Abstract
Small intestinal CD103+ dendritic cells (DCs) have the selective ability to promote de novo generation of regulatory T cells via the production of retinoic acid (RA). Considering that aldehyde dehydrogenase (ALDH) activity controls the production of RA, we used a flow cytometry–based assay to measure ALDH activity at the single-cell level and to perform a comprehensive analysis of the RA-producing DC populations present in lymphoid and nonlymphoid mouse tissues. RA-producing DCs were primarily of the tissue-derived, migratory DC subtype and can be readily found in the skin and in the lungs as well as in their corresponding draining lymph nodes. The RA-producing skin-derived DCs were capable of triggering the generation of regulatory T cells, a finding demonstrating that the presence of RA-producing, tolerogenic DCs is not restricted to the intestinal tract as previously thought. Unexpectedly, the production of RA by skin DCs was restricted to CD103− DCs, indicating that CD103 expression does not constitute a “universal” marker for RA-producing mouse DCs. Finally, Toll-like receptor (TLR) triggering or the presence of a commensal microflora was not essential for the induction of ALDH activity in the discrete ALDH+ DC subsets that characterize tissues constituting environmental interfaces.
Introduction
The vitamin A metabolite retinoic acid (RA) is a lipophilic molecule that controls the activity of a constellation of genes via binding to nuclear receptors.1 Vitamin A is derived from the diet, and the liver constitutes a large reservoir of vitamin A in the form of retinyl esters. Retinyl esters are hydrolyzed to retinol and released into the blood. Once retinol enters cells expressing appropriate enzymes, it is converted successively into retinal and RA. The first step of the conversion is catalyzed by alcohol dehydrogenases and by microsomal retinol dehydrogenases that are expressed by most cells, including dendritic cells (DCs). The second step consists of the oxidation of retinal into RA and is catalyzed by 3 aldehyde dehydrogenases (ALDHs), known as RALDH1, 2, and 3 and encoded by the Aldh1a1, -2, and -3 genes, respectively. RALDH expression is limited to certain cell types and, despite the widespread availability of retinol, only cells expressing one of the RALDHs can oxidize retinaldehyde to RA.
Conventional DCs (cDCs) and plasmacytoid DCs (pDCs) reside throughout their life cycle in secondary lymphoid organs and are denoted as lymphoid tissue–resident DCs to distinguish them from tissue-derived, migratory DCs (mDCs).2 cDCs express intermediate levels of major histocompatibility complex (MHC) class II molecules and high levels of CD11c (MHCIIinterCD11chigh), whereas mDCs express high levels of MHC class II molecules and intermediate to high levels of CD11c (MHCIIhighCD11cinter to high). Recently, DCs that are located in gut-associated lymphoid tissue (GALT) and express Aldh1a2 have gained considerable attention because of their ability to produce RA. On migration to mesentric lymph nodes (MLNs), this exclusive property allows them to promote the expression of the gut-tropic α4β7 integrin and CCR9 chemokine receptor on antigen-responsive T cells and in turn confer them gut-seeking properties.3,4 Importantly, RA production by GALT-associated DCs is also involved in the generation of induced Foxp3+ regulatory T cells (iTregs).5-7 iTregs can be distinguished from “naturally occurring” Foxp3+ regulatory T cells (nTregs) on the basis of their development.8 Whereas nTregs develop in the thymus, iTregs develop de novo in secondary lymphoid organs from conventional, naive CD4+ T cells. This conversion that is triggered by DCs requires submitogenic dose of antigen and low costimulation, high levels of transforming growth factor-β (TGF-β) and is greatly enhanced by the presence of RA. The exact mechanism through which DC-produced RA impacts on the generation of iTregs is still a matter of debate.9-11 For instance, RA has been proposed to enhance the TGF-β–dependent differentiation of naive CD4+ T cells into Foxp3+ iTregs by blocking their differentiation into proinflammatory T cells.5-7,12,13 Alternatively, RA may indirectly affects iTreg generation by preventing memory CD4+ T cells from producing cytokines (interleukin-4 [IL-4], IL-21, and interferon-γ), which inhibit the differentiation of iTregs.9 RA production by GALT-associated DCs has been proposed to maintain the balance between effector and Tregs in the gastrointestinal tract and to constitute a major mechanism underlying oral tolerance.14,15 The production of RA by gut DCs is restricted to mDCs expressing the integrin αE chain CD1035,6 and requires the presence of both granulocyte-macrophage colony-stimulating factor (GM-CSF) and RA in the lamina propria (LP).16
Considering that, under physiologic conditions, ALDH expression constitutes the only parameter that limits RA production, we used a flow cytometry–based assay to measure ALDH activity at the single-cell level17,18 and performed a comprehensive analysis of the RA-producing DC populations present in various lymphoid and nonlymphoid mouse tisssues under both steady-state conditions and on viral infection. We demonstrated that RA-producing DCs can be readily found in the skin and in the lungs as well as in their corresponding draining lymph nodes. The RA-producing skin-derived DCs were capable of triggering the generation of iTregs, a finding demonstrating that the presence of RA-producing, tolerogenic DCs is not restricted to the intestinal tract. Unexpectedly, the production of RA by skin DCs was restricted to the CD103− CD11b+ DC subset, a result indicating that CD103 expression does not constitute a “universal” marker for RA-producing mouse DCs. Finally, rather than being essential for the induction of ALDH activity, Toll-like receptor (TLR) triggering or the presence of a commensal microflora only slightly enhanced the constitutive levels of ALDH activity in DCs.
Methods
Mice
Mice were housed under specific pathogen–free (SPF) conditions and handled in accordance with French and European directives with ethical approval from Center d'Immunologie de Marseille-Luminy. C57BL/6 (B6) female mice were purchased from Charles River. OT-II mice19 were kept on a RAG-2-deficient background. MyD88−/−TrifLps2/Lps2 double-deficient mice were obtained by crossing mice deficient in MyD8820 and in Trif (Ticam1).21 Germ-free B6 mice obtained from TAAM-UPS44 were transported under sterile conditions and immediately used on arrival.
DC isolation
DCs were isolated from lymphoid organs, lung, and liver as previously described.22 Briefly, organs were first cut in small pieces and incubated with a mixture of type II collagenase (Worthington Biochemical) and of DNase (Sigma-Aldrich). Light density cells were purified by centrifugation on a Nycoprep solution (d = 1.068; Abcys). Skin DCs were extracted from mouse ears. Briefly, ears were split in 2 parts (dorsal and ventral) and incubated during 45 minutes at 37°C in phosphate-buffered saline (PBS) containing 2.5 mg/mL dispase II (Roche Diagnostics) to allow separation of dermal and epidermal sheets. The separated epidermal and dermal sheets were then cut into small pieces and incubated for 45 minutes at 37°C with a solution of RPMI containing 1 mg/mL DNase (Sigma-Aldrich) and 1 mg/mL collagenase type IV (Worthington Biochemical) to obtain a homogeneous cell suspension. Intestinal LP DCs were extracted from the small intestine. Briefly, the small intestine was collected and the Peyer patches were removed. Intestines were opened longitudinally, cut into pieces of 5 mm, and washed extensively in PBS. Intestinal epithelium lymphocytes were then eliminated by 2 incubations of intestinal pieces for 20 minutes at 37°C in PBS containing 5mM ethylenediaminetetraacetic acid, 15mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, and 10% (vol/vol) fetal calf serum. Intestinal pieces were then washed in RPMI medium containing 10% fetal calf serum, then cut into small pieces and incubated for 30 minutes at 37°C with a solution of RPMI containing 1 mg/mL DNase (Sigma-Aldrich) and 1 mg/mL collagenase type IV (Worthington Biochemical) to obtain a homogeneous cell suspension. Light density cells were then purified by centrifugation on a Nycoprep solution (d = 1.068; Abcys).
CCL21-mediated explant cultures of epidermal and dermal sheets
Ears were split in 2 parts (dorsal and ventral) and incubated during 45 minutes at 37°C in PBS containing 2.5 mg/mL dispase II (Roche Diagnostics) to allow separation of dermal and epidermal sheets. The separated epidermal and dermal sheets were then cultured in the presence of CCL21 (2.5 μg/mL) for 48 hours at 37°C in complete medium. The CD11c+ cells migrating out of the epidermis and dermis explants were separately harvested and analyzed for ALDH activity.
Flow cytometry
Before staining, cells were preincubated on ice for at least 10 minutes with the 2.4G2 antibody to block Fc receptors. Multiparameter fluorescence-activated cell sorter (FACS) analysis was performed using a FACSCanto system (BD Biosciences). Analysis was performed using FlowJo software (TreeStar). Autofluorescent cells were gated out using the AmCyan channel, and neutrophils were gated out according to their Ly6G+ or Gr-1 (Ly6C/Ly6G)+ phenotype (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Cell viability was evaluated using TO-PRO-3 (Invitrogen) according to the manufacturer's protocol. Foxp3 expression in CD4 T cells was evaluated using the Foxp3 staining set (eBioscience) following the manufacturer's protocol. Antibodies used in this study were: anti-CD11c (HL3 or N418), anti-CD8α (53-6.7), anti-MHCII (M5/114 or AF6-120.1), anti-CD45.2 (104), anti-CD24 (M1/69), anti-CD11b (M1/70), anti-SIRPa (P84), anti-CD103 (M290), anti-Ly6C (AL21), anti-Ly6G (1A8), Gr-1 (RB6-8C5), F4/80 (CI:A3-1), B220 (RA3-6B2), anti-CD4 (L3T4), NK1.1 (PK136), anti-CD5 (53-7.3), CCR9 (242503) and anti-TCR-Vα2 (B20.1). Antibodies were all purchased from BD Biosciences PharMingen, except for anti-CD11c (N418) and anti-B220 from eBioscience and anti-F4/80 and anti-Ly6C Ab from Serotec.
Analysis of ALDH activity at the single-cell level
The presence of cells displaying aldehyde dehydrogenase activity (ALDH+ cells) was determined using an ALDEFLUOR staining kit (StemCell Technologies). The following modifications were introduced into the manufacturer's protocol. Briefly, cells (1 × 106 cells/mL) were incubated in the dark for 45 minutes at 37°C in ALDEFLUOR assay buffer containing activated ALDEFLUOR substrate, with or without the ALDH inhibitor diethylaminobenzaldehyde (DEAB). Cells were subsequently stained using the specified antibodies in ice-cold ALDEFLUOR assay buffer. Cells were subsequently washed in ALDEFLUOR assay buffer, resuspended in ALDEFLUOR assay buffer, and kept on ice before analysis on a FACSCanto system (BD Biosciences).
Sorting of ALDH+ and ALDH− DCs
Light density cells from mesenteric and cutaneous lymph nodes were purified and incubated with ALDEFLUOR substrate (“Analysis of ALDH activity at the single-cell level”). Cells were then stained with anti-MHC class II and anti-CD11c antibodies, and MHCIIhighCD11cinter to high tissue-derived DCs were sorted in 2 subpopulations based on ALDEFLUOR staining as shown in Figure 5A. Sorted cells were then put in culture and their ALDH activity evaluated 18 hours later using the ALDEFLUOR assay.
Culture of CFSE-labeled OT-II T cells with DC subpopulations
CD4+ T cells were isolated from the spleen of OT-II Rag-2−/− mice using a CD4+ T cell− isolation kit (Dynal; Invitrogen). Purity was determined by staining with CD4, CD5, and TCR Vα2. For carboxyfluorescein diacetate succinimidyl ester (CFSE) labeling, purified OT-II Rag-2−/− T cells were resuspended in PBS containing 2.5mM CFSE (Invitrogen) for 10 minutes at 37°C. A total of 3 × 103 DCs were cocultured with 2 × 104 CFSE-labeled OT-II Rag-2−/− T cells in 150 μL in the presence of ovalbumin (257-264) peptide (0.06 μg/mL) and of TGF-β (1 ng/mL) or LE540 (1μM) as indicated. After 5 days of culture, proliferation was measured by flow cytometry as a loss of CFSE staining and expression of Foxp3 and CCR9 was evaluated.
TLR stimulation of DCs
DCs were purified from the light density cell fraction of the spleen or lymph nodes using positive CD11c MACS isolation (Miltenyi Biotec) and cultured in vitro at 3 × 104 DCs per well in 150 μL of complete medium. DCs were stimulated with zymosan (10 μg/mL), lipopolysaccharide (1 μg/mL), or CpG ODN1826 (5μM), all purchased from Invivogen. After18 hours of culture, DCs were incubated with ALDEFLUOR substrate, stained for surface antigens, and analyzed by flow cytometry.
Mouse cytomegalovirus infection
Infections were initiated at day 0 by intraperitoneal injection of 5 × 104 PFU of the V70 strain of mouse cytomegalovirus (MCMV). Infection experiments were conducted in accordance with the regional ethical committee and institutional guidelines. The MCMV plaque assay was performed as described.23 Briefly, at various times after MCMV infection, spleens were harvested and mechanically disrupted in 1 mL of complete Dulbecco modified Eagle medium on ice using a Power Gen 125 tissue homogenizer (Fisher Scientific). Viral loads were measured in spleen homogenates using a plaque assay protocol based on NIH-3T3 cells.
Statistical analysis
Comparative experiments were tested for statistical significance using the unpaired Student t test in GraphPad Prism software (Version 4.0; GraphPad).
Results
Identification of cells displaying ALDH activity
By combining multiparameter flow cytometry and staining with ALDEFLUOR, a fluorescent substrate allowing detection of ALDH activity at the single-cell level,18 we aimed at identifying cells endowed with RA-producing capacity within lymphoid and nonlymphoid tissues. To validate this approach, cells were isolated from the MLNs of B6 mice that were kept under SPF conditions and the frequency of ALDH+ cells determined (supplemental Figure 1). In these pilot experiments, we sought to obtain an unbiased view of the cells endowed with ALDH activity. Accordingly, after digesting the MLNs with collagenase, we did not enrich for light density cells using centrifugation on density gradient. Importantly, in each experiment involving ALDEFLUOR staining, controls were performed in the presence of diethylamino-benzaldehyde (DEAB), an ALDH inhibitor that permits to accurately define the ALDH+ gate (supplemental Figure 1).
ALDH+ cells were readily identified in the cell suspension isolated from MLNs and consisted of Ly6G+ MHCII− and Ly6G− MHCII+ cells that corresponded to CD11b+ Ly6G+ neutrophils and to mDCs, respectively (supplemental Figure 1). Compared with mDCs, neutrophils expressed lower ALDH signals that barely merged into the ALDH+ gate. However, use of the DEAB inhibitor demonstrated the specificity of such low intensity signals. The high ALDH signals found in the MLN DCs confirmed that mDCs from the GALT are endowed with constitutive RA-producing capacity.5,6,16 Having validated our experimental approach, we undertook a comprehensive analysis of various lymphoid and nonlymphoid tissues to determine which antigen-presenting cells (APCs) were capable of constitutive ALDH activity. To increase the resolution of such analysis, we systematically gated out neutrophils based on their Ly6G+ or Gr-1(Ly6G/C)+ phenotype and systematically used centrifugation on density gradient to enrich for light-density APCs.
ALDH activity in gut-associated lymphoid tissues
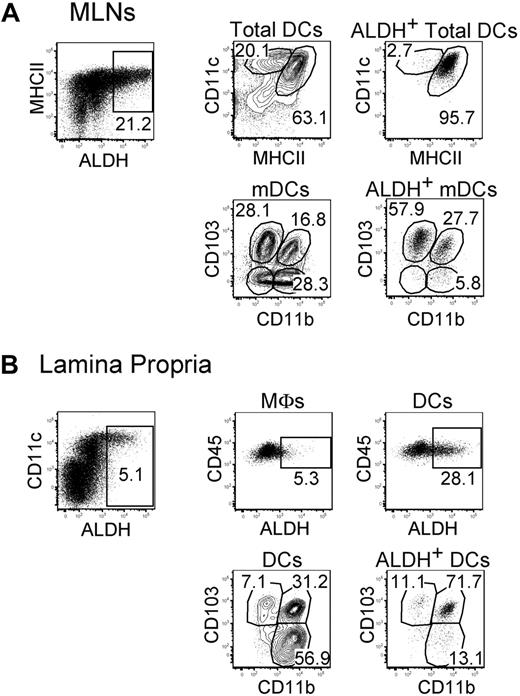
In steady-state MLNs, expression of the RALDH2 isoenzyme is largely restricted to CD103+ mDCs.5,6,16 Consistent with those studies, CD103+ mDCs freshly isolated from MLNs displayed high levels of ALDH activity (Figure 1A). MLN CD103+ mDCs can be segregated into a CD11b+ and a CD11b− fraction and ALDH was expressed on approximately 50% of both fractions (Figures 1–2). Moreover, a small fraction of the CD103−CD11b+ and CD103−CD11b− mDCs present in MLNs also expressed some ALDH activity (Figure 2). We analyzed next whether ALDH activity can be detected in the DCs and the macrophages that are found in the LP. Consistent with a previous study measuring expression of the Aldh1a2 gene,24 LP macrophages were found to express ALDH (Figure 1B). The percentage of ALDH+ cells and the intensity of the ALDH signal were, however, considerably higher in LP DCs compared with LP macrophages, suggesting that DCs have a higher RA-producing capacity on a per-cell basis. As shown in Figure 1B, the LP contains 3 DC subsets that include CD103+CD11b+ DCs, CD103−CD11b+ DCs, and CD103+CD11b− DCs.6,25 Within LP DCs, the majority of ALDH+ DCs had a CD103+CD11b+ phenotype and probably constitutes the source of the ALDH+ CD103+CD11b+ mDCs present in MLNs. Therefore, these data support the view that CD103+CD11b+ DCs acquire their ALDH activity in the LP before their migration to the MLNs.14,15 Based on the measurement of ALDH activity (Figure 2), it appears, however, that only half of the CD103+CD11b+ DCs have undergone such conditioning by the intestine environment.
ALDH+ DCs from the MLNs and the LP display a heterogeneous surface phenotype. Light-density cells isolated from MLNs (A) and from the small intestine LP (B) were incubated with ALDEFLUOR and analyzed by flow cytometry for expression of CD11c, MHCII, CD11b, CD103, and CD45 and for ALDH activity. Dot plots correspond to nonautofluorescent cells with a FSChighSSChigh profile characteristic of DCs and from which neutrophils were gated out using Ly6G and Gr-1 staining. Positioning of the ALDH+ gate is based on incubation in the presence of DEAB (supplemental Figure 3). For MLNs, the CD11cinter to highMHCIIhigh and the CD11chighMHCIIinter gates correspond to mDCs and lymphoid tissue–resident DCs, respectively. In the case of LP, the gates corresponding to macrophages (MFs) and to DCs are specified in supplemental Figure 2A. Numbers in outlined areas indicate percentage of cells. Data are representative of at least 3 separate experiments involving groups of 3 to 6 mice.
ALDH+ DCs from the MLNs and the LP display a heterogeneous surface phenotype. Light-density cells isolated from MLNs (A) and from the small intestine LP (B) were incubated with ALDEFLUOR and analyzed by flow cytometry for expression of CD11c, MHCII, CD11b, CD103, and CD45 and for ALDH activity. Dot plots correspond to nonautofluorescent cells with a FSChighSSChigh profile characteristic of DCs and from which neutrophils were gated out using Ly6G and Gr-1 staining. Positioning of the ALDH+ gate is based on incubation in the presence of DEAB (supplemental Figure 3). For MLNs, the CD11cinter to highMHCIIhigh and the CD11chighMHCIIinter gates correspond to mDCs and lymphoid tissue–resident DCs, respectively. In the case of LP, the gates corresponding to macrophages (MFs) and to DCs are specified in supplemental Figure 2A. Numbers in outlined areas indicate percentage of cells. Data are representative of at least 3 separate experiments involving groups of 3 to 6 mice.
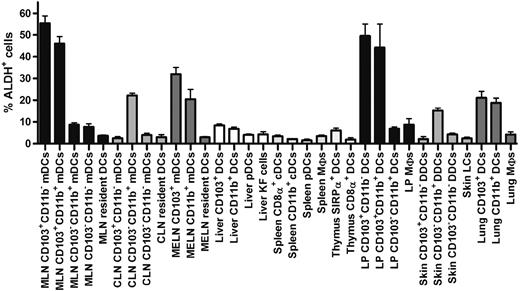
Comparison of the percentage of ALDH+ cells found in DCs and MF subsets in various lymphoid and nonlymphoid tissues. DCs and MFs were isolated from the skin, the LP of the small intestine, liver, thymus, spleen, and the MLNs, MELNs, and CLNs using the gates specified in Figures 1, 3, and 4 and supplemental Figures 2 and 3 and analyzed for ALDH activity. Data correspond to mean ± SD of results from 3 separate experiments.
Comparison of the percentage of ALDH+ cells found in DCs and MF subsets in various lymphoid and nonlymphoid tissues. DCs and MFs were isolated from the skin, the LP of the small intestine, liver, thymus, spleen, and the MLNs, MELNs, and CLNs using the gates specified in Figures 1, 3, and 4 and supplemental Figures 2 and 3 and analyzed for ALDH activity. Data correspond to mean ± SD of results from 3 separate experiments.
ALDH activity among DCs associated with the skin and the lungs
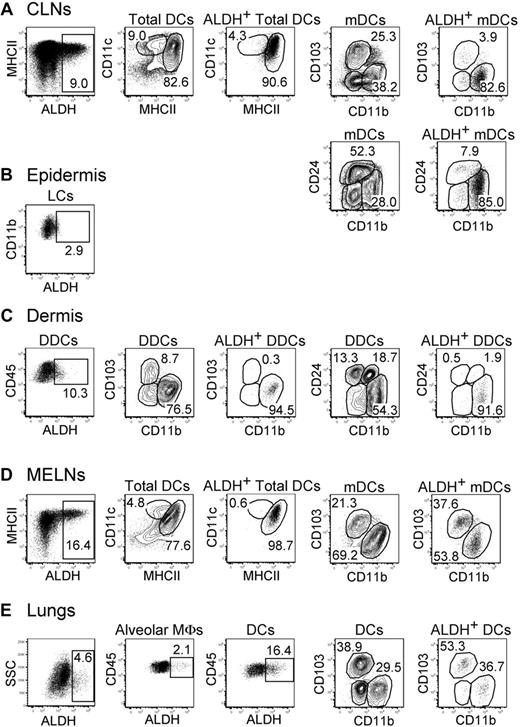
In contrast to DCs present in the GALT, those found in the skin and the lungs have been thought to be devoid of ALDH activity and the intestine considered as a privileged site for iTreg induction.15 ALDH+ DCs can be, however, readily detected within skin-draining cutaneous lymph nodes (CLNs) and lung-draining mediastinal lymph nodes (MELNs; Figures 2–3). In the CLNs, the majority of ALDH+ cells were mDCs with a CD103−CD11b+CD24int phenotype. Note that ALDEFLUOR staining, which requires viable cells, was not compatible with the simultaneous detection of CD207 (Langerin) expression using either anti-CD207 staining, which requires cell permeabilization, or mice expressing enhanced green fluorescence protein under the control of the Langerin gene (both enhanced green fluorescence protein and ALDEFLUOR emit in the first channel of a flow cytometer). However, based on their phenotype, the ALDH+CD103−CD11b+CD24int mDCs can be unambiguously identified as migratory CD207−CD11b+ dermal DCs (DDCs) in that they differ from both migratory Langerhans cells and migratory CD207+CD11b− DDCs that are both CD11b− and CD24+.26-29 RNA-PCR analysis showed that the ALDH isoform expressed by the CD103−CD11b+CD24int mDDCs was encoded by the Aldh1a2 gene (data not shown). Measurement of ALDH activity in cell suspensions isolated from ear dermis indicated that the CD103−CD11b+CD24int DDCs that constitute the precursor of the CD103−CD11b+CD24int mDCs found in the CLNs30 have already acquired their RA-producing capacity in the dermis before their migration to the CLNs (Figure 3C). In addition, analysis of cell suspensions isolated from ear epidermis showed that epidermal Langerhans cells displayed only marginal levels of ALDH activity (Figure 3B). Consistent with this last view, the Langerhans cells that crawled out of epidermal explants in the presence of the CCL21 chemokine showed only low levels of ALDH activity compared with the CD103−CD11b+CD24int DDCs crawling out of dermal explants in the presence of CCL21 (supplemental Figure 4). Therefore, emigration from the skin does not modify the pattern of ALDH activity that is imprinted within this tissue before migration to the CLNs.
Identification of ALDH+ DCs in the skin, lungs, and corresponding draining lymph nodes. Light-density cells isolated from the CLNs (A), the ear epidermis (B), the ear dermis (C), the MELNs (D), and the lungs (E) were incubated with ALDEFLUOR and analyzed by flow cytometry for expression of CD11c, MHCII, CD11b, CD103, CD45, and CD24 and for ALDH activity. Dot plots correspond to nonautofluorescent cells (skin and LNs) or to both autofluorescent and nonautofluorescent cells (lungs) with a FSChigh SSChigh profile characteristic of DCs and from which neutrophils were gated out using Ly6G and Gr-1 staining. Positioning of the ALDH+ gate is based on incubation in the presence of DEAB (supplemental Figure 3). For the CLNs and MELNs, the CD11cinter to highMHCIIhigh and the CD11chighMHCIIinter gates correspond to mDCs and lymphoid tissue–resident DCs, respectively. The gates corresponding to epidermal Langerhans cells, DDCs, alveolar MFs, and lung DCs are specified in supplemental Figure 2B through D. Numbers in outlined areas indicate percentage of cells. Data are representative of 3 separate experiments involving groups of 3 to 6 mice.
Identification of ALDH+ DCs in the skin, lungs, and corresponding draining lymph nodes. Light-density cells isolated from the CLNs (A), the ear epidermis (B), the ear dermis (C), the MELNs (D), and the lungs (E) were incubated with ALDEFLUOR and analyzed by flow cytometry for expression of CD11c, MHCII, CD11b, CD103, CD45, and CD24 and for ALDH activity. Dot plots correspond to nonautofluorescent cells (skin and LNs) or to both autofluorescent and nonautofluorescent cells (lungs) with a FSChigh SSChigh profile characteristic of DCs and from which neutrophils were gated out using Ly6G and Gr-1 staining. Positioning of the ALDH+ gate is based on incubation in the presence of DEAB (supplemental Figure 3). For the CLNs and MELNs, the CD11cinter to highMHCIIhigh and the CD11chighMHCIIinter gates correspond to mDCs and lymphoid tissue–resident DCs, respectively. The gates corresponding to epidermal Langerhans cells, DDCs, alveolar MFs, and lung DCs are specified in supplemental Figure 2B through D. Numbers in outlined areas indicate percentage of cells. Data are representative of 3 separate experiments involving groups of 3 to 6 mice.
In MELNs, ALDH+ DCs segregated into 2 subsets that correspond to CD103+CD11b− and CD103−CD11b+ mDCs (Figures 2,3D). In the lungs, the precursors of these DC subsets31 were already ALDH+, suggesting that this activity was acquired before the migration to the MELNs (Figures 2,3E). Although alveolar macrophages showed some ALDH activity, both the percentage of ALDH+ cells and the intensity of the ALDH signal were considerably higher in DCs, suggesting that lung DCs have an increased RA-producing capacity compared with alveolar macrophages (Figures 2,3E).
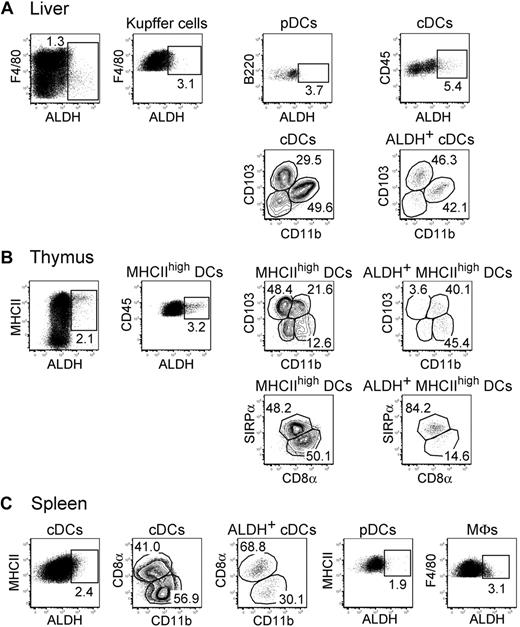
The thymus, liver, and spleen contained reduced numbers of ALDH+ APCs compared with the gut, skin, lungs, and their corresponding draining lymph nodes (Figures 2,4). A small percentage of ALDH+ cells could be found within liver pDCs, cDCs, and Kupffer cells (Figure 4A). The liver ALDH+ cDCs corresponded to both CD103+CD11b− and CD103−CD11b+ subsets. As previously noted for LP and alveolar macrophages, both the percentage of ALDH+ cells and the intensity of the ALDH signal were higher in liver cDCs compared with Kupffer cells. In the thymus, the small population of ALDH+ DC cells showed a homogeneous SIRPα expression (Figure 4B). Finally, the rare ALDH+ cDCs that can be detected in the spleen were evenly distributed among the CD8α+ and CD11b+ subsets. In addition, splenic macrophages and pDCs displayed only marginal levels of ALDH activity (Figure 4C). Therefore, DCs expressing high levels of ALDH activity can be readily found in the skin and the lungs and in their corresponding draining lymph nodes, demonstrating that the presence of RA-producing DCs is not restricted to the intestinal tract.
Reduced levels of ALDH activity in DCs from liver, thymus, and spleen. Light-density cells isolated from liver (A), thymus (B), and spleen (C) were incubated with ALDEFLUOR and analyzed by flow cytometry for the expression of CD11c, MHCII, CD11b, CD103, CD45, SIRPa, and CD8a and for ALDH activity. Dot plots correspond to nonautofluorescent cells from which neutrophils were gated out using Ly6G and Gr-1 staining. Positioning of the ALDH+ gate is based on incubation in the presence of DEAB (supplemental Figure 3). The gates corresponding to liver Kupffer cells, liver DCs, thymic MHCIIhigh DCs, splenic cDCs, splenic pDCs, and splenic MFs are specified in supplemental Figure 2E through G. Numbers in outlined areas indicate percentage of cells. Data are representative of at least 3 separate experiments involving groups of 3 to 6 mice.
Reduced levels of ALDH activity in DCs from liver, thymus, and spleen. Light-density cells isolated from liver (A), thymus (B), and spleen (C) were incubated with ALDEFLUOR and analyzed by flow cytometry for the expression of CD11c, MHCII, CD11b, CD103, CD45, SIRPa, and CD8a and for ALDH activity. Dot plots correspond to nonautofluorescent cells from which neutrophils were gated out using Ly6G and Gr-1 staining. Positioning of the ALDH+ gate is based on incubation in the presence of DEAB (supplemental Figure 3). The gates corresponding to liver Kupffer cells, liver DCs, thymic MHCIIhigh DCs, splenic cDCs, splenic pDCs, and splenic MFs are specified in supplemental Figure 2E through G. Numbers in outlined areas indicate percentage of cells. Data are representative of at least 3 separate experiments involving groups of 3 to 6 mice.
ALDH+ DCs from cutaneous lymph nodes induce Tregs
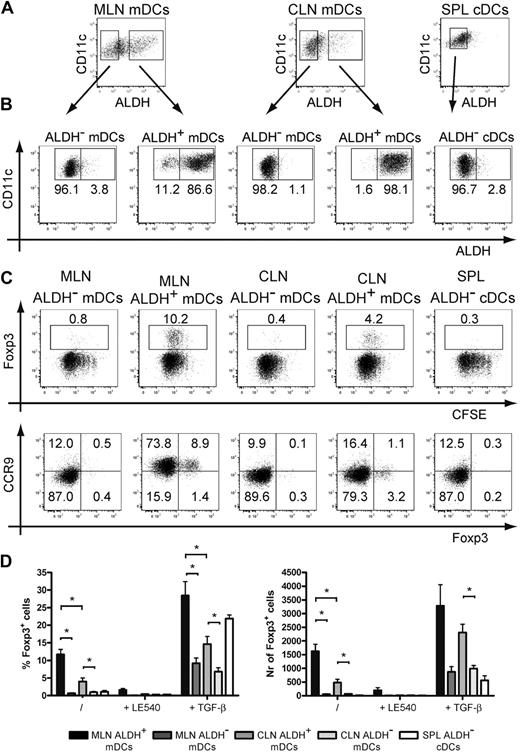
We evaluated next whether ALDH+ DCs isolated from CLNs were capable of generating iTregs akin to the RA-producing CD103+ DCs isolated from the GALT.5,6 When maintained on a Rag-2−/− background, transgenic mice that express a TCR specific for I-Ab-OVA complexes (OT-II Rag-2−/− mice) contain only conventional (Foxp3−) CD4+ T cells in their periphery, a situation that facilitates the measurement of their conversion into iTregs. Such conversion requires I-Ab+ DCs and the presence of the OVA-derived peptide specifically recognized by OT-II CD4+ T cells. It also depends on the secretion by the antigen-presenting DCs of TGF-β and of RA. Accordingly, CD4+ T cells isolated from OT-II Rag-2−/− mice were incubated with freshly sorted ALDH− and ALDH+ mDCs from CLNs and MLNs and cDCs from spleen. Before assessing the iTreg-inducing ability of the sorted DCs, we first demonstrated that sorted ALDH+ DCs kept in culture for 18 hours retained their ALDH activity and conversely that ALDH− DCs kept under identical conditions did not acquire any ALDH activity (Figure 5A-B). Peptide-pulsed ALDH+ and ALDH− DC subsets were both capable of inducing intense proliferation of OT-II Rag-2−/− CD4+ T cells as measured by CFSE dilution (Figure 5C). However, only the ALDH+ DCs from CLNs and MLNs induced the expression of Foxp3 in 11.7% (± 1.5%) and 4.0% (± 1.1%) of OT-II Rag-2−/− CD4+ T cells, respectively. Importantly, generation of iTregs by ALDH+ DCs from CLNs and MLNs was inhibited by addition of LE540, an inhibitor of RA receptors, indicating that RA produced by ALDH+ DCs is indeed involved in the generation of iTregs. Consistent with the view that RA acts primarily as a cofactor during TGF-β–mediated iTreg induction,5,6,10,11 addition of exogenous TGF-β to the culture conferred to ALDH− DCs the ability to generate low numbers of iTregs and enhanced the constitutive iTreg-inducing capacity of ALDH+ DCs (Figure 5D).
ALDH+ DCs isolated from MLNs and CLNs trigger the differentiation of iTregs. (A) Light-density cells isolated from spleen and from MLNs and CLNs were incubated with ALDEFLUOR and stained for CD11c and MHCII. mDCs from MLNs and CLNs and cDCs from spleen were then sorted into ALDH− and ALDH+ fractions. (B) To verify the quality of the sort and assess for the stability of ALDH activity, sorted cells kept in culture for 18 hours were incubated with ALDEFLUOR, stained for CD11c and MHCII, and analyzed by flow cytometry. Dead cells were gated out before analysis using TO-PRO-3 staining. Positioning of the ALDH+ gate is based on incubation in the presence of DEAB (not shown). (C-D) Freshly sorted ALDH− and ALDH+ mDCs from MLNs and CLNs and cDCs from spleen were incubated with CFSE-labeled OT-II Rag-2−/− T cells in the presence of the OVA257-264 peptide (0.06 μg/mL). After 5 days of culture, T cells were analyzed for the expression of Foxp3 and of CCR9. Numbers in outlined areas indicate percentage of cells. Data are representative of at least 3 separate experiments. (D) Comparison of the percentage and the number of converted Foxp3+ Tregs induced by ALDH− and ALDH+ mDCs from MLNs, CLNs, and cDCs from spleen. Also shown is the effect resulting from the addition of the LE540 (1μM) inhibitor and of exogenous TGF-β (1 ng/mL). Data are representative of 3 independent experiments. Bars represent mean ± SD of triplicate wells within 1 of the 3 independent experiments. *P < .05
ALDH+ DCs isolated from MLNs and CLNs trigger the differentiation of iTregs. (A) Light-density cells isolated from spleen and from MLNs and CLNs were incubated with ALDEFLUOR and stained for CD11c and MHCII. mDCs from MLNs and CLNs and cDCs from spleen were then sorted into ALDH− and ALDH+ fractions. (B) To verify the quality of the sort and assess for the stability of ALDH activity, sorted cells kept in culture for 18 hours were incubated with ALDEFLUOR, stained for CD11c and MHCII, and analyzed by flow cytometry. Dead cells were gated out before analysis using TO-PRO-3 staining. Positioning of the ALDH+ gate is based on incubation in the presence of DEAB (not shown). (C-D) Freshly sorted ALDH− and ALDH+ mDCs from MLNs and CLNs and cDCs from spleen were incubated with CFSE-labeled OT-II Rag-2−/− T cells in the presence of the OVA257-264 peptide (0.06 μg/mL). After 5 days of culture, T cells were analyzed for the expression of Foxp3 and of CCR9. Numbers in outlined areas indicate percentage of cells. Data are representative of at least 3 separate experiments. (D) Comparison of the percentage and the number of converted Foxp3+ Tregs induced by ALDH− and ALDH+ mDCs from MLNs, CLNs, and cDCs from spleen. Also shown is the effect resulting from the addition of the LE540 (1μM) inhibitor and of exogenous TGF-β (1 ng/mL). Data are representative of 3 independent experiments. Bars represent mean ± SD of triplicate wells within 1 of the 3 independent experiments. *P < .05
We also analyzed whether the DC subsets sorted on the basis of ALDH activity were capable of inducing expression of the CCR9 gut-tropic chemokine receptor on OT-II Rag-2−/− CD4+ T cells. Congruent with previous data,4,32 ALDH+ DCs sorted from MLNs were able to induce the expression of CCR9 on the majority of OT-II Rag-2− CD4+ T cells (Figure 5C). However, consistent with the view that skin-derived DCs do not imprint gut-tropic properties on T cells, neither ALDH+ nor ALDH− DCs isolated from CLNs were able to induce CCR9 expression on OT-II Rag-2− CD4+ T cells (Figure 5C). Collectively, these data indicate that the ALDH+ mDCs identified in the CLNs constitute bona fide RA-producing DCs that possess the capacity to generate iTregs, a property previously thought to be exclusively restricted to gut DCs.
Modest increase in ALDH activity after in vitro and in vivo TLR stimulation
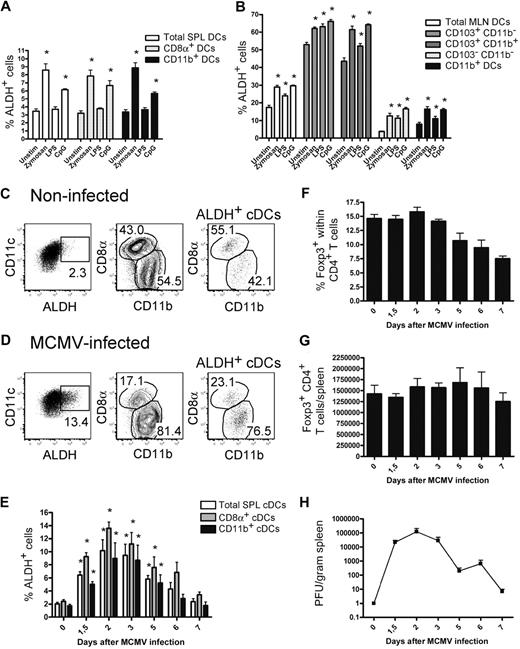
It has been recently described that TLR stimulation of whole splenic DCs induces the expression of the Aldh1a2 gene.16,33 To determine the type of DCs that acquire ALDH activity on TLR triggering and the magnitude of such activity, DCs isolated from the spleen and MLNs were stimulated with TLR ligands and analyzed for ALDH activity. In the case of DC subsets displaying a constitutive ALDH activity, a slight but significant increase in the percentage of ALDH+ DCs was noted on zymosan and CpG stimulation; whereas in the case of DC subsets that were ALDH− under steady-state conditions, zymosan and CpG stimulation was capable of inducing low levels of ALDH activity (Figure 6A-B).
ALDH activity is moderately affected by in vitro TLR triggering and by in vivo MCMV infection. (A-B) CD11c+ DCs were purified from spleen (A) and MLNs (B) using MACS isolation and cultured in the presence or absence of zymosan (10 μg/mL), lipopolysaccharide (1 μg/mL), or CpG (5μM). After 18 hours of culture, cells were incubated with ALDEFLUOR and analyzed by flow cytometry. Histograms show the percentages of ALDH+ cells found in the specified subsets. Data correspond to mean ± SD of 3 separate experiments. The values corresponding to unstimulated and TLR-stimulated cultures were subjected to statistical comparison. *P < .05. (C-G) Mice were infected with 5 × 104 PFU of MCMV. Light-density cells from the spleen of noninfected (C) or mice infected for 2 days with MCMV (D) were incubated with ALDEFLUOR and analyzed by flow cytometry for CD11c, CD8α, and CD11b expression and ALDH activity. Positioning of the ALDH+ gate is based on incubation in the presence of DEAB (not shown). Dot plots correspond to conventional DCs of individual animals, and numbers in outlined areas indicate percentage of cells. The percentage of ALDH+ cells within the cDCs (E), the percentage of Foxp3+ cells within the CD4+ T cells (F), the total number of Foxp3+ T cells per spleen (G), and the viral load present in spleens (H) are shown at different time points after infection. Time point “0” corresponds to noninfected control mice. Data correspond to mean ± SD of at least 2 separate experiments per time point. The values corresponding to noninfected and MCMV-infected mice were subjected to statistical comparison. *P < .05.
ALDH activity is moderately affected by in vitro TLR triggering and by in vivo MCMV infection. (A-B) CD11c+ DCs were purified from spleen (A) and MLNs (B) using MACS isolation and cultured in the presence or absence of zymosan (10 μg/mL), lipopolysaccharide (1 μg/mL), or CpG (5μM). After 18 hours of culture, cells were incubated with ALDEFLUOR and analyzed by flow cytometry. Histograms show the percentages of ALDH+ cells found in the specified subsets. Data correspond to mean ± SD of 3 separate experiments. The values corresponding to unstimulated and TLR-stimulated cultures were subjected to statistical comparison. *P < .05. (C-G) Mice were infected with 5 × 104 PFU of MCMV. Light-density cells from the spleen of noninfected (C) or mice infected for 2 days with MCMV (D) were incubated with ALDEFLUOR and analyzed by flow cytometry for CD11c, CD8α, and CD11b expression and ALDH activity. Positioning of the ALDH+ gate is based on incubation in the presence of DEAB (not shown). Dot plots correspond to conventional DCs of individual animals, and numbers in outlined areas indicate percentage of cells. The percentage of ALDH+ cells within the cDCs (E), the percentage of Foxp3+ cells within the CD4+ T cells (F), the total number of Foxp3+ T cells per spleen (G), and the viral load present in spleens (H) are shown at different time points after infection. Time point “0” corresponds to noninfected control mice. Data correspond to mean ± SD of at least 2 separate experiments per time point. The values corresponding to noninfected and MCMV-infected mice were subjected to statistical comparison. *P < .05.
Although of rather low magnitude, the significant increase in ALDH activity observed on in vitro TLR triggering raises the possibility that pathogens might engage TLR to induce RA production by the DCs they encounter and thereby stimulate the generation of pathogen-specific iTregs in vivo. Along that line, MCMV infection has recently been shown to trigger the generation of iTregs in mice that are deficient in the Carma1 adaptor and lack nTregs.34 To evaluate whether MCMV infection results in an increase in DC ALDH activity, B6 mice were infected with MCMV and splenic DCs were analyzed at various times after infection. As shown in Figure 6C through E, MCMV infection induced a significant increase in ALDH activity in both CD8α+ and CD11b+ splenic DCs, the amplitude of which correlated with the viral load present in the spleen (Figure 6H). RT-PCR analysis indicated that MCMV infection specifically induced the expression of the Aldh1a2 isoform (not shown). The modest increase in ALDH activity was, however, not associated with a significant increase in the number of splenic Foxp3+ T cells (Figure 6G). Therefore, although TLR triggering enhances ALDH activity of DC subsets that were either ALDHhigh or ALDHlow in steady-state conditions, such changes remained, however, of rather limited magnitude.
Constitutive ALDH activity by DCs from the skin and the gut does not depend on TLR signaling or on the presence of commensal microflora
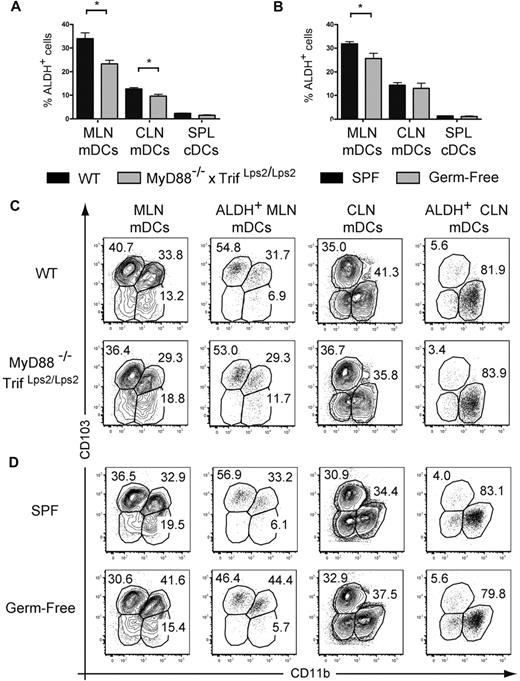
The fact that TLR signaling leads to a small increase in ALDH activity under in vitro and in vivo conditions suggests that, because of their capacity to engage TLRs, the commensal microflora may participate in the conditioning of gut CD103+ DCs into RA-producing, tolerogenic DCs.15,16,33 Therefore, we evaluated ALDH activity in mDCs isolated from MyD88−/−TrifLps2/Lps2 double-deficient mice that lack central adaptors used by all TLRs and thus lack the capacity to signal via TLRs. mDCs from the MLNs and CLNs of MyD88−/−TrifLps2/Lps2 displayed significantly lower ALDH activity compared with age-matched wild-type mice (Figure 7A). However, this corresponded at most to a 30% decrease in the percentage of ALDH+ DCs; and as a result, mDCs originating from the gut and the skin of MyD88−/−TrifLps2/Lps2 mice still possess high levels of ALDH activity compared with splenic cDCs (Figure 7A). To exclude the possibility that the commensal flora present in MyD88−/−TrifLps2/Lps2 mice induces ALDH activity in a TLR-independent manner, we assessed the ALDH activity of DC subsets from the skin and gut-draining lymph nodes of germ-free mice. As shown in Figure 7B, the absence of commensal microflora modestly affected the ALDH activity of mDCs from MLNs and CLNs and the relative representation of the MLN and CLN subsets, including those endowed with ALDH activity, was strikingly similar in the presence or absence of TLR signaling or of commensal microflora (Figure 7C-D). Therefore, the local commensal microflora and TLR signaling are dispensable for the imprinting of a constitutive RA-producing capacity on the discrete subsets of ALDH+ mDCs found in the gut and in the skin.
Absence of TLR triggering or of commensal flora moderately diminishes the percentage of ALDH+ DCs. Light-density cells isolated from spleen (SPL) and from MLNs and CLNs of MyD88−/−TrifLps2/Lps2 double-deficient mice and from wild-type C57BL/6 mice kept under SPF or germ-free conditions were incubated with ALDEFLUOR in the absence or presence of DEAB (not shown) and analyzed by flow cytometry for expression of CD11c, MHCII, CD11b, and CD103 and for ALDH activity. (A-B) The percentage of ALDH+ cells within mDCs from MLNs, CLNs, and cDCs from spleen is indicated. Data correspond to mean ± SD of at least 2 separate experiments involving 3 separate animals. Statistical comparison of the values corresponding to wild-type SPF mice with germ-free or MyD88−/−Trif Lps2/Lps2 double-deficient mice. *P < .05. (C) Expression of CD103 and CD11b on total and on ALDH+ mDCs from MLNs and CLNs isolated from wild-type and MyD88−/−TrifLps2/Lps2 double-deficient mice. (D) Expression of CD103 and CD11b on total and on ALDH+ mDCs from MLNs and CLNs isolated from SPF and germ-free mice. In panels C and D, numbers in outlined areas indicate percentage of cells, and FACS profiles are representative of 1 of 3 individual animals from at least 2 separate experiments.
Absence of TLR triggering or of commensal flora moderately diminishes the percentage of ALDH+ DCs. Light-density cells isolated from spleen (SPL) and from MLNs and CLNs of MyD88−/−TrifLps2/Lps2 double-deficient mice and from wild-type C57BL/6 mice kept under SPF or germ-free conditions were incubated with ALDEFLUOR in the absence or presence of DEAB (not shown) and analyzed by flow cytometry for expression of CD11c, MHCII, CD11b, and CD103 and for ALDH activity. (A-B) The percentage of ALDH+ cells within mDCs from MLNs, CLNs, and cDCs from spleen is indicated. Data correspond to mean ± SD of at least 2 separate experiments involving 3 separate animals. Statistical comparison of the values corresponding to wild-type SPF mice with germ-free or MyD88−/−Trif Lps2/Lps2 double-deficient mice. *P < .05. (C) Expression of CD103 and CD11b on total and on ALDH+ mDCs from MLNs and CLNs isolated from wild-type and MyD88−/−TrifLps2/Lps2 double-deficient mice. (D) Expression of CD103 and CD11b on total and on ALDH+ mDCs from MLNs and CLNs isolated from SPF and germ-free mice. In panels C and D, numbers in outlined areas indicate percentage of cells, and FACS profiles are representative of 1 of 3 individual animals from at least 2 separate experiments.
Discussion
Congruent with a recent study that focused on intestinal DCs and used ALDEFLUOR staining,16 we showed that, among the DCs found in the LP of the small intestine and in the MLNs, the CD103+ DC subset is preferentially endowed with ALDH activity. The possibility to detect ALDH activity at the single-cell level further revealed that, in the LP and the MLNs, ALDH activity was only expressed by approximately one-half of the CD103+ DCs. Our comprehensive analysis of ALDH activity in cDCs and mDCs present in various lymphoid and nonlymphoid tissues yielded several unexpected findings. RA-producing DCs were not limited to intestinal tissues and were primarily associated with tissues that constitute environmental interfaces (intestine, lung, skin). Moreover, ALDH activity was primarily found in mDCs. The presence of DCs expressing high constitutive ALDH activity in several tissues that constitute body barriers suggests that such activity results from cues that are delivered by those tissues and that exist under steady-state conditions. In support of that view, it has been recently demonstrated that, in the intestinal tissues, the presence of ALDH+ DCs depends on the production of GM-CSF by LP macrophages and of RA by intestinal epithelial cells.16 Considering that the Aldh1a2 gene that is expressed in DCs contains multiple signal transducers and activators of transcription-binding motifs and putative RA-response elements, RA and GM-CSF may thus synergistically induce ALDH activity within some DC subtypes.16 Here, we analyzed whether TLR signaling contributes to promote ALDH activity in DCs and showed that DCs from mice deficient in TLR signaling still display a substantial ALDH activity. The analysis of germ-free mice indicated that the commensal microflora was also dispensable for the generation of ALDH+ DCs. What are the cellular sources of GM-CSF and of RA in the skin and the lungs? Airway epithelial cells express Aldh1a1 and might also constitute a source of GM-CSF.31,35,36 In the skin, the RALDH2 and RALDH3 enzymes are highly expressed by hair follicles,37,38 whereas GM-CSF is produced by keratinocytes and dermal fibroblasts.39,40 Therefore, these recurrent features may explain the presence of ALDH+ DCs in the small intestine, lungs, and skin and point to the existence of cellular niches capable of inducing ALDH activity in DCs. The fact that RA-producing capacity is not evenly distributed among the DC subtypes found associated with a given tissue suggests that these niches are only accessible to particular subset of DCs.
The generation of iTregs by ALDH+ GALT DCs has been proposed to contribute to oral tolerance.14,15 Here, we demonstrated that ALDH+ DCs freshly isolated from the CLNs are also capable of inducing iTregs. Such potential depended on the production of RA and was somewhat lower than that of ALDH+ DCs isolated from MLNs. Therefore, the ability to convert naive CD4+ T cells into iTregs is not exclusive to the DCs associated with the MLNs. In the CLNs, there exists a discrete subset of CD103+ mDCs that originates from CD103+ DDCs.41 Unexpectedly, the ALDH+ mDCs found in the CLNs do not belong to this CD103+ DDC subset and have a CD11b+CD207−CD103− phenotype. Therefore, CD103 expression does not constitute a generic mark of DCs endowed with ALDH activity, and ALDEFLUOR staining still remains the most direct way of identifying RA-producing DCs at the single-cell level and of sorting them under vital conditions. The ability of CD11b+CD207−CD103− mDDCs to induce the conversion of naive CD4+ T cells into iTregs in an antigen- and RA-dependent manner is also interesting to consider in view of the large numbers of Foxp3+ T regulatory cells that are present in the dermis both under steady-state and inflammatory conditions and that are thought to balance the antimicrobial but potentially toxic effect of interferon-γ produced by effector T cells.42-44 In the case of the skin, it cannot be excluded that Langerhans cells that produce TGF-β45 also play a role in iTreg induction. Considering that epidermal and migratory Langerhans cells showed only marginal levels of ALDH activity, this putative iTreg-inducing ability would, however, be independent of RA production and require conditions that remain to be defined. In that context, lymphoid tissue–resident CD8α+ DCs and pDCs have been proposed to participate in the generation of iTregs in a TGF-β-dependent manner that, based on our data, probably does not depend on RA.46-48 Therefore, depending on the tissue considered, RA production may not be a prerequisite for iTreg generation. In the case of the lymph nodes draining the skin and the gut, 2 organs in constant contact with bacteria and fungi, RA production by DCs may be required to counteract the proinflammatory cues such tissues project to the draining lymph nodes49 and thereby allow the generation of iTregs necessary to prevent or dampen immune responses. The production of RA by GALT DCs has also been shown to induce the expression of gut-homing receptors on T cells.4,32 However, in contrast to the ALDH+ DCs isolated from MLNs, the ALDH+ DCs from CLNs failed to induce CCR9 expression on OT-II Rag-2−/− CD4+ T cells after antigen triggering. As described for vitamin D3 in the case of human skin DCs,50 it is probable that factors produced by RA-producing DCs from CLNs counterbalance the CCR9-inductive role of RA.
In conclusion, several unexpected findings emerged from our global survey of RA-producing mouse DCs. We showed that the skin and the lungs resemble the gut in that they contain DCs that constitutively produce RA and induce Foxp3+ Tregs. Moreover, we demonstrated that CD103, a marker that has become recently a focus of intensive investigation because it allows isolation of the RA-producing, tolerogenic DCs found in the LP and the MLNs loses its discriminatory power outside the gut. For instance, in both the skin and the lungs, CD103−CD11b+ DCs are endowed with high ALDH activity. Importantly, we showed that the induction of ALDH activity within particular DC subsets associated with tissues that constitute environmental interfaces occurred regardless of the presence of commensal bacteria and of TLR triggering. Finally, the possibility to identify RA-producing DCs at the single-cell level and of sorting them under vital conditions using ALDEFLUOR staining may contribute to understanding the function of ALDH+ DCs present in various tissues under steady-state and inflammatory conditions and may shed light on the way these cells initiate tolerogenic immune responses.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Marie Malissen, Emmanuelle Charafe-Jauffret, Adrien Kissenpfennig, and Lee Leserman for discussion and Bruce Beutler and Shizuo Akira for mice.
This work was supported by Centre National de la Recherche Scientifique, Inserm, European Communities (MUGEN Network of Excellence and MASTERSWITCH Integrating Project, B.M.), Institut National du Cancer (INCA; Melan-Imm project), Agence Nationale de la Recherche (ANR; DC in vivo, B.M.; pDC physiology, M.D.), postdoctoral fellowships from Ministère de la Recherche (S.T.), Association pour la Recherche sur le Cancer (ARC; M.G., K.C.), and a Marie Curie Fellowship from the European Communities (project no. 237109; M.G.).
Authorship
Contribution: M.G. designed and performed experiments, analyzed and interpreted data, and cowrote the manuscript; K.C., S.H., S.T., P.G., E.D., and B.d.B. performed experiments and collected data; L.A. provided key reagents; M.D. supervised the experiments involving MCMV; and B.M. directed the study and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bernard Malissen, Centre d'Immunologie de Marseille-Luminy, Campus de Luminy, Case 906, 13288 Marseille cedex 09, France; e-mail: bernardm@ciml.univ-mrs.fr.