Abstract
Histone modifications play an important role in the process of transcription. However, in contrast to lysine methylation, the role of arginine methylation in chromatin structure and transcription has been underexplored. The globin genes are regulated by a highly organized chromatin structure that juxtaposes the locus control region (LCR) with downstream globin genes. We report here that the targeted recruitment of asymmetric dimethyl H4R3 catalyzed by PRMT1 (protein arginine methyltransferase 1) facilitates histone H3 acetylation on Lys9/Lys14. Dimethyl H4R3 provides a binding surface for P300/CBP-associated factor (PCAF) and directly enhances histone H3 acetylation in vitro. We show that these active modifications are essential for efficient interactions between the LCR and the βmaj-promoter as well as transcription of the β-globin gene. Furthermore, knockdown (KD) of PRMT1 by RNA interference in erythroid progenitor cells prevents histone acetylation, enhancer and promoter interaction, and recruitment of transcription complexes to the active β-globin promoter. Reintroducing rat PRMT1 into the PRMT1 KD MEL cells rescues PRMT1 binding, β-globin transcription, and erythroid differentiation. Taken together, our data suggest that PRMT1-mediated dimethyl H4R3 facilitates histone acetylation and enhancer/promoter communications, which lead to the efficient recruitment of transcription preinitiation complexes to active promoters.
Introduction
Covalent modifications of N-terminal histone tails are critically involved in transcriptional activation and repression.1 The interplay between individual modifications may exert distinct regulatory effects on different gene loci during development and cellular differentiation. For example, H3K9 and H3K27 methylations are generally linked to gene repression, whereas methylation of H3K4 correlates with transcriptionally active euchromatin.2 However, in the β-globin locus H3K9 methylation was also detected in the active globin genes.3 Arginine methylation of histones is associated with both transcriptional repression and activation.4 PRMT6-mediated H3R2 dimethylation negatively regulates deposition of H3K4 trimethylation at active promoters,5 whereas dimethyl H4R3 correlates with transcriptional activation.6,7 Asymmetric dimethylation of H4R3 residues by protein arginine methyltransferase PRMT1 is essential in vitro and in vivo for the establishment or maintenance of active histone acetylation patterns.7,8 The interdependence of these modifications appears to be important for the transcription of a p53-dependent reporter gene in a cell-free system with reconstituted chromatin templates.9 Furthermore, PRMT1 was associated with the activation of HNF4 and HoxA9 genes during tissue development and oncogenesis, respectively.10,11 We showed recently that PRMT1 directly interacts with transcription factor USF1 (upstream regulatory factor 1), which has been implicated in chromatin barrier function and β-globin gene regulation.12-15 The cross-communication between different histone modifications might provide regulatory potentials to many biologic processes including transcription. However, in contrast to lysine methylation, little is known about how PRMT1-mediated asymmetric dimethyl H4R3 modulates the process of transcription.
The βmaj-globin gene is expressed only in adult erythrocytes and is located approximately 40 kbp downstream of the locus control region (LCR) at the 3′ end of the murine β-globin locus. The LCR is a powerful erythroid-specific enhancer that orchestrates the recruitment of transcription factors and cofactors, as well as the establishment of the active epigenetic marks, such as H3K4 methylation and histone acetylation, and contributes to the tissue- and stage-specific expression of the globin genes.16-18 The LCR plays a critical role in maintaining the appropriate spatial chromatin configuration of this locus at the definitive erythroid stage.19 Consistent with the roles of the LCR, the transcription of the β-globin gene in the repressive environment of a differentiating erythroid cell requires a domain-wide open chromatin structure and formation of an active chromatin hub, bringing the LCR in close proximity to the βmaj-promoter.20,21 In addition, formation of the active chromatin hub requires the binding of the erythroid-expressed activators, GATA-1, EKLF, FOG-1, TAL1, and the widely expressed coregulator Ldb1.22-24 Loops between regulatory elements and genes have also been observed in the imprinted Igf2/H19 locus, the Ig Kappa gene, the Kit gene, the olfactory receptor, and the TH2 cytokine loci.25-28 These examples illustrate that proper looped chromatin configurations that place DNA regulatory elements into close proximity to the genes are important for controlling gene expression. However, a central question following the discovery of long-range chromatin interactions is whether active histone modifications directly affect the formation of active chromatin hubs.
Here, we show that the establishment of asymmetric dimethyl H4R3 at HS2 (DNase I hypersensitive site 2) of the LCR and the βmaj-promoter potentiates histone H3 acetylation on Lys9 and Lys14 (AcH3K9/K14) and strongly correlates with the activation of adult β-globin gene transcription. The inhibition of dimethyl H4R3 by suppression of PRMT1 in hematopoietic cells led to a decrease in the recruitment of histone acetyltransferases (HATs) and subsequent histone acetylation, to a disruption of a chromatin architecture that normally brings the LCR in close proximity to the β-globin gene, and ultimately to a reduced recruitment of transcription complexes to the adult βmaj-promoter. Furthermore, the rescue of PRMT1 expression in PRMT1 knockdown (KD) cells led to the reactivation of β-globin transcription and erythroid differentiation. Taken together, our data demonstrate that PRMT1-mediated asymmetric dimethyl H4R3 regulates enhancer/promoter communications, which are required for the efficient recruitment of transcription complexes to transcriptionally active promoters.
Methods
Cell culture and shRNA knockdown
MEL cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. For differentiation, 2 × 105 cells/mL were incubated with 1.5% dimethyl sulfoxide (DMSO) for indicated time periods. The hemoglobin was visualized by benzidine reagent (Sigma) as described.23 The fraction of hemoglobin-expressing cells was determined from counting 250 cells in 3 independent experiments. Murine embryonic stem (ES) cells were maintained and induced to differentiate with erythropoietin (EPO) as described.29 The yolk sac, fetal liver, and fetal brain cells were prepared from mouse embryos and single-cell suspensions were prepared and cross-linked with a final concentration of 0.5% formaldehyde for the chromatin immunoprecipitation (ChIP) assay.
The PRMT1-shRNA construct was obtained from Dr Eric So (The Institute of Cancer Research).11 Infectious viruses were produced by calcium phosphate transfection of Phoenix cells (collected 48 hours after transfection) and infected MEL and ES cells, and selected by puromycin (1 μg/mL; Sigma) resistance. All protocols were approved by the Institutional Animal Care and Use Committee of the University of Florida.
Quantitative reverse-transcription–PCR
Total RNA was prepared using the SV total RNA isolation kit according to the manufacturer's instruction (Promega). RNA (1.5 μg) was treated with RNase-free DNase I and reverse transcribed using the Superscript II reverse Transcriptase (Invitrogen). The quantity of cDNA was determined by quantitative polymerase chain reaction (qPCR) analysis using a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad). Primer sequences and PCR conditions are listed in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
ChIP and sequential ChIP
ChIP assays were performed as described previously15 using antibodies specific for transcription factors, modifying enzymes, and various histone modifications (anti-p300, anti-CBP, anti-USF1, anti–GATA-1, anti–NF-E2, and anti–TATA-binding protein (TBP) were from Santa Cruz Biotechnology; anti–RNA PolII, anti-PRMT1, anti–dimethyl H4R3 (dimeH4R3), anti-H3K9K14ac, and anti-H4ac were from Upstate Biotechnology; anti–P300/CBP-associated factor [PCAF] was from Abcam). The relative enrichment was determined using the following equation: 2Ct(ref) − Ct(IP), where Ct(ref) is PCR cycles of the input DNA and Ct(IP) is PCR cycles of ChIP DNA. The sequential ChIP assays were carried out essentially as described,17 with some modifications. Briefly, chromatin prepared from 1 × 108 cells was immunoprecipitated with USF1 antibody. The USF1-chromatin complexes were eluted, dialyzed, and subsequently immunoprecipitated with PRMT1 antibody. The bound protein-DNA complexes were reverse cross-linked, purified, and analyzed by qPCR.
HMT and in vitro histone binding assay
[35S]methionine-labeled PCAF and p300 were synthesized using the TnT-coupled reticulocyte lysate system (Promega). The in vitro histone methyltransferase (HMT) assay was previously described.8 Briefly, 10 μg of biotin-conjugated H4 peptide (Upstate) was incubated with 0.5 μg of PRMT1 (Upstate) in the buffer containing 20mM Tris (tris(hydroxymethyl)aminomethane)–HCl (pH 8.0), 4mM ethylenediaminetetraacetic acid, 1mM phenylmethylsulphonyl fluoride, 0.5mM dithiothreitol, and 2 μL of S-adenosyl methionine (SAM, 32mM; New England Biolabs [NEB]) at 30°C for 1 hour, and immobilized to the Dynabeads M-280 streptavidin (Dynal) according to the manufacturer's direction. The immobilized methylated or unmethylated H4 tails were washed 3 times with the binding buffer (50mM Tris-HCl, pH 8.0, 2mM ethylenediaminetetraacetic acid, 200mM NaCl, and 0.1% Nonidet P-40) to remove PRMT1 and incubated with 35S-labeled PCAF or p300. The reaction mixture was washed, resolved by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis, and analyzed on a Typhoon 9410 imager (Molecular Dynamics) using ImageQuant (GE Healthcare).
Purification of oligonucleosomes and HAT assay
The dimethyl H4R3–depleted oligonucleosomes were purified from the PRMT1 KD MEL cells and methylated by adding 1 μg of PRMT1 in the presence and absence of SAM (32mM; NEB) as described previously.8,15 PRMT1 was then removed by immunoprecipitation with 200 μg of PRMT1 antibody. The oligonucleosome-containing supernatant was collected for HAT assays using purified p300 and PCAF. The enrichment of dimeH4R3 and acH3K9/K14 was detected by Western blot using antibodies specific to dimeH4R3 and acH3K9/K14.
Chromatin conformation capture
The chromatin conformation capture (3C) assay was performed as described previously,21,24 with minor modifications. In brief, 4 × 107 cells were cross-linked with 2% formaldehyde for 10 minutes and stopped by the addition of glycine at a final concentration of 0.125M. Cells were pelleted and washed twice with cold PBS and lysed in lysis buffer (10mM Tris, pH 8.0, 10mM NaCl, 0.2% Nonidet P-40, and protease inhibitors) at 4°C for 90 minutes with gentle rotation. Nuclei were collected and washed with appropriate 1× restriction enzyme buffer (Buffer 3; NEB) and then resuspended in restriction buffer containing 0.3% of SDS at 37°C for 1 hour with shaking. Triton X-100 was then added to a final concentration of 1.8% to sequester SDS at 37°C for another 1 hour with shaking. Chromatin was digested with BglII (NEB) at 37°C overnight with shaking and stopped by adding SDS to a final concentration of 1.6% at 65°C for 30 minutes. The digestion reaction was diluted into 1 mL of ligation reaction buffer containing 1% Triton X-100, 50mM Tris, pH 7.5, 10mM MgCl2, 10mM dithiothreitol, 0.1 mg/mL BSA, and 1mM adenosine triphosphate at 37°C for 1 hour. T4 DNA ligase (NEB) was added and the reaction incubated at 16°C for 5 hours followed by ligation for 30 minutes at room temperature. After reversal, cross-linked DNA was isolated, amplified, and cloned into pCR4-TOPO vector (Invitrogen) for sequencing. To control the PCR amplification efficiency of different primer pairs, equimolar amounts of plasmids containing PCR products were mixed to generate the control templates. PCR reactions were performed with the control templates in parallel with experimental templates. Furthermore, to control for differences in template quality, the interactions at the β-globin locus from an experimental template were normalized to a control interaction at the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) locus and quantified with ImageQuant. The detailed calculation of the relative cross-linking frequency between 2 loci is listed in the supplemental data.
Viral transduction and the rat PRMT1 rescued cell line
The reintroduction of the shPRMT1-resistant rat PRMT1 cDNA was performed as previously described,30 with minor modifications. Briefly, the rat PRMT1 cDNA was mutated in the third codon within the shRNA targeting site by site-directed mutagenesis and cloned into the pOZ-FH-N vector.30 The infectious virus was produced and infected with 5 × 105 exponentially growing PRMT1 KD MEL cells. The transduced cells were sorted by anti–interleukin-2 receptor–conjugated Dynabeads.
Results
Dimethyl H4R3 correlates with the activation of the β-globin gene
To investigate the role of PRMT1 and dimethyl H4R3 in the regulation of β-globin gene expression, we first compared the patterns of asymmetric dimethylation of H4R3 across the mouse β-globin locus (Figure 1A) in fetal liver and fetal brain cells isolated from embryonic day 14.5 (E14.5) embryos using ChIP-qPCR assays. The β-globin gene is highly transcribed in the fetal liver after E12 and is silenced in the fetal brain.31 There is a marked enrichment of dimethyl H4R3 at the βmaj-promoter in fetal liver cells (Figure 1B), but not in transcriptionally silent fetal brain cells (Figure 1C). A small but significant enrichment of dimethyl H4R3 is also found between HS2 and HS3 of the LCR in the fetal liver (Figure 1B). Furthermore, H4R3 methylation is not detected at the ϵy and βh1 genes (Figure 1B), which are transcribed only at the yolk sac stage of erythropoiesis before E12 and are silent in the fetal liver.31 In addition, there is no enrichment of dimethyl H4R3 at the promoter regions of the housekeeping gene GAPDH and the silenced MyoD gene (Figure 1B), suggesting that dimethyl H4R3 is important for activation of β-globin gene.
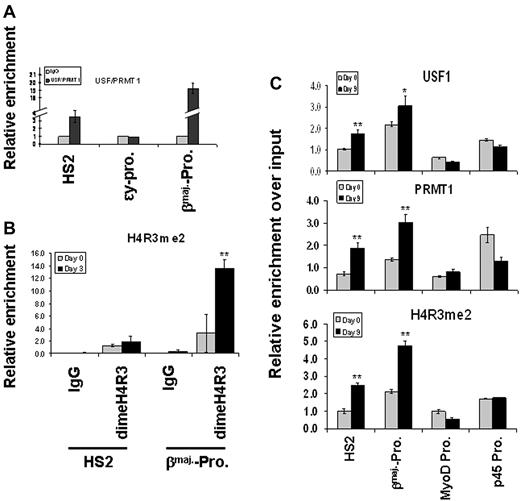
Asymmetric dimethyl H4R3 correlates with transcriptional activation of the β-globin gene. (A) Schematic representation of the 135-kbp mouse β-globin locus. The globin genes are indicated by red boxes, and DNase I hypersensitive sites (HS) are indicated by arrows. The locations and names of primers used for the ChIP-quantitative PCR (qPCR) assay are indicated below the locus. (B) Fetal livers were prepared from E14.5 mouse embryos and isolated as single-cell suspensions. The cells were cross-linked with formaldehyde and sonicated for ChIP-qPCR analysis using α-dimethyl H4R3 antibody or normal rabbit immunoglobulin G (IgG) as a control. The relative enrichment of dimethyl H4R3 for each amplicon was determined by 3 independent experiments. (C) ChIP analysis of dimethyl H4R3 at the endogenous mouse β-globin locus in E14.5 fetal brain. (D-E) Yolk sacs were prepared from wild-type or dominant-negative USF1-expressing transgenic embryos at E10.5 and subjected to ChIP-qPCR across the globin locus using PRMT1- (D) or dimethyl H4R3–specific (E) antibodies. Significant difference by Student t test: **P < .01.
Asymmetric dimethyl H4R3 correlates with transcriptional activation of the β-globin gene. (A) Schematic representation of the 135-kbp mouse β-globin locus. The globin genes are indicated by red boxes, and DNase I hypersensitive sites (HS) are indicated by arrows. The locations and names of primers used for the ChIP-quantitative PCR (qPCR) assay are indicated below the locus. (B) Fetal livers were prepared from E14.5 mouse embryos and isolated as single-cell suspensions. The cells were cross-linked with formaldehyde and sonicated for ChIP-qPCR analysis using α-dimethyl H4R3 antibody or normal rabbit immunoglobulin G (IgG) as a control. The relative enrichment of dimethyl H4R3 for each amplicon was determined by 3 independent experiments. (C) ChIP analysis of dimethyl H4R3 at the endogenous mouse β-globin locus in E14.5 fetal brain. (D-E) Yolk sacs were prepared from wild-type or dominant-negative USF1-expressing transgenic embryos at E10.5 and subjected to ChIP-qPCR across the globin locus using PRMT1- (D) or dimethyl H4R3–specific (E) antibodies. Significant difference by Student t test: **P < .01.
PRMT1 does not recognize a specific DNA element. However, it interacts with transcription factor USF1 that regulates β-globin gene expression by interacting with LCR HS2 and the βmaj-promoter.12,13,15 We tested whether inhibition of USF1 DNA binding would lead to a loss of PRMT1 recruitment and reduction in dimethyl H4R3 at the globin locus in transgenic mice expressing dominant-negative USF1. These mice exhibit reduced globin gene transcription and die during transition from primitive to definitive hematopoiesis between E11.5 and E12.5.32 Suppression of USF1 binding resulted in a significant decrease of PRMT1 recruitment and reduction in dimethyl H4R3 at LCR HS2 and the βmaj-promoter (Figure 1D-E). Next, we examined whether PRMT1 and USF simultaneously co-occupy the β-globin gene locus. A sequential ChIP analysis was performed in which the cross-linked chromatin from MEL cells was first precipitated with USF1 antibody and the USF1 selected chromatin was then immunoprecipitated with PRMT1 antibody. Both USF1 and PRMT1 co-occupy HS2 of the LCR and the βmaj-promoter, but not the transcriptionally silenced ϵy promoter (Figure 2A, supplemental Figure 1A-B).
PRMT1 and dimethyl H4R3 are targeted to the β-globin locus by transcription factor USF1. (A) Cross-linked chromatin prepared from MEL cells was immunoprecipitated with α-USF1 antibody, and USF1-DNA complexes from the first ChIP were subjected to a second ChIP using α-PRMT1 antibody. The protein/DNA complexes were reversed and the purified DNA fragments were amplified using primers specific for the HS2 core, ϵy-globin promoter and βmajor-globin promoter. (B) MEL cells were cultured in the presence or absence of 1.5% DMSO. The cross-linked chromatin was analyzed by ChIP assay using antibodies specific to dimethyl H4R3. The protein/DNA complexes were then reversed and the purified DNA fragments were amplified using primers specific for HS2 and βmaj-globin promoter. (C) ES cells were cultured in the presence or absence of 2 U/mL EPO. The cross-linked chromatin was analyzed by ChIP assay using antibodies specific for USF1 (top), PRMT1 (middle), and dimethyl H4R3 (bottom). The MyoD promoter and the p45 promoter were also analyzed as inactive and active promoter controls, respectively. Significant difference by Student t test: *P < .05, **P < .01. Shown are the mean ± SDM of 3 independent experiments.
PRMT1 and dimethyl H4R3 are targeted to the β-globin locus by transcription factor USF1. (A) Cross-linked chromatin prepared from MEL cells was immunoprecipitated with α-USF1 antibody, and USF1-DNA complexes from the first ChIP were subjected to a second ChIP using α-PRMT1 antibody. The protein/DNA complexes were reversed and the purified DNA fragments were amplified using primers specific for the HS2 core, ϵy-globin promoter and βmajor-globin promoter. (B) MEL cells were cultured in the presence or absence of 1.5% DMSO. The cross-linked chromatin was analyzed by ChIP assay using antibodies specific to dimethyl H4R3. The protein/DNA complexes were then reversed and the purified DNA fragments were amplified using primers specific for HS2 and βmaj-globin promoter. (C) ES cells were cultured in the presence or absence of 2 U/mL EPO. The cross-linked chromatin was analyzed by ChIP assay using antibodies specific for USF1 (top), PRMT1 (middle), and dimethyl H4R3 (bottom). The MyoD promoter and the p45 promoter were also analyzed as inactive and active promoter controls, respectively. Significant difference by Student t test: *P < .05, **P < .01. Shown are the mean ± SDM of 3 independent experiments.
Consistent with PRMT1 localization at the globin locus, asymmetric dimethyl H4R3 is increased slightly at HS2 and significantly at the βmaj-promoter upon DMSO-induced MEL differentiation (Figure 2B, P < .01). In addition, we examined the binding of USF1 and PRMT1 as well as dimethyl H4R3 modification in erythropoietin (EPO)–induced differentiated mouse embryonic stem (ES) cells. The binding of USF1 (P < .05 at the βmaj-promoter), PRMT1, and dimethyl H4R3 modification is significantly increased at HS2 and the βmaj-promoter upon EPO induction (Figure 2C, P < .01). They are not enriched at the inactive myogenic differentiation (MyoD) promoter, and the dimethyl H4R3 signal is also low in the active erythroid-specific p45 promoter (Figure 2C). The combined data suggest that dimethyl H4R3 targeted by the USF1/PRMT complex might play an important role in the activation of the β-globin gene during hematopoiesis.
Knockdown of PRMT1 inhibits erythroid differentiation and β-globin transcription
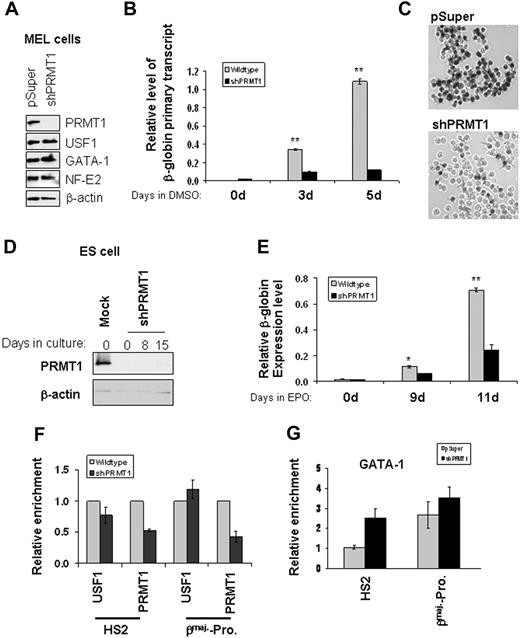
PRMT1 is the only known enzyme that asymmetrically methylates H4R3.6,7 To address the role of dimethyl H4R3 in the transcription of globin genes, we generated a stable PRMT1 knockdown (KD) MEL line using a pSuper retroviral vector encoding a PRMT1 shRNA (Figure 3A). Suppression of PRMT1 led to a decrease of β-globin primary transcripts by more than 75% compared with cells transfected with the control vector (Figure 3B) and strongly inhibited MEL differentiation (Figure 3C). The effects are likely due to a decrease in the recruitment of PRMT1 to the β-globin locus because the knockdown of PRMT1 did not affect the expression of ubiquitous or hematopoietic-specific transcription factors, such as USF1, GATA-1, EKLF, and NF-E2, which regulate the transcription of globin genes (Figure 3A and supplemental Figure 1C). Furthermore, the binding of USF1/2 and GATA-1 to HS2 of the LCR and the βmaj-promoter was unaffected or increased by the knockdown (Figure 3F-G).
PRMT1 is critical for erythroid differentiation and βmaj-globin transcription. (A) Western blotting of MEL cells stably transduced with pSuper vector or PRMT1 shRNA was performed with the indicated antibodies. (B) Stable MEL clones harboring control or shPRMT1-expressing constructs were incubated with 1.5% DMSO and total RNA was extracted at days 0, 3, and 5. The βmaj-globin primary transcripts were analyzed by quantitative reverse-transcription (RT)–qPCR and normalized to β-actin. (C) MEL cell clones stably transduced with retrovirus encoded pSuper vector or PRMT1 shRNA (shPRMT1) were treated with 1.5% DMSO and hemoglobinization was assayed by benzidine staining at day 5. (D) PRMT1 expression of stable ES cell clones transduced with the control or shPRMT1 retroviral vectors was analyzed by Western blot analysis. (E) Stable ES cell clones transduced with the control or shPRMT1 retroviral vectors were treated with 2 U/mL EPO for 9 and 11 days and total RNA was isolated. βmaj-globin mRNA expression was analyzed by qRT-PCR and normalized using β-actin as a control. (F) ChIP analysis of USF1 and PRMT1 binding to HS2 and the βmaj-promoter in the parental and PRMT1 KD ES cells. (G) Analysis of the binding of GATA-1 to the mouse β-globin locus in control and PRMT1 KD cells. ChIP assay was carried out using antibodies specific to GATA-1 comparing pSuper vector control-transfected cells with the PRMT1 KD MEL cells. Precipitated DNA fragments were amplified by real-time quantitative PCR using primers specific to the HS2 core and the βmaj-promoter. Significant difference by Student t test: *P < .05, **P < .01. Shown are the mean ± SDM of 3 independent ChIP experiments.
PRMT1 is critical for erythroid differentiation and βmaj-globin transcription. (A) Western blotting of MEL cells stably transduced with pSuper vector or PRMT1 shRNA was performed with the indicated antibodies. (B) Stable MEL clones harboring control or shPRMT1-expressing constructs were incubated with 1.5% DMSO and total RNA was extracted at days 0, 3, and 5. The βmaj-globin primary transcripts were analyzed by quantitative reverse-transcription (RT)–qPCR and normalized to β-actin. (C) MEL cell clones stably transduced with retrovirus encoded pSuper vector or PRMT1 shRNA (shPRMT1) were treated with 1.5% DMSO and hemoglobinization was assayed by benzidine staining at day 5. (D) PRMT1 expression of stable ES cell clones transduced with the control or shPRMT1 retroviral vectors was analyzed by Western blot analysis. (E) Stable ES cell clones transduced with the control or shPRMT1 retroviral vectors were treated with 2 U/mL EPO for 9 and 11 days and total RNA was isolated. βmaj-globin mRNA expression was analyzed by qRT-PCR and normalized using β-actin as a control. (F) ChIP analysis of USF1 and PRMT1 binding to HS2 and the βmaj-promoter in the parental and PRMT1 KD ES cells. (G) Analysis of the binding of GATA-1 to the mouse β-globin locus in control and PRMT1 KD cells. ChIP assay was carried out using antibodies specific to GATA-1 comparing pSuper vector control-transfected cells with the PRMT1 KD MEL cells. Precipitated DNA fragments were amplified by real-time quantitative PCR using primers specific to the HS2 core and the βmaj-promoter. Significant difference by Student t test: *P < .05, **P < .01. Shown are the mean ± SDM of 3 independent ChIP experiments.
To further examine the biologic role of PRMT1 in β-globin transcription, we also generated a stable PRMT1 knockdown mouse ES cell line (Figure 3D) that was differentiated into erythroid cells with EPO for 9 and 11 days.29 Reduction of PRMT1 resulted in a profound decrease in β-globin expression by 65% and 76% at day 9 and day 11, respectively (Figure 3E). Together, these data indicate that PRMT1 is required for efficient β-globin gene transcription.
KD of PRMT1 leads to a disruption of long-range enhancer/promoter interactions in the β-globin locus
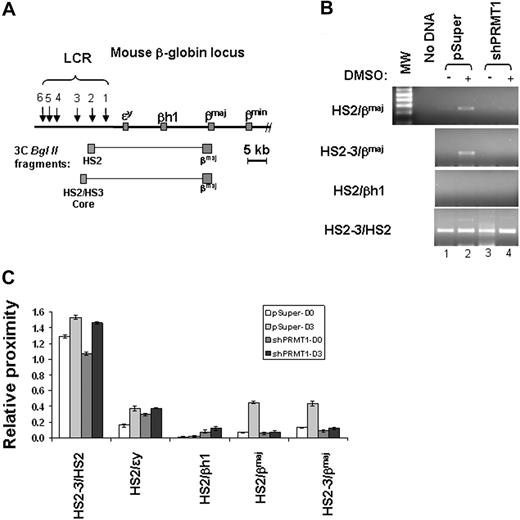
The globin gene locus adopts an active chromatin hub configuration in erythroid cells that brings distant regulatory regions into close proximity.19,21,33 The change in configuration may be associated with the recruitment of the β-globin gene to engaged transcription factories.34 We hypothesize that the inhibition of β-globin transcription by PRMT1 knockdown may correlate with the impairment of long-range chromatin interactions in the mouse β-globin locus. To test this hypothesis, we performed chromatin conformation capture (3C) assays comparing the vector control and the PRMT1 KD MEL cells using primers specific for HS2 or HS2-3 core and for the βmaj-promoter, respectively. The association of HS2 and the βmaj-promoter should amplify a 250-bp fragment that includes both HS2 and βmaj-promoter sequences (Figure 4A). Consistent with stimulation of β-globin expression in differentiated cells, the loop between HS2 or HS2-3 core and the downstream βmaj-promoter was increased upon DMSO-induced differentiation (Figure 4B-C compare lanes 1 and 2). As a negative control, HS2 interacted weakly or not at all with the βh1 gene (Figure 4B-C HS2/βh1). The 250-bp fragment was cloned and sequenced. It constitutes a fusion of sequences from HS2 and the βmaj-promoter (supplemental Figure 2). The loss of dimethyl H4R3 by PRMT1 KD resulted in a specific inhibition of looping between HS2-3 or HS2 and the βmaj-promoter (Figure 4B-C compare lanes 2 and 4) but did not affect the ligation of the positive control fragments (Figure 4B-C HS2-3/HS2). PRMT1 KD did not affect BglII cleavage efficiency (supplemental Figure 3). The finding suggests that the dimethylation of H4R3 is one of the important events required for the long-range interaction between 2 distantly located regulatory elements.
Loss of dimethyl H4R3 leads to disruption of long-range chromatin interactions between HS2 and the βmaj-globin promoter in the globin locus. (A) Schematic representation of the mouse β-globin locus. The globin genes are indicated by red boxes and DNase I hypersensitive sites (HSs) are indicated by arrows. The predicted 3C BglII fragments from HS2 and the βmaj-promoter are shown in gray. (B) Shown is 3C analysis of the interaction between HS2/3 and the βmaj-promoter in the mouse β-globin locus in the vector control or shPRMT1-transduced clones treated with or without DMSO for 3 days. The predicted 250-bp product was then subcloned and analyzed by DNA sequencing. (C) A total of 3 independent 3C experiments were quantitated by densitometry. Shown are the mean ± SDM of 3 independent experiments.
Loss of dimethyl H4R3 leads to disruption of long-range chromatin interactions between HS2 and the βmaj-globin promoter in the globin locus. (A) Schematic representation of the mouse β-globin locus. The globin genes are indicated by red boxes and DNase I hypersensitive sites (HSs) are indicated by arrows. The predicted 3C BglII fragments from HS2 and the βmaj-promoter are shown in gray. (B) Shown is 3C analysis of the interaction between HS2/3 and the βmaj-promoter in the mouse β-globin locus in the vector control or shPRMT1-transduced clones treated with or without DMSO for 3 days. The predicted 250-bp product was then subcloned and analyzed by DNA sequencing. (C) A total of 3 independent 3C experiments were quantitated by densitometry. Shown are the mean ± SDM of 3 independent experiments.
Methylation of H4R3 cross talks with histone acetylation
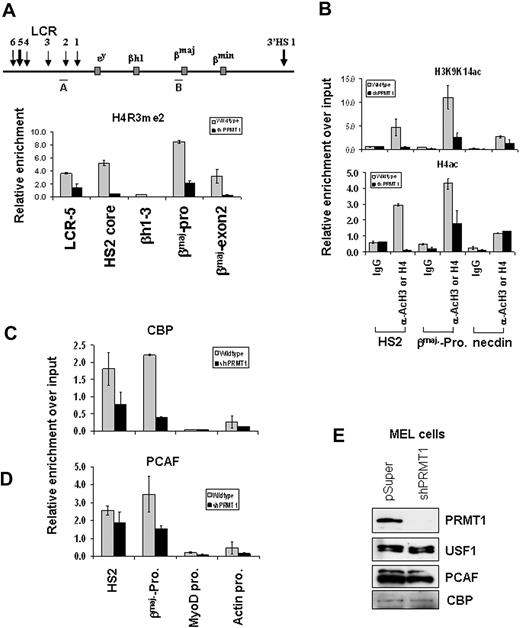
The role of PRMT1 in transcriptional activation and maintenance of an active chromatin environment depends on its H4R3 methyltransferase activity.7,8 Similarly, histone acetylation is strongly linked to transcriptional activation and plays an important function in LCR and βmaj-promoter activity.35 We asked whether loss of dimethyl H4R3 at the enhancer and promoter results in the disruption of active histone modifications at the β-globin locus. We examined changes of histone acetylation in the PRMT1 knockdown and control cells. Consistent with a loss of dimethyl H4R3 at HS2 and the β-globin promoter (Figure 5A), acH3K9/K14 and acH4 are significantly reduced at the same sites (Figure 5B) in the PRMT1 knockdown MEL cells. Furthermore, both dimethyl H4R3 and acH3K9K14 are also significantly inhibited in the PRMT1 KD murine ES cells (supplemental Figure 4A-B), confirming that PRMT1 controls asymmetric dimethyl H4R3 and subsequent histone acetylation.8 The effect of dimethyl H4R3 on histone acetylation was not due to the loss of USF or GATA-1 binding. As noted in Figure 3, the binding only of PRMT1, but not that of USF or GATA-1, is affected by the expression of the PRMT1 shRNA (Figure 3F-G). The data indicate that asymmetric dimethyl H4R3 affects histone acetylation and establishment of an active chromatin conformation in the globin locus.
Methylation of Arg3 on H4 tails is required for the formation of an active chromatin domain. (A) ChIP analysis of dimethyl H4R3 at the β-globin locus in stable MEL cell lines transduced with pSuper vector or shPRMT1 vector in the presence of 1.5% DMSO for 3 days. The precipitated DNA was analyzed by real-time qPCR. (B) ChIP analysis of the H3K9/K14ac and acH4 patterns at HS2, the βmaj-promoter, and the inactive necdin promoter comparing the parental and the shPRMT1-transduced MEL cells treated with 1.5% DMSO for 3 days. (C-D) ChIP assays of CBP and PCAF at the HS2 enhancer, the βmaj-globin promoter, the MyoD promoter, and the actin promoter in stable MEL cell clones transduced with the vector control or shPRMT1 in the presence of 1.5% DMSO for 3 days. (E) Western blot analysis of proteins from MEL cells stably transduced with pSuper vector or PRMT1 shRNA vector with antibodies as indicated. Shown are the mean ± SDM of 3 independent experiments.
Methylation of Arg3 on H4 tails is required for the formation of an active chromatin domain. (A) ChIP analysis of dimethyl H4R3 at the β-globin locus in stable MEL cell lines transduced with pSuper vector or shPRMT1 vector in the presence of 1.5% DMSO for 3 days. The precipitated DNA was analyzed by real-time qPCR. (B) ChIP analysis of the H3K9/K14ac and acH4 patterns at HS2, the βmaj-promoter, and the inactive necdin promoter comparing the parental and the shPRMT1-transduced MEL cells treated with 1.5% DMSO for 3 days. (C-D) ChIP assays of CBP and PCAF at the HS2 enhancer, the βmaj-globin promoter, the MyoD promoter, and the actin promoter in stable MEL cell clones transduced with the vector control or shPRMT1 in the presence of 1.5% DMSO for 3 days. (E) Western blot analysis of proteins from MEL cells stably transduced with pSuper vector or PRMT1 shRNA vector with antibodies as indicated. Shown are the mean ± SDM of 3 independent experiments.
P300/CBP and PCAF are transcriptional coactivators possessing HAT activities on histone H3 and H4 substrates. PCAF is present in USF1/PRMT1 complexes.15 The loss of histone acetylation by PRMT1 KD might result from the decreased binding of HATs to chromatin. To investigate this possibility, we examined the binding of p300/CBP and PCAF at the regulatory elements of the mouse β-globin locus. Knockdown of PRMT1 markedly reduced the recruitment of CBP (Figure 5C) and PCAF (Figure 5D). These HATs did not interact with the silent MyoD gene (Figure 5C-D). The reduction of HAT activity at the LCR and the βmaj-promoter is not due to decreased expression of PCAF and CBP (Figure 5E). The results establish a functional link between PRMT1 and HATs, perhaps initiated by the binding of USFs to regulatory DNA elements (“Discussion”), in regulating β-globin gene expression.
PCAF binds to dimethyl Arg3 at histone H4 tails
The fact that PRMT1 KD affects subsequent histone acetylation in vivo does not necessarily establish a direct link between H4R3 methylation and HAT-mediated acetylation. HATs could interact directly with dimethyl H4R3 or they could be recruited indirectly by PRMT1 or other proteins that recognize the dimethyl H4R3 mark. To distinguish between these 2 possibilities, biotin-conjugated histone H4 tails were methylated with PRMT1 in the presence or absence of S-adenosyl methionine and incubated with 35S-labeled PCAF (Figure 6A). PCAF consistently binds more efficiently with the Arg3 methylated H4 tails compared with the unmodified tails (Figure 6B). The binding of methylated tails is increased 73% compared with that of unmethylated H4 tails (Figure 6C, P < .05). The result indicates that PCAF can directly recognize asymmetric dimethylated H4R3 marks and provides a mechanistic insight into how PRMT1-mediated H4R3 methylation potentiates histone H3 and H4 acetylation.
Dimethyl Arg3 at histone H4 tails is bound by HATs and potentiates H3 acetylation in vitro. (A) Outline of the PCAF in vitro binding experimental procedure. (B) Biotin-conjugated histone H4 tails were incubated with purified PRMT1 in the presence or absence of S-adenosyl methionine (SAM) and PRMT1 was subsequently removed from the reaction. After methylation, histone tails were incubated with 35S-labeled PCAF. The reaction mixtures were then resolved on SDS–polyacrylamide gel electrophoresis and visualized by autoradiography. (C) A total of 4 independent experiments were quantitated by densitometry. Shown are the mean ± SDM of 4 independent experiments. Significant difference by Student t test: *P < .05. (D) Outline of the experimental procedure for analyzing cross talk between dimethyl H4R3 and acH3K9/K14. (E) Purified oligonucleosomes from the PRMT1 KD MEL cells were incubated with PRMT1 in the presence or absence of SAM. After arginine methylation, PRMT1 was removed and then purified p300 and PCAF were added to the reaction mixture for 1 hour at 30°C as shown. The enrichment of dimethyl H4R3 and acH3K9/K14 was analyzed by Western blot analysis using antibodies against dimethyl H4R3, acH3K9/K14, and H3.
Dimethyl Arg3 at histone H4 tails is bound by HATs and potentiates H3 acetylation in vitro. (A) Outline of the PCAF in vitro binding experimental procedure. (B) Biotin-conjugated histone H4 tails were incubated with purified PRMT1 in the presence or absence of S-adenosyl methionine (SAM) and PRMT1 was subsequently removed from the reaction. After methylation, histone tails were incubated with 35S-labeled PCAF. The reaction mixtures were then resolved on SDS–polyacrylamide gel electrophoresis and visualized by autoradiography. (C) A total of 4 independent experiments were quantitated by densitometry. Shown are the mean ± SDM of 4 independent experiments. Significant difference by Student t test: *P < .05. (D) Outline of the experimental procedure for analyzing cross talk between dimethyl H4R3 and acH3K9/K14. (E) Purified oligonucleosomes from the PRMT1 KD MEL cells were incubated with PRMT1 in the presence or absence of SAM. After arginine methylation, PRMT1 was removed and then purified p300 and PCAF were added to the reaction mixture for 1 hour at 30°C as shown. The enrichment of dimethyl H4R3 and acH3K9/K14 was analyzed by Western blot analysis using antibodies against dimethyl H4R3, acH3K9/K14, and H3.
Methylation of H4R3 cross talks with HATs in directly controlling histone acetylation in vitro
Given that PCAF directly binds to asymmetric dimethyl H4R3 tails, we asked whether dimethyl H4R3 could directly potentiate HAT-mediated histone H3 acetylation on K9/K14 in the context of a nucleosomal substrate. To test this possibility, we purified oligonucleosomes from a PRMT1 KD MEL cell line to measure the ability of dimethyl H4R3, but not PRMT1, to control H3 acetylation in trans (Figure 6D). The nucleosomes were methylated with PRMT1 in the presence of cofactor SAM (or incubated in its absence as a control) and PRMT1 was then removed from the reaction to eliminate the possibility that PRMT1 mediates direct recruitment of HATs. p300 or PCAF was then added to the reaction system (Figure 6D). Whereas unmethylated nucleosomes were acetylated at a low level (Figure 6E lane 1), the H4R3 dimethylated nucleosomes potentiate acetylation by p300 and PCAF on K9/K14 at H3 tails (Figure 6E lane 2), demonstrating that PRMT1-mediated asymmetric dimethyl H4R3 can facilitate H3K9/K14 acetylation in trans and suggesting the possibility of cross talk of different modifications between histone H3 and H4 tails.
Knockdown of PRMT1 inhibits the recruitment of the transcription preinitiation complex to the βmaj-globin gene
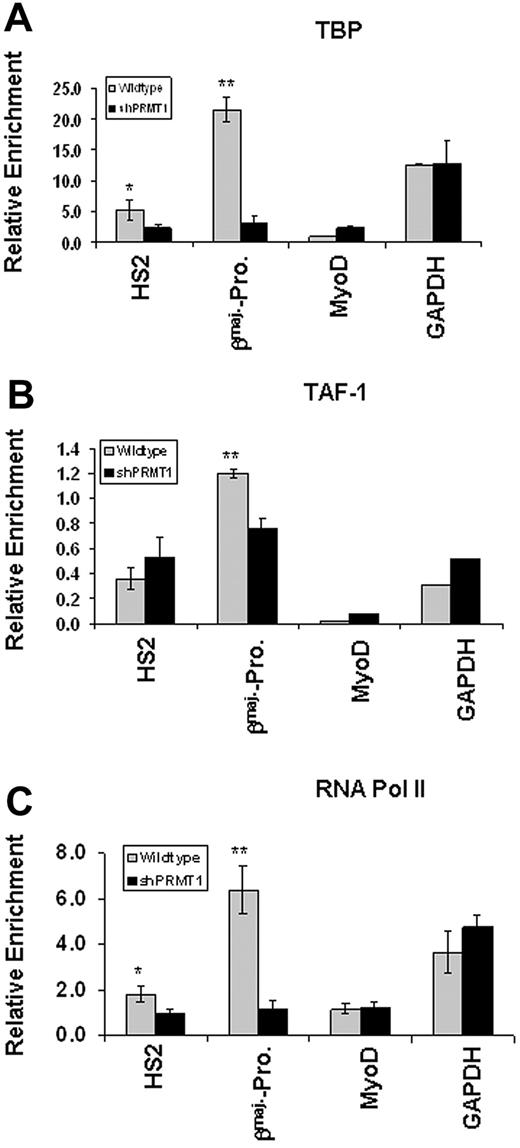
It was reported that acH3K9/K14 targets TAF1 to transcriptionally active promoters.36 We have shown above that disruption of asymmetric methylation of H4R3 inhibits acetylation of H3K9/K14 at the LCR and at the globin gene promoter (Figure 5A-B). We reasoned that loss of acH3K9/K14 and subsequent disruption of looping might affect the recruitment of the transcription complex to the βmaj-promoter. To test this possibility, we investigated the binding of components of the basal transcription apparatus, TBP, TAF1, and Pol II, to HS2 and to the promoter comparing the vector control and the PRMT1 knockdown MEL cells. Although PRMT1 knockdown reduces the binding of TBP by 50% at LCR HS2 (P < .05), the binding of TBP is inhibited even more strongly—by more than 80%—at the βmaj-promoter (Figure 7A, P < .01). The binding of TAF1 is reduced by 40% at the βmaj-promoter (Figure 7B, P < .05), but the protein levels of TAF1 are not (supplemental Figure 4C). Coincident with the reduction of the preinitiation complex at the promoter, the recruitment of RNA Pol II is significantly reduced at HS2 (P < .05) and even more profoundly at the βmaj-promoter (Figure 7C, P < .01). As a control, neither the binding of TFIID complex, TBP and TAF1, nor RNA Pol II was detected at the silenced MyoD gene promoter (Figure 7A-C). Furthermore, the recruitment of transcription preinitiation complex to active GAPDH gene promoter is not affected by loss of PRMT1 and its mediated dimethyl H4R3 (Figure 7). The results suggest that cross talk between dimethyl H4R3 and histone acetylation is important for modulating active chromatin configuration and for the efficient assembly of the transcription machinery at the βmaj-globin gene promoter.
Knockdown of PRMT1 inhibits the recruitment of the transcription preinitiation complex to the β-globin locus. (A) ChIP assays of TBP at the HS2 enhancer and βmaj-globin promoter in MEL cell lines transduced with the vector control or shPRMT1 in the presence of 1.5% DMSO for 3 days. The precipitated DNA was analyzed by real-time qPCR. The MyoD promoter and the GAPDH promoter were also analyzed as inactive and active promoter controls, respectively. (B) ChIP assays of TAF1 at the HS2 enhancer and βmaj-globin promoter in MEL cell lines transduced with the vector control or shPRMT1 in the presence of 1.5% DMSO for 3 days. (C) ChIP assays of RNA Pol II at the HS2 enhancer and βmaj-promoter comparing the vector control and shPRMT1-transduced MEL cell lines in the presence of 1.5% DMSO for 3 days. Significant difference by Student t test: *P < .05, **P < .01. Shown are the mean ± SDM of 3 independent experiments.
Knockdown of PRMT1 inhibits the recruitment of the transcription preinitiation complex to the β-globin locus. (A) ChIP assays of TBP at the HS2 enhancer and βmaj-globin promoter in MEL cell lines transduced with the vector control or shPRMT1 in the presence of 1.5% DMSO for 3 days. The precipitated DNA was analyzed by real-time qPCR. The MyoD promoter and the GAPDH promoter were also analyzed as inactive and active promoter controls, respectively. (B) ChIP assays of TAF1 at the HS2 enhancer and βmaj-globin promoter in MEL cell lines transduced with the vector control or shPRMT1 in the presence of 1.5% DMSO for 3 days. (C) ChIP assays of RNA Pol II at the HS2 enhancer and βmaj-promoter comparing the vector control and shPRMT1-transduced MEL cell lines in the presence of 1.5% DMSO for 3 days. Significant difference by Student t test: *P < .05, **P < .01. Shown are the mean ± SDM of 3 independent experiments.
The restoration of PRMT1 expression in PRMT1 KD cells rescues erythroid differentiation and β-globin transcription
If the effects of PRMT1 KD on β-globin transcription are the result of the disruption of PRMT1 function but not a typical off-target effect of the shRNA, the restoration of PRMT1 expression should rescue the KD phenotypes. The rat PRMT1 cDNA, which was mutated in the third codon positions within the shRNA target site to escape shRNA silencing, was introduced into the PRMT1 KD MEL cells (Figure 8A). The restoration of PRMT1, but not the enzymatic compromised PRMT1 mutant (supplemental Figure 4D), resulted in the rescue of erythroid differentiation and β-globin transcription compared with that of the vector control and the PRMT1 KD line (Figure 8B). Furthermore, levels of PRMT1 binding, dimethyl H4R3, and acH3K9/K14 at HS2 and the βmaj-promoter were comparable between rescued cells and wild-type MEL cells, whereas the levels in the PRMT1 KD MEL cells were significantly lower (Figure 8C-D, P < .01). Therefore, these data suggest that the effects of PRMT1 KD on globin transcription are most likely due to reduced recruitment of PRMT1 and diminished histone modifications at the βmaj-promoter region.
Rescue of erythroid differentiation and β-globin transcription by expression of rat PRMT1 in PRMT1 KD cells. (A) Detection of exogenous shRNA-resistant Flag-tagged rat PRMT1 expressed in stably transfected PRMT1 KD MEL cell clones by Western blotting using anti-Flag and anti-PRMT1 antibodies. (B) Stable MEL clones transfected with control, shPRMT1, or shRNA vectors plus PRMT1 rescue-construct were incubated with 1.5% DMSO, and total RNA was extracted at days 0, 3, and 5. The βmaj-globin mRNA expression was analyzed by qRT-PCR and normalized to β-actin (top), and hemoglobinization was assayed by benzidine staining at day 5 (bottom). (C) ChIP assays of PRMT1 at the HS2 enhancer and βmaj-promoter in MEL cell lines harboring the vector control, shPRMT1, and shPRMT1 plus PRMT1 rescue-construct. (D) ChIP assays of dimethyl H4R3 and H3K9/K14ac at the HS2 enhancer and βmaj-promoter in MEL cell lines harboring the vector control, shPRMT1, and shPRMT1 plus PRMT1 rescue-construct. Significant differences by Student t test: *P < .05, **P < .01. (E) A simplified model illustrating the role of asymmetric dimethyl H4R3, introduced by PRMT1, in transcription regulation. PRMT1 is initially recruited to specific gene loci by sequence-specific transcription factors including USF1. Subsequently, H4R3 dimethylation of neighboring nucleosomes serves as a signal for the recruitment of HATs and the transcription machinery. Shown are the mean ± SDM of 3 independent experiments.
Rescue of erythroid differentiation and β-globin transcription by expression of rat PRMT1 in PRMT1 KD cells. (A) Detection of exogenous shRNA-resistant Flag-tagged rat PRMT1 expressed in stably transfected PRMT1 KD MEL cell clones by Western blotting using anti-Flag and anti-PRMT1 antibodies. (B) Stable MEL clones transfected with control, shPRMT1, or shRNA vectors plus PRMT1 rescue-construct were incubated with 1.5% DMSO, and total RNA was extracted at days 0, 3, and 5. The βmaj-globin mRNA expression was analyzed by qRT-PCR and normalized to β-actin (top), and hemoglobinization was assayed by benzidine staining at day 5 (bottom). (C) ChIP assays of PRMT1 at the HS2 enhancer and βmaj-promoter in MEL cell lines harboring the vector control, shPRMT1, and shPRMT1 plus PRMT1 rescue-construct. (D) ChIP assays of dimethyl H4R3 and H3K9/K14ac at the HS2 enhancer and βmaj-promoter in MEL cell lines harboring the vector control, shPRMT1, and shPRMT1 plus PRMT1 rescue-construct. Significant differences by Student t test: *P < .05, **P < .01. (E) A simplified model illustrating the role of asymmetric dimethyl H4R3, introduced by PRMT1, in transcription regulation. PRMT1 is initially recruited to specific gene loci by sequence-specific transcription factors including USF1. Subsequently, H4R3 dimethylation of neighboring nucleosomes serves as a signal for the recruitment of HATs and the transcription machinery. Shown are the mean ± SDM of 3 independent experiments.
Discussion
Although many studies have established links between histone modifications and gene activity,1 the molecular mechanisms by which arginine methylation directs transcription events are unknown. Here, we demonstrated how asymmetric dimethylation of H4R3 by PRMT1 activates β-globin transcription through trans stimulation of H3 acetylation on Lys9 and Lys14. We showed that the inhibition of dimethyl H4R3 disrupts the intrachromosomal interaction between the distal HS2 enhancer and the β-globin promoter, as well as the efficient formation of transcription complexes at the promoter of the transcribed globin gene.
The communication between genes and cis regulatory elements by intrachromosomal or interchromosomal folding has been demonstrated for several other gene loci and is recognized as an important mechanism for gene activation over long distances.25-28 It is still unclear how an active chromatin configuration is established and maintained. Several lines of evidence support the notion that promoter-localized hyperacetylated histone H3 and H4 are required for transcriptional activation of metazoan genes.37 During erythropoiesis, the β-globin locus adopts a nonuniform pattern of hyperacetylated H3 and H4 being enriched at the LCR and at the transcribed β-globin gene.38,39 Both the LCR and the βmaj-promoter play important roles in generating an active chromatin territory (Hub) for maximal activity.33,34,40 The LCR can serve as a primary site to recruit transcription factors and chromatin modifying or remodeling factors and to stably alter topology of the β-globin locus in erythrocytes.41 However, deletion of HS2 led to only a 30% decrease in β-globin transcription.41,42 In contrast, KD of PRMT1 has a more drastic effect, more similar to deletion of the entire LCR. The effect is likely because PRMT1 is recruited not only to the LCR but also to the βmaj-promoter, and possibly to other genes as well. It is interesting to note, however, that similar to loss of the whole LCR, loss of dimethyl H4R3 leads to a decrease in H3 and H4 acetylation as well as subsequent recruitment of the transcription preinitiation complex at HS2 and at the βmaj-promoter. This suggests that loss of dimethyl H4R3 might affect total LCR function and lead to subsequent disruption of long-range looping in the globin locus (Figures 4–5). Together, the data provide a potential mechanism linking active histone modifications and long-range chromatin loop formation in the globin gene locus.
Individual histone marks may be read by a variety of effectors, each in principle associated with a different effect on function. For example, H3K4 dimethylation or trimethylation is recognized by several effectors including adenosine triphosphate–dependent chromatin remodeling factors CHD1 and NURF,43,44 as well as histone covalent modifying enzyme JMJD2A.45 Dimethylation of Arg3 at H4 tails, but not PRMT1 itself, can directly communicate with p300 and PCAF to increase acetylation on H3K9/K14 (Figure 8E). Intriguingly, although USF1 and PRMT1 are present in the same protein complex, both of them can individually associate with HATs, p300, and PCAF.15,46 It is conceivable that HATs are initially recruited by USF1 or via its association with PRMT1. Subsequently, H4R3 dimethylation of neighboring nucleosomes by PRMT1 may further stabilize or recruit additional HATs to bind to chromatin templates. It is possible that the arginine methylation generated by PRMT1, which is recruited to chromatin by USF1 or other transcription factors,7,9,10,15 functions as a signal to facilitate an extended family of other modifications that initiate a cascade of histone modifications.
In vitro (Figure 6) and in vivo (Figure 5) evidence suggests that the recruitment of HATs, p300/CBP, and PCAF is dependent on the asymmetric dimethylation of H4R3 catalyzed by PRMT1 at LCR element HS2 and at the βmaj promoter. How does histone acetylation correlate with LCR function in activating transcription of globin genes? It is known that enhancer targeted HATs (p300/CBP) are able to interact with TBP and other components of TFIID,47 thereby bridging transcription factors with the Pol II holoenzyme to regulate assembly of a tissue- or cell type–specific transcription complex at the promoter. Furthermore, acetylation of H3K9/K14 was shown to augment the binding of TFIID (via TAF1) to histone H3 tails at transcriptionally active promoters.36 H4 acetylation on Lys5 and Lys12 augments the binding of TAF1, a major component of the TFIID complex.48 It is thus possible that specific histone acetylation patterns target TFIID to specific chromatin structures at the promoters to mediate transcription. Finally, a thymocyte-specific enhancer has been shown to direct the recruitment of CBP and Pol II during promoter activation in the TCRβ locus.49 These results strongly suggest that transcription factor–mediated histone acetylations direct interactions between enhancer and promoter. In addition, a looping configuration may result from a transcriptional machinery shared by the LCR and promoters. Histone acetylation renders chromatin more accessible so that transcription factors and Pol II components can anchor to these sites in the nucleus and this, in turn, could stabilize loop formation. In this way, LCRs are capable of activating target genes over substantial distances and of establishing autonomously regulated chromatin domains.17,18
It was predicted that cross-regulatory interactions among individual epigenetic marks influence transcription efficiencies.1 This idea is supported by several recent observations. Studies from yeast and mammalian cells demonstrated that asymmetric dimethylation of histone H3 arginine 2 mediated by PRMT6 correlates with repressive heterochromatin and abrogates the trimethylation of H3K4 by the SET1/MLL complexes.5 In contrast, our data demonstrate that asymmetric dimethylation of H4R3 facilitates the recruitment of HATs and subsequent histone acetylation. Whereas asymmetric dimethylation of H3R2 inhibits TFIID binding to trimethyl H3K4,36 asymmetric dimethylation of Arg3 at H4 tails mediates efficient recruitment of the transcription preinitiation complex to the active promoter (Figure 7). It is particularly interesting that PRMT5, an enzyme that symmetrically methylates exactly the same H4R3 residue and subsequently induces DNA methylation, silences the globin locus by establishing repressive marks at the LCR and globin gene promoter.50 Both PRMT1 and PRMT5 can modify the same arginine residue in different ways and exert the exact opposite effects on the transcription of globin genes, suggesting that arginine methylation plays a critical regulatory role in chromatin structure and gene regulation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to members of the Huang laboratory for their suggestions and comments. We thank Eric So (The Institute of Cancer Research) for reagents.
This work was supported by grants from the National Institutes of Health (S.H., HL090589, HL091929, and HL091929-01A1S1-the ARRA Administrative Supplement; J.B., DK52356) and by grants from the Bankhead-Coley Cancer Research (Y.Q. and S.H.). G.F. is supported by the Intramural Research program, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
National Institutes of Health
Authorship
Contribution: X.L. designed research, performed experiments, and analyzed results; X.H., B.P., and R.Y. performed experiments; Z.Z. and S.L. contributed vital reagents; Y.Q. and J.B. designed research and analyzed results; G.F. designed research; and S.H. designed research, analyzed results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Suming Huang, Department of Biochemistry and Molecular Biology, University of Florida College of Medicine, PO Box 103633, Gainesville, FL 32610; e-mail: sumingh@ufl.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal