ naive T-cell population. (A) With the use of flow cytometry, T cells from 3 CB units were gated on CD3+ cells and then sorted for cells that were either double positive for both CD45RA and CCR7 or double negative for CD45RA and CCR7. Each of these populations was then stimulated with transduced autologous antigen-presenting cells. Indicated are percentage of CD45RA−/CCR7− T cells versus percentage of CD45RA+/CCR7+ T cells that were selected. (B) After 3 stimulations, the phenotype of the CTL lines derived from cells that were either double positive for both CD45RA and CCR7 (CD45RA+/CCR7+) or double negative for CD45RA and CCR7 (CD45RA−/CCR7−) was determined by flow cytometry. (C) Virus-specific activity of the CTL lines was determined after 3 stimulations by the IFN-γ ELISPOT assay in response to direct stimulation with CMVpp65 PepMix (CMVpp65), CMVIE1 PepMix (CMVIE1), Adenovirus hexon PepMix (Adv hexon), irradiated autologous EBV-LCL at an E/T ratio of 4:1 (EBV) or PMA-ionomycin (PMA-I). ELISPOT analysis of a T-cell line derived from the CD45RA and CCR7 double-positive fraction versus a T-cell line derived from the double-negative fraction from a representative CB unit from a CMV-seronegative mother is shown. Mean values (± SDs) of triplicate experiments are reported.
Virus-specific cytotoxic T lymphocytes from CB are derived from the CD45RA+/CCR7+ naive T-cell population. (A) With the use of flow cytometry, T cells from 3 CB units were gated on CD3+ cells and then sorted for cells that were either double positive for both CD45RA and CCR7 or double negative for CD45RA and CCR7. Each of these populations was then stimulated with transduced autologous antigen-presenting cells. Indicated are percentage of CD45RA−/CCR7− T cells versus percentage of CD45RA+/CCR7+ T cells that were selected. (B) After 3 stimulations, the phenotype of the CTL lines derived from cells that were either double positive for both CD45RA and CCR7 (CD45RA+/CCR7+) or double negative for CD45RA and CCR7 (CD45RA−/CCR7−) was determined by flow cytometry. (C) Virus-specific activity of the CTL lines was determined after 3 stimulations by the IFN-γ ELISPOT assay in response to direct stimulation with CMVpp65 PepMix (CMVpp65), CMVIE1 PepMix (CMVIE1), Adenovirus hexon PepMix (Adv hexon), irradiated autologous EBV-LCL at an E/T ratio of 4:1 (EBV) or PMA-ionomycin (PMA-I). ELISPOT analysis of a T-cell line derived from the CD45RA and CCR7 double-positive fraction versus a T-cell line derived from the double-negative fraction from a representative CB unit from a CMV-seronegative mother is shown. Mean values (± SDs) of triplicate experiments are reported.
Virus-specific CTLs derived from CB recognize immunodominant CD4-restricted Adv-hexon epitopes
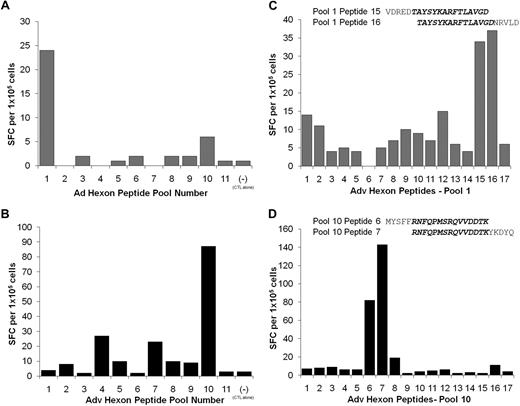
To map the Adv-hexon epitope specificity of the CTL lines, we used an overlapping peptide library representing the entire sequence of the Adv serotype 5 hexon protein.18,24 For the initial screening, CTL lines were stimulated by overnight incubation with the 188 overlapping hexon peptides, divided into 11 pools, containing 17 or 18 peptides per pool.
The results from Adv-hexon peptide pool screening of 6 evaluable CTL lines are summarized in Table 2. Of the 6 CTL lines, 3 had reactivity against one Adv-hexon epitope, and 2 lines had reactivity against multiple hexon epitopes. Three lines had reactivity against Adv penton. The hexon epitopes identified in these CB CTL lines were the same CD4-restricted epitopes as those identified in the multivirus CTL lines generated from adult peripheral blood.18
Adenovirus specificity of CB virus-specific T-cell lines
| Cord ID . | HLA typing cord . | Adv hexon epitope . |
|---|---|---|
| CB2 | A2;24/B7 | Penton specificity only |
| CB3 | A2;29/B35;44 | 556-580: VPFHIQVPQKFFAIKNLLLLPGSYT |
| CB6 | A2;33/B40(60);44 | 76-95: TAYSYKARFTLAVGDNRVLD |
| CB7 | A11;30/B18;52 | Hexon pools 1, 2, 3, 4 and penton specificity |
| CB8 | A2;3/B8;18 | Hexon pools 6, 10 and penton specificity |
| CB9 | A29;31/B7;51 | 796-815:RNFQPMSRQVVDDTKYKDYQ |
| Cord ID . | HLA typing cord . | Adv hexon epitope . |
|---|---|---|
| CB2 | A2;24/B7 | Penton specificity only |
| CB3 | A2;29/B35;44 | 556-580: VPFHIQVPQKFFAIKNLLLLPGSYT |
| CB6 | A2;33/B40(60);44 | 76-95: TAYSYKARFTLAVGDNRVLD |
| CB7 | A11;30/B18;52 | Hexon pools 1, 2, 3, 4 and penton specificity |
| CB8 | A2;3/B8;18 | Hexon pools 6, 10 and penton specificity |
| CB9 | A29;31/B7;51 | 796-815:RNFQPMSRQVVDDTKYKDYQ |
Figure 3 shows the details of such analyses from 2 of these CB-derived CTL lines. One line reacts predominantly against pool 1 (Figure 3A) and the other against pool 10 (Figure 3B). Subsequent analysis with individual 20mer peptides contained in these pools shows that the CTL reactivity in pool 1 could be mapped to overlapping peptides 15 (aa 71-90 VDREDTAYSYKARFTLAVGD) and 16 (aa 76-95 TAYSYKARFTLAVGDNRVLD; Figure 3C), whereas CTL reactivity directed against pool 10 could be mapped to overlapping peptide 6 (aa 791-814 MYSFFRNFQPMSRQVVDDTK) and peptide 7 (aa 796-815 RNFQPMSRQVVDDTKYKDYQ; Figure 3D). Thus, CB-derived CTLs have a similar pattern of epitope recognition to adult blood–derived CTLs. Furthermore, these epitopes are CD4-restricted with apparent immunodominance because they have been previously detected in multiple CTL lines derived from peripheral blood.18
Specificity of CB CTL lines for adenovirus hexon. CB-derived CTL lines were screened by ELISPOT assay against 11 pools of hexon peptides (20mers overlapping by 15 aa) to identify which peptide pools were being recognized by each CTL line. (A) The CTL line CB6 showed specific IFN-γ release specifically against peptide pool 1. (B) The CTL line CB9 showed specific IFN-γ release specifically against peptide pool 10. Results are expressed as spot-forming cells (SFCs)/105 CTLs. (C) To identify the stimulating 20mer peptides, CTL line CB6 was rescreened against the individual peptides contained in pool 1. The CTL reactivity against pool 1 were mapped to the overlapping peptides 15 (VDREDTAYSYKARFTLAVGD) and 16 (TAYSYKARFTLAVGDNRVLD). (D) In contrast, the reactivity in CTL line CB9 was mapped to peptides 6 (MYSFFRNFQPMSRQVVDDTK) and 7 (RNFQPMSRQVVDDTKYKDYQ) in pool 10. Results are expressed as SFCs per 105 CTLs.
Specificity of CB CTL lines for adenovirus hexon. CB-derived CTL lines were screened by ELISPOT assay against 11 pools of hexon peptides (20mers overlapping by 15 aa) to identify which peptide pools were being recognized by each CTL line. (A) The CTL line CB6 showed specific IFN-γ release specifically against peptide pool 1. (B) The CTL line CB9 showed specific IFN-γ release specifically against peptide pool 10. Results are expressed as spot-forming cells (SFCs)/105 CTLs. (C) To identify the stimulating 20mer peptides, CTL line CB6 was rescreened against the individual peptides contained in pool 1. The CTL reactivity against pool 1 were mapped to the overlapping peptides 15 (VDREDTAYSYKARFTLAVGD) and 16 (TAYSYKARFTLAVGDNRVLD). (D) In contrast, the reactivity in CTL line CB9 was mapped to peptides 6 (MYSFFRNFQPMSRQVVDDTK) and 7 (RNFQPMSRQVVDDTKYKDYQ) in pool 10. Results are expressed as SFCs per 105 CTLs.
Virus-specific CTLs derived from CB recognizes unconventional CD4- and CD8-restricted CMVpp65 epitopes
We characterized the CMVpp65 epitope specificity of the CTLs by incubating them with CMVpp65 peptide pools20 and measuring IFNγ release by ELISPOT assays. Table 3 shows that evaluable virus-specific lines derived from umbilical CB can secrete IFNγ in response to several different CMVpp65 peptides, representing discrete epitopes on the CMVpp65 antigen.
CMVpp65 epitope specificity of cord blood virus-specific T-cell lines
| Cord ID . | HLA typing cord . | CMVpp65 epitope(s) . | Predicted immunodominant CMVpp65 epitope(s) not identified . |
|---|---|---|---|
| CB1 | A2;3/B44;51 | 31-51: DTPVLPHETRLLQTGIHVRV126-146: INVHHYPSAAERKHRHLPVA | 495-503: NLVPMVATV |
| CB2 | A2;24/B7 | 120-129: MLNIPSINV351-371: LLQRGPQYSEHPTFT | 495-503: NLVPMVATV341-349: QYDPVAALF417-426: TPRVTGGGAM 265-275: RPHERNGFTVL |
| CB3 | A2;29/B35;44 | 31-51: DTPVLPHETRLLQTGIHVRV120-129: MLNIPSINV347-358: ALFFFDIDLLL | 495-503: NLVPMVATV |
| CB4 | A3/B7;44 | Pools 14,16,19 (epitope not identified) | 417-426: TPRVTGGGAM 265-275: RPHERNGFTVL |
| CB5 | A3;29/B44;45 | Pools 6,7,10,14,18 (epitope not identified) | N/A |
| CB6 | A2;33/B40(60);44 | 221-241: DQYVKVYLESFCEDVPSGKL256-276: MTRNPQPFMRPHERNGFTVL | 495-503: NLVPMVATV |
| CB7 | A11;30/B18;52 | 11-31:MISVLGPISGHVLKAVFSRG151-171:ASGKQMWQARLTVSGLAWTR | N/A |
| CB8 | A2;3/B8;18 | Not identified | 495-503: NLVPMVATV |
| CB9 | A29;31/B7;51 | 116-131: LPLKMLNIPSINVHHYPSAA | 417-426: TPRVTGGGAM265-275: RPHERNGFTVL |
| Cord ID . | HLA typing cord . | CMVpp65 epitope(s) . | Predicted immunodominant CMVpp65 epitope(s) not identified . |
|---|---|---|---|
| CB1 | A2;3/B44;51 | 31-51: DTPVLPHETRLLQTGIHVRV126-146: INVHHYPSAAERKHRHLPVA | 495-503: NLVPMVATV |
| CB2 | A2;24/B7 | 120-129: MLNIPSINV351-371: LLQRGPQYSEHPTFT | 495-503: NLVPMVATV341-349: QYDPVAALF417-426: TPRVTGGGAM 265-275: RPHERNGFTVL |
| CB3 | A2;29/B35;44 | 31-51: DTPVLPHETRLLQTGIHVRV120-129: MLNIPSINV347-358: ALFFFDIDLLL | 495-503: NLVPMVATV |
| CB4 | A3/B7;44 | Pools 14,16,19 (epitope not identified) | 417-426: TPRVTGGGAM 265-275: RPHERNGFTVL |
| CB5 | A3;29/B44;45 | Pools 6,7,10,14,18 (epitope not identified) | N/A |
| CB6 | A2;33/B40(60);44 | 221-241: DQYVKVYLESFCEDVPSGKL256-276: MTRNPQPFMRPHERNGFTVL | 495-503: NLVPMVATV |
| CB7 | A11;30/B18;52 | 11-31:MISVLGPISGHVLKAVFSRG151-171:ASGKQMWQARLTVSGLAWTR | N/A |
| CB8 | A2;3/B8;18 | Not identified | 495-503: NLVPMVATV |
| CB9 | A29;31/B7;51 | 116-131: LPLKMLNIPSINVHHYPSAA | 417-426: TPRVTGGGAM265-275: RPHERNGFTVL |
N/A indicates not applicable.
A representative example is detailed in Figure 4. This CTL line (HLA type: A2,29;B35,44) had a detectable T-cell response to 20mer peptides from pools 7, 8, 9, 10, 11, 13, 14, and 18 (Figure 4A). The 20mer peptides common to these 3 pools were peptides 7, 8, 22, 23, 69, and 70 (Figure 4B). As shown in Figure 4C, screening of these 20mer peptides showed T cells specific for at least 2 epitopes. They were the known HLA A0201-restricted epitope MLNIPSINV (confirmed by tetramer as shown in Figure 4D) and 2 other epitopes (ALFFFDIDLLLQRGPQYSE and DTPVLPHETRLLQTGIHVRV) spanning the regions 347 to 358 and 31 to 51, respectively. By contrast, analysis of this CTL line for the known HLA A2–restricted immunodominant CMVpp65 peptide (NLVPMVATV) was negative. Furthermore, 7 of the 7 CTL lines that were HLA A2 and/or A24 and/or B7 positive lacked T cells specific for the immunodominant A2 NLVPMVATV epitope and/or the B7 TPRVTGGGAM and RPHERNGFTVL epitopes and/or the A24 QYDPVAALF epitope (Table 3). These results are in contrast to CTL lines generated from peripheral blood donors11 with these HLA types in which 11 of 11 CTL lines exhibited specific activity against these immunodominant epitopes (Table 4).
Identification of CMVpp65 epitopes with the use of a CMVpp65-peptide library. (A) CTLs (105/well) from CB3 (HLA-A2, A29, B35, B44) were stimulated with a CMVpp65-peptide library pooled into 22 pools. Responses were measured in an 18-hour IFN-γ ELISPOT assay. Shown is mean and SD of duplicate wells. (B) All peptides were divided into 22 pools in such a way that each peptide is present in 2 pools. This method allows determining those single peptides that probably induced responses to the peptide pools. Thus, responses to pools 7, 8, and 13 or 9, 10, and 18 or 10, 11, and 14 may be induced by single peptides 7 and 8 or 69 and 70 or 22 and 23, respectively (C) Testing of these individual 20mers identifies the sequence of peptides 7 and 8 or 69 and 70 as most probably the overlapping 15 amino acids, as the CTL epitope. In addition, the T-cell line mapped to a known HLA A2–restricted epitope (MLNIPSINV) contained in peptides 23 and 24. (D) The polyclonal CB-derived virus-specific CTL line in which this epitope had been identified was stained with an HLA-A*0201 MLNIPSINV tetramer. Indicated is the percentage of tetramer-positive cells within the CD8+ population.
Identification of CMVpp65 epitopes with the use of a CMVpp65-peptide library. (A) CTLs (105/well) from CB3 (HLA-A2, A29, B35, B44) were stimulated with a CMVpp65-peptide library pooled into 22 pools. Responses were measured in an 18-hour IFN-γ ELISPOT assay. Shown is mean and SD of duplicate wells. (B) All peptides were divided into 22 pools in such a way that each peptide is present in 2 pools. This method allows determining those single peptides that probably induced responses to the peptide pools. Thus, responses to pools 7, 8, and 13 or 9, 10, and 18 or 10, 11, and 14 may be induced by single peptides 7 and 8 or 69 and 70 or 22 and 23, respectively (C) Testing of these individual 20mers identifies the sequence of peptides 7 and 8 or 69 and 70 as most probably the overlapping 15 amino acids, as the CTL epitope. In addition, the T-cell line mapped to a known HLA A2–restricted epitope (MLNIPSINV) contained in peptides 23 and 24. (D) The polyclonal CB-derived virus-specific CTL line in which this epitope had been identified was stained with an HLA-A*0201 MLNIPSINV tetramer. Indicated is the percentage of tetramer-positive cells within the CD8+ population.
CMVpp65 epitope specificity of peripheral blood virus-specific T-cell lines
| Peripheral blood CTL ID . | HLA typing . | CMVpp65 epitope(s) . |
|---|---|---|
| PB1 | A1;24/B51;57 | QYDPVAALF 369-379: FTSQYRIQGKL 113-121:VYALPLKML |
| PB2 | A1;80/B7;53 | TPRVTGGGAMRPHERNGFTVL |
| PB3 | A2/B8;49 | NLVPMVATV |
| PB4 | A2402,3201/B1302,3501 | QYDPVAALF |
| PB5 | A2;68/B35;40 | NLVPMVATV |
| PB6 | A1;2/B8;15 | NLVPMVATV |
| PB7 | A2,66/B27(w6),52(w6) | NLVPMVATV |
| PB8 | A2,3/B7,44 | NLVPMVATVTPRVTGGGAMRPHERNGFTVL |
| PB9 | A2/B40(61);46 | NLVPMVATV |
| PB10 | A2/B35;39 | NLVPMVATV |
| PB11 | A2;3/B1501;5301 | NLVPMVATV |
| Peripheral blood CTL ID . | HLA typing . | CMVpp65 epitope(s) . |
|---|---|---|
| PB1 | A1;24/B51;57 | QYDPVAALF 369-379: FTSQYRIQGKL 113-121:VYALPLKML |
| PB2 | A1;80/B7;53 | TPRVTGGGAMRPHERNGFTVL |
| PB3 | A2/B8;49 | NLVPMVATV |
| PB4 | A2402,3201/B1302,3501 | QYDPVAALF |
| PB5 | A2;68/B35;40 | NLVPMVATV |
| PB6 | A1;2/B8;15 | NLVPMVATV |
| PB7 | A2,66/B27(w6),52(w6) | NLVPMVATV |
| PB8 | A2,3/B7,44 | NLVPMVATVTPRVTGGGAMRPHERNGFTVL |
| PB9 | A2/B40(61);46 | NLVPMVATV |
| PB10 | A2/B35;39 | NLVPMVATV |
| PB11 | A2;3/B1501;5301 | NLVPMVATV |
To characterize the EBV-specific T-cell response, we screened the CTL lines for known immunodominant epitopes in EBNA 3, BZLF1, and LMP2.25 We found, however, no evidence for such dominant epitope-specific T cells (data not shown).
Virus-specific CTLs derived from CB are expanded from neonatal and not maternal T cells
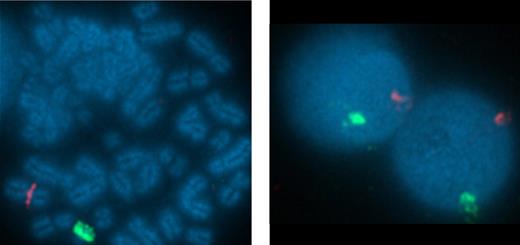
To confirm that the virus-specific T-cell responses observed in the CB-derived CTL lines were of neonatal and not maternal origin we used fluorescence in situ hybridization with sex chromosome-specific probes. X and Y chromosomes were identified by red and green fluorescence, respectively (Figure 5). Of the 200 metaphases analyzed, 199 were positive for X and Y chromosomes, confirming that the cells were of CB origin. For an additional 3 CTL lines from female neonates, the HLA types of the mother were known. As shown in Table 1 the maternal HLA types were compared with the CTL lines generated from cords C1000.4, C1001.4, and C1002.4. In all 3, only a single maternal HLA haplotype was found, so that the CTL line was derived from neonatal not maternal T cells.
Virus-specific CTL derived from CB are of neonatal and not maternal origin. To confirm the origin of the virus-specific T-cell responses observed in the CB-derived CTL line CB3 were of neonatal and not maternal origin we used fluorescence in situ hybridization (FISH) with sex chromosome–specific probes. FISH probes specific to α satellite centromeric region of chromosome X labeled with spectrum orange and the satellite III (Yp12) region of chromosome Y labeled with spectrum green were obtained from Abbott Laboratories. Hybridization was performed according to the manufacturer's protocols. The slides were counterstained with 4,6-diamidino-2-phenylindole (DAPI) and the images were captured using 10×/25 aperture at magnification of 100×/1.40 oil R on Nikon E800 microscope equipped with a cooled-charge coupled device (CCD) camera. The cells were analyzed using Quips Pathvysion (Applied Imaging). A total of 200 interphase nuclei and 5 metaphase spreads were analyzed for sex chromosome signal pattern.
Virus-specific CTL derived from CB are of neonatal and not maternal origin. To confirm the origin of the virus-specific T-cell responses observed in the CB-derived CTL line CB3 were of neonatal and not maternal origin we used fluorescence in situ hybridization (FISH) with sex chromosome–specific probes. FISH probes specific to α satellite centromeric region of chromosome X labeled with spectrum orange and the satellite III (Yp12) region of chromosome Y labeled with spectrum green were obtained from Abbott Laboratories. Hybridization was performed according to the manufacturer's protocols. The slides were counterstained with 4,6-diamidino-2-phenylindole (DAPI) and the images were captured using 10×/25 aperture at magnification of 100×/1.40 oil R on Nikon E800 microscope equipped with a cooled-charge coupled device (CCD) camera. The cells were analyzed using Quips Pathvysion (Applied Imaging). A total of 200 interphase nuclei and 5 metaphase spreads were analyzed for sex chromosome signal pattern.
Discussion
This study provides the first direct evidence that antigen-specific T cells targeting multiple viral epitopes from Adv, CMV, and EBV can be primed simultaneously from CB T cells with a naive phenotype. In addition, the multivirus-specific T cells recognize an array of epitopes after only 2 weeks of expansion in vivo. Notably, the pattern of CMV epitopes recognized appears to be different to “adult”-derived virus-specific T cells, whereas the recognition of Adv epitopes is the same.
Park et al26 have previously shown that CMV-specific T cells can be generated in vitro from umbilical CB, but the antigen and epitope specificity of the CMV-specific CTL was largely uncharacterized. Moreover, it was unclear whether the CMV-specific CTLs they detected were derived from T cells with a naive CD45RA+/CCR7+ phenotype.27 Questions also remained as to whether their CB-derived CTL came from contaminating maternal blood cells. Our current study addresses these limitations and also establishes the technology for simultaneously generating CTLs that are specific for all 3 of the commonest viral infections after CBT, using a clinically relevant and GMP-compliant method.
CB-derived T cells can respond to endogenous antigens, such as minor histocompatibility antigens that are disparate between mother and fetus, and to exogenous antigens, such as parasites and environmental allergens.28,29 They may also respond to viral antigens if the mother is seropositive or has been vaccinated. Hence, antigens in the maternal environment may prime fetal T cells transplacentally and elicit antigen-specific T-cell responses.30,31 In our study, however, it was evident that the responses we obtained were derived by in vitro priming of the naive population of T cells and could be obtained irrespective of whether the mother was CMV seropositive or consistently seronegative, before and after term. We showed that these cells were not of maternal origin; moreover, placental tissue from our donors was Adv and CMV negative by polymerase chain reaction) analysis and by immunohistochemistry, further excluding the possibility of priming in utero (data not shown).12
Although our ex vivo–expanded CB-derived multivirus-specific T cells developed a memory phenotype profile that was similar to peripheral blood–derived multivirus-specific T cells,11 their epitope specificity could be strikingly different. Thus, none of the CB-derived CTL lines recognized the strongly immunodominant epitopes that are universally identified in the peripheral blood CMVpp65 CTL lines generated from CMV-seropositive donors (Table 4), with a preference instead for unconventional and even novel CMVpp65 epitopes. CB T cells may be a transitional population between thymocytes and adult T cells,28 so that the specificities we describe may thus represent a “default response” by recent thymic emigrants, which have not undergone postnatal selection for more specific and, perhaps, higher affinity antiviral reactivity.22,32 This hypothesis is supported by the broad Vβ repertoire we observed in CB-derived CTLs with the use of spectratyping (data not shown). Murine studies showing that promiscuous low-affinity TCR/MHC-peptide interactions on the surface of functionally immature neonatal T cells leads to an intensive “burst” of short-lived cellular immunity before the establishment of conventional T-cell memory mediated by high-affinity T cells.33 Our own human in vitro and the murine in vivo studies appear to reflect physiologic events in humans, because CMV-infected fetuses are also deficient in CD8+ T cells responding to the canonical, immunodominant, B7-restricted RPH and A2-restricted NLV epitopes.31
Notwithstanding the recognition of noncanonical epitopes, CB-derived CMV-specific CTLs are capable of specifically lysing CMVpp65-pulsed targets. Moreover, these usually “subdominant” CMV epitopes still successfully compete against Adv-hexon epitopes, because CTLs responding to CMV-pp65 continue to exceed in number the CTLs responding to Adv-hexon, a characteristic they share with memory cell–derived CTLs directed to canonical pp65 epitopes.
Although the specificity of CB-naive T-cell responses to CMV were markedly different from adult seropositive peripheral blood T cells, the responses of the 2 T-cell populations to Adv were, perhaps surprisingly, indistinguishable. With the use of pools of overlapping 20mer peptides for hexon and pp65, we identified T cells specific for the same immunodominant CD4-restricted hexon epitopes found in adult seropositive donors, such as the hexon peptides that span amino acids 791 to 815 (MYSFFRNFQPMSRQVVDDTKYKDYQ).18 We do not know why CB and peripheral blood T cells should share epitope recognition for Adv but not CMV, but the difference probably reflects the profoundly different biology of the 2 viruses. Adv is an acute infection that elicits strong innate immunity, followed by an adaptive CD4-restricted hexon-specific T-cell response that in turn maximizes the range of epitopes recognized by CD4+ and CD8+ effector cells.18 As a latent virus, however, CMV recruits a dominant and oligoclonal CD8+ response.34 CB T cells are biased toward a T helper type 2 phenotype,28 which may support development of a conventional adenoviral CD4-specific T-cell response, but which, in combination with the immaturity of the responding CB T cells, may redirect the CMV response away from the canonical epitopes.
Our explanation leads us to predict that our CB T-cell responses to EBV, a latent virus like CMV, would also be to nondominant epitopes. An evaluation for noncanonical responses is more difficult for EBV than for CMV, because T cells from most responders recognize epitopes from multiple latent and early lytic cycle EBV antigens. Nevertheless, our screen of CTL lines for known immunodominant epitopes in EBNA 3, BZLF1, and LMP225 failed to show any of the anticipated epitope-specific T cells (data not shown) despite a functional cytokine and cytotoxic response to HLA-matched EBV-LCLs (Figure 1C-D). These observations are at least consistent with the differences we propose between CB naive T-cell responses to acute and latent viral antigens.
Although we were able to generate multivirus-specific CTLs from CB, success was only achieved when we supplemented the cultures with IL-7, IL-12, and IL-15. Such supplementation is unnecessary for the generation of virus-specific CTLs from adult memory T cells11 and may simply indicate a lack of memory T-cell precursors a deficiency of CB APC secretion of these cytokines, or both. Neonatal DCs certainly appear to have a deficiency in IL-12 production, and we have shown how cytokine supplementation can overcome these limitations.35 In the current studies, it is possible that the altered epitope recognition we observed in the CMV responses is a consequence of the addition of these supplementary cytokines. This seems unlikely because the sole explanation for the phenomenon not only in view of the supporting data from murine and human studies described earlier but also because the responses to Adv-hexon epitopes were unchanged. Hence, we believe these differences more likely reflect intrinsic differences in the responsiveness of naive CB versus memory T cells derived from CMV-seropositive donors.
Although antiviral pharmacotherapy may help prevent or treat CMV36,37 and CD20-specific antibodies may control EBV-associated lymphoproliferation,38 these drugs are expensive, toxic, and often ineffective because of primary or secondary resistance.39 Moreover, infections with adenoviruses are increasingly reported after both allogeneic HSCT and CBT, and effective treatments are not currently available.37 Strategies to reduce the incidence of viral infections after CBT include using grafts with the best HLA match and higher TNC, changes in conditioning, and graft-versus-host disease prophylaxis regimens. However, adoptive transfer of peripheral blood–derived T-cell lines enriched in cells recognizing CMV, EBV, and Adv can reproducibly control infections resulting from all 3 viruses after allogeneic HSCT11 in a cost-effective manner. We suggest that our ability to generate virus-specific CTLs from CB against a plethora of epitopes recognized by both CD4+ and CD8+ T cells should minimize the risk of viral escape and maximize therapeutic benefit on administration of these cells to CB recipients at risk of severe viral disease.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a Dan L. Duncan collaborative research grant (C.M.B. and E.J.S.), the National Heart, Lung, and Blood Institute (U54HL081007), an Amy Strelzer Manasevits Award from the National Marrow Donor Program (A.M.L.), a Dan L. Duncan chair (H.E.H.), a Fayez Sarofim chair (M.K.B.), a Leukemia & Lymphoma Society Clinical Research Scholar award (C.M.B.), and the National Cancer Institute (RO1 CA061508-16; E.J.S.).
National Institutes of Health
Authorship
Contribution: P.J.H. and C.R.Y.C. conducted the in vitro studies and contributed to the writing of the paper; B.S., A.M.L., M.K., M.S., and W.D. helped develop the protocol and conduct the in vitro studies; J.J.M. performed Vβ spectratyping analysis; H.L. provided statistical support; A.P.G. helped with scale up for Good Manufacturing Practice (GMP); C.M.R., H.E.H., and G.D. helped with data analysis and provided advice on experimental design; M.K.B. helped with data analysis and contributed to the writing of the paper; E.J.S. provided advice on ex vivo use of cord blood and contributed to the writing of the paper; and C.M.B. developed the study, supervised the experiments, and contributed to the writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine Bollard, Center for Cell and Gene Therapy, Baylor College of Medicine, 6621 Fannin St, MC 3-3320, Houston, TX 77030; e-mail: cmbollar@txccc.org.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal