Abstract
Insulin-like growth factor-1 (IGF-1) is an important growth and survival factor in multiple myeloma (MM). Here, we demonstrate that IGF-1 induces significant down-regulation of the proapoptotic BH3-only protein Bim in MM cells. Reduced Bim levels by RNA interference (RNAi) protected cells from drug-induced cell death. The IGF-1–mediated down-regulation of Bim was the result of (1) reduced transcription by activation of the Akt pathway and inactivation of the transcription factor FoxO3a, (2) increased proteasome-mediated degradation of the Bim extra-long protein by activation of the mitogen-activated protein kinase pathway, and (3) epigenetic regulation of both the Bim and the FoxO3a promoter. Treatment of cells with the histone deacetylase inhibitor LBH589 resulted in a clear up-regulation in the expression of Bim. Furthermore, the methylation inhibitor 5-aza-2′deoxycytidine (decitabine) significantly increased the effects of LBH589. On IGF-1 treatment, the Bim promoter region was found to be unmethylated, whereas chromatin immunoprecipitation analysis of the IGF-1–treated cells showed both a reduced histone H3 tail Lys9 (H3K9) acetylation and an increased H3K9 dimethylation, which contributed actively to its silencing. These data identify a new mechanism in the IGF-1–dependent survival of MM cells and emphasize the need for IGF-1–targeted drug therapy.
Introduction
Multiple myeloma (MM) is a lethal plasma cell malignancy hallmarked by uncontrolled accumulation of monoclonal plasma cells in the bone marrow (BM).1 There, the MM cells receive signals to survive and proliferate because of the existence of functional, mutual interactions through growth factors and adhesion molecules.2 Importantly, these interactions also confer resistance to conventional therapies.2
One of the most important growth factors involved in MM progression is insulin-like growth factor-1 (IGF-1).3–5 This cytokine induces proliferation of both interleukin-6 (IL-6)–independent and -dependent cell lines, acts synergistically with IL-6, and protects MM cells from dexamethasone-induced apoptosis.3 Moreover, IGF-1 also stimulates homing and production of angiogenic factors.4,6,7 Binding to its receptor results in the activation of the Ras/Raf/MAPK (mitogen-activated protein kinase)/Erk (mainly triggering proliferation and secretion of vascular endothelial growth factor) and the PI-3K (phosphatidylinositol-3 kinase)/Akt pathways (involved mainly in antiapoptotic stimuli and mediating migration).6,8 Both IGF-1 serum levels and IGF-1 receptor (IGF-1R) expression by the MM cells are important negative prognostic factors in MM.9,10 The central role of IGF-1 in the MM pathophysiology is further supported by the encouraging preclinical results observed after targeting the IGF-1R with neutralizing antibodies or small molecule inhibitors. Previously, we demonstrated the efficacy of the IGF-1R tyrosine kinase inhibitor picropodophyllin to block the function of IGF-1R in vitro and in vivo and this both in murine and human MM cells.11,12 The anti-MM activity of selective IGF-1R inhibitors was also confirmed with the use of the selective kinase inhibitor NVP-ADW742 in an orthotopic xenograft MM model.4 Clinical trials examining the effect of both blocking antibodies and small molecule inhibitors are currently ongoing.3
Bim (Bcl2like11) is a member of the BH3-only group of the Bcl-2 protein family and is a mediator of apoptosis.13,14 Bim is a sensor of cell stress (withdrawal or inhibition of growth factor signaling or treatment with numerous cytotoxic agents)13,15 and promotes apoptosis either indirectly by antagonizing the inhibitory function of the Bcl-2/Bcl-Xl members and thus liberating the innate cytotoxic Bax-like members or directly by activating the Bax-like members.13 Three major isoforms of Bim are known: extra-long (BimEL), long (BimL), and short (BimS). All 3 isoforms induce apoptosis; the shortest being the most potent but the longest the most preponderant.16 The proapoptotic activity of Bim seems to be controlled in various ways. In healthy cells, Bim is sequestered in its inactive form to the microtubular cytoskeleton or exists as inactive heterodimers (with antiapoptotic Bcl-2 family members) sequestered to the mitochondria.17 In B and MM cells, Bim appears to be constitutively associated with Mcl-1. At induction of apoptosis, Bim is released from Mcl-1, thus activating its proapoptotic function.13,17,18 Another major regulatory mechanism of Bim-dependent apoptosis is driven by phosphorylation.19 Bim transcription is down-regulated on phosphorylation of Akt and the downstream transcription factor FoxO3a.20 Furthermore, Bim expression is regulated on the posttranslational level through phosphorylation of Erk, leading to proteasomal degradation of the BimEL isoform.21 More recently, Bim expression was also found to be epigenetically regulated. Indeed, histone deacetylase inhibitors (HDACi) are known to induce Bim expression in various tumors, including MM.22,23 Moreover, silencing of the Bim promoter by hypermethylation has been shown in B-cell lymphomas.24
In this study, we used gene expression profiling to determine the effect of IGF-1 treatment on the gene expression of MM cells. We demonstrated that Bim is down-regulated after IGF-1 treatment. In MM, Bim expression was shown to be down-regulated by IL-6 and adhesion to fibronectin.18,25 However, the mechanisms behind this altered expression have not yet been fully elucidated. This prompted us to investigate the mechanism involved. Finally, the relevance of the IGF-1–mediated Bim down-regulation in terms of MM progression was shown.
Methods
Mice
C57Bl/KaLwRij mice were purchased from Harlan CPB. Mice were housed and treated following the conditions approved by the Ethical Committee for Animal Experiments, Vrije Universiteit Brussel (license no. LA1230281).
Cells
The 5T33MMvv cells originated from elderly C57Bl/KaLwRij mice that spontaneously developed MM. The cells have since been propagated into syngeneic, young mice by transplantation of diseased BM cells. The clinical and molecular characteristics of these models are very similar to those of the human disease, making them suitable models to study MM progression.26 On showing clear signs of morbidity, mice were killed, and isolation of the MM cells was performed as previously described.27 Cells obtained from end-stage mice were 95% pure. These cells should be considered as primary cells because they only survive for a short term in vitro.
The human myeloma cell lines Karpas707,28 OPM-2,29 and LP-130 were cultured and maintained in RPMI-1640 medium (BioWhittaker) supplemented with 10% fetal calf serum (Hyclone), 100 U/mL penicillin/streptomycin, and 2mM l-glutamine (supplements from BioWhittaker). The Karpas707 cell line harbors a bcr-abl translocation, the LP-1 cell line a (4;14) translocation and the OPM-2 cell line a (4;14), (8;14), and (?;20) translocation.31 The myeloma origin of the Karpas707 cell line was recently confirmed by performing comprehensive protein expression profiling in 48 human tissues and 45 human cell lines.32
The HEK293T cells were cultured in DMEM medium (BioWhittaker) with the same supplements as mentioned before.
Oligonucleotide microarrays
The mRNA expression profile of 5T33MMvv cells (10 × 106) treated or not with IGF-1 (100 ng/mL; Novozymes GroPep) for 12 hours was analyzed with the use of mouse 430A microarrays from Affymetrix. Total RNA was extracted with the use of the RNeasy mini kit (QIAGEN) as described by the manufacturer. Next, total RNA was used to prepare biotinylated cRNA according to the standard Affymetrix protocol. Briefly, total RNA was reverse transcribed with the use of the SuperScript Choice System (Invitrogen) by oligo-dT primers containing a T7 RNA polymerase promotor site. Complementary DNA was in vitro transcribed and labeled with the use of the RNA transcript labeling kit (Enzo). The concentration of labeled cRNA was measured with the use of the NanoDrop ND-1000 spectrophotometer. Labeled cRNA was fragmented in a fragmentation buffer during 35 minutes at 94°C. The quality of labeled and fragmented cRNA was analyzed with the use of the Bioanalyzer 2100 (Agilent). Fragmented cRNA was hybridized to the array during 16 hours at 45°C. The arrays were washed and stained in a fluidics station (Affymetrix) and scanned with the use of the Affymetrix 3000 GeneScanner. Raw data were analyzed with the use of the Affymetrix GCOS software. Raw data of the experiments can be found at the GEO Datasets website (accession no. GSE17140).
Real-time reverse transcription polymerase chain reaction analysis
Cells (5 × 106) were lysed in 250 μL of TRIzol and maintained at −80°C. Total RNA was extracted with the use of the RNeasy mini kit (QIAGEN) as described by the manufacturer. The concentration and purity of the RNA were determined by spectrophotometric measurement (Gene Quant II; Pharmacia Biotech). Total RNA (2 μg) was converted into cDNA by the Superscript first-strand synthesis system (Invitrogen) with the use of random hexamers as primers. Real-time reverse transcription polymerase chain reaction (RT-PCR) analysis was performed with the use of the ABI PRISM 7700 Sequence Detector. For the detection of the target gene, Assays on Demand (Applied Biosystems) were used as described by the manufacturer. The primers are designed in such a way that they detect the mRNA sequence shared by the 3 isoforms. To standardize the amount of sample RNA, we amplified an endogenous reference gene, β-glucuronidase with the use of an Assay on Demand. The reporter and quencher of the probe are, respectively, FAM and a nonfluorescent quencher. TaqMan RT-PCR was performed in a 50-μL reaction mix containing 25 μL of TaqMan Universal PCR master mix (Applied Biosystems); 2.5 μL of Assays on Demand (primers and probe), and 50 ng of cDNA or RNA. Samples were amplified by 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. Each sample was amplified in triplicate. The relative standard curve method was used to quantify the relative target gene expression.
Western blot
Cell pellets were lysed in lysis buffer containing 50mM Tris, 150mM NaCl, 1% Nonidet P40, and 0.25% sodium deoxycholate. The following protease and phosphatase inhibitors were added: 4mM Na3VO4, 1mM Na4P2O7, 2 μg/mL aprotinin, 50 μg/mL leupeptin, 500 μg/mL trypsin inhibitor, 10μM benzamidine, 2.5mM pnp benzoate (all from Sigma-Aldrich), 50mM NaF, 5mM ethylenediaminetetraacetic acid (both from VWR International), 1mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, and 50 μg/mL pepstatin A (both from ICN). The cell debris was removed by centrifugation (5 minutes, 13 000g), and sample buffer was added. After boiling, the samples were separated on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene diflouride membranes (Bio-Rad). The membranes were blocked with phosphate-buffered saline containing 5% low-fat milk and 0.1% Tween 20. Antibodies against p-Thr202/Tyr204 ERK (E10; no. 9106), p-Thr308 Akt (244F9; no. 4056), total ERK (no. 9102), total Akt (no. 9272), Bim (no. 2819), actin (no. 4967), p-Ser253 FoxO3a (no. 9466), total FoxO3a (no. 9467), acetyl-histone H3 (no. 06-599), and acetyl-histone H4 (no. 06-866) were used. Most antibodies were obtained from Cell Signaling Technology. Acetyl-histone H3 and acetyl-histone H4 were purchased from Upstate Biotechnology.
Treatment of the cells
Cells were treated, respectively, with the demethylation agent decitabine (5-aza-2′deoxycytidine, 500nM; Sigma-Aldrich) for 72 hours, the HDACi LBH589 (15nM; Novartis) for 24 hours, and a combination of both inhibitors. Decitabine (DAC) is a cytosine analog with a 5′ modification, whereas LBH589 is a cinnamic hydroxamic acid. Cells were replenished with fresh complete growth medium containing DAC after 48 hours and for the combination therapy LBH589 was added the last 24 hours. For these experiments, 5T33MMvv cells were excluded because they only survive 24 hours in vitro.
For Western blot analysis, 5T33MM and Karpas707 cells were medium starved, respectively, for 1 hour or overnight and pretreated or not with the indicated concentrations of LY294002 or MG132 (both from Sigma-Aldrich) for 1 hour. Cultures either were stimulated with 100 ng/mL IGF-1 for the indicated times in serum-free medium or were not stimulated.
For IGF-1 experiments studying the epigenetic regulation of the Bim and FoxO3a promoter, cells were treated 4 days in serum-free medium. Cells were replenished with fresh medium containing IGF-1 every 24 hours.
Methylation pattern of the Bim gene promoter region
Genomic DNA was isolated with the use of the QIAamp DNA Mini Kit (QIAGEN). Bisulfite treatment, PCR amplification, and pyrosequencing were done by Varionostic GmbH. From the 785-base pair (bp) fragment of the human Bim promoter published earlier,33 the 304-bp region immediately upstream of the Bim transcription start was analyzed identifying 32 CpG sites.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) analysis was conducted as described by the SimpleChIP Enzymatic Chromatin IP Kit (magnetic beads) from Cell Signaling Technology. The chromatin was immunoprecipitated with normal immunoglobulin G (IgG; negative control; no. 2729), anti–acetyl-histone H3 (Lys9) (C5B11; no. 9649), anti–trimethyl-histone H3 (Lys4) (C42D8; no. 9751), anti–dimethyl-histone H3 (Lys9; no. 9753), and anti–histone H3 (positive control; no. 2650) antibodies. All antibodies were obtained from Cell Signaling Technology. Two percent of the supernatant fraction from the chromatin lacking primary antibody was saved as the “input sample.” Real-time RT-PCRs were performed with the use of the Maxima SYBR Green qPCR Master Mix (Fermentas) with specific designed primers amplifying promoter regions of the selected genes. Primers used: Bim sense 5′-TAGGTGAGCGGGAGGCTAGGGTACA-3′, Bim antisense 5′-GTGCAGGCTCGGACAGGTAAAGGC-3′, FoxO3A sense 5′-CCTTCTCGCCCGCTAGTGTTTTTA-3′, and FoxO3A antisense 5′-GGGAGCGAGTCGGAACATAAATCT-3′. Primers were ordered at Integrated DNA Technologies.
Lentiviral construction, production, and transduction
The pSuper, pLVH-CMV-eGFP, and pLVH-CMV-eGFP-Scrambled constructs were kind gifts from Stefan Bonné (Beta Cell Neogenesis [BENE], Vrije Universiteit Brussel [VUB], Brussels, Belgium). To generate shRNA sequences against human Bim, oligos with BamHI/HindIII overhangs were designed and ordered at Integrated DNA Technologies. The selected target sequence for human Bim was 5′-AAAGCAACCTTCTGATGTA-3′.34 The oligos were annealed and cloned into the pSuper. The H1 promoter together with the short hairpin cassette were excised by EcoRI/ClaI digestion and cloned into the pLVH-CMV-eGFP to yield pLVH-CMV-eGFP-shBim. Lentiviral vector particles were produced in 293T cells by transient cotransfection of the transfer, envelope (pMD.G), and packaging (pCMVΔR8.9) plasmid as previously described.35 The vector stock was collected 48 and 72 hours after transfection and concentrated by ultracentrifugation as described.35 The viral titer was determined by infection of 293T cells with serial dilutions of the vector stock. Seventy-two hours after infection, the number of eGFP-positive cells was determined by fluorescence-activated cell sorting analysis.
For every transduction, 5 × 105 cells were transduced. Two weeks after transduction, cells were eGFP-sorted (Becton Dickinson FACSVantage that used CellQuest Software). Cells were allowed to adapt overnight before identifying specific knockdown and performing functional studies.
Cell viability and apoptosis
Cell viability and apoptosis were measured in the presence or absence of drug treatment with the use of, respectively, the CellTiter-Glo and Caspase-Glo 3/7 assay from Promega according to the manufacturer's instructions. Karpas707 cells were seeded in 96-well optical plates at a density of 30 000 cells (CellTiter-Glo assay) or 10 000 cells (Caspase-Glo 3/7 assay). A preliminary series of experiments was conducted to determine the IC50 for bortezomib (Velcade; Janssen Pharmaceutica NV, Johnson & Johnson Pharmaceutical Research & Development), LBH589, and melphalan (Sigma-Aldrich) with the use of the CellTiter-Glo assay (data not shown). Cells were serum starved for 24 hours followed by treatment with LBH589 (15nM), bortezomib (10nM), or melphalan (15μM) for 24 and 48 hours. Each experimental condition was performed in triplicate.
Statistical analysis
For statistical analysis of the in vitro tests the Mann-Whitney U test was used. For the microarrays selection criteria were as follows: (1) present either in all of the control samples and/or in all of the experimental samples, (2) autofilter in all columns is equal on increased or decreased, (3) statistical analysis by a 2-tailed, unpaired t test, and (4) performance of a Bonferroni correction, taking P less than .05 divided by the number of genes obtained after the 3 above-mentioned criteria as threshold for significance.
Results
IGF-1 down-regulates Bim expression in the murine 5T33MMvv myeloma cells
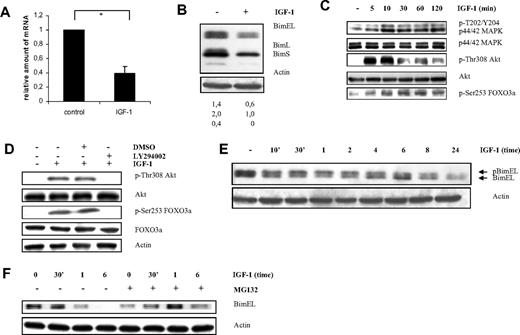
Using gene expression profiling we ascertained the effect of IGF-I treatment on the gene expression of the murine 5T33MMvv cells (supplemental Microarray Data 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Statistical analysis of the microarray data resulted in 79 consistently down-regulated and 80 up-regulated genes (supplemental Microarray Data 2). A list of the 10 most significant up- and down-regulated genes is shown in Table 1. One of the genes significantly and consistently down-regulated was the proapoptotic Bim gene. The murine 5T33MMvv cells showed a down-regulation of 55% on microarray analysis on treatment with IGF-1 for 12 hours (Figure 1A). This down-regulation was confirmed by quantitative RT-PCR analysis that showed an inhibition of 63% at mRNA level for the BimEL, BimL, and BimS isoforms (data not shown). Consistent with the mRNA expression, Western blot analysis showed that protein levels were also down-regulated after 24 hours of IGF-1 treatment (Figure 1B).
The 10 most significant up- and down-regulated genes after treatment with IGF-1 for 12 hours
| Gene name . | Abbreviation . | Process . |
|---|---|---|
| Genes significantly up-regulated | ||
| Cystathionase (cystathionine gamma-lyase) | Cth | Amino acid/nucleotide synthesis |
| Dynactin 5 | Dctn5 | Transport |
| Glycogen synthase 3, brain | Gys3 | Glucose uptake/ metabolism |
| Squalene epoxidase | Sqle | Lipogenesis |
| Tspy-like 4 | Tspyl4 | Chromatin assembly |
| 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1 | Hmgcs1 | Lipogenesis |
| Low-density lipoprotein receptor | Ldlr | Lipogenesis |
| Solute carrier family 2 (facilitated glucose transporter), member 1 | Slc2a1 | Nutrient uptake |
| Stanniocalcin 2 | Stc2 | Signaling |
| Translocase of inner mitochondrial membrane 8 homolog a (yeast) | Timm8a | Glucose uptake/metabolism |
| Genes significantly down-regulated | ||
| Yippee-like3 | Ypel3 | Apoptosis |
| Transcription factor 2 | Tcf2 | Transcription/ RNA processing |
| Hypothetical protein R07G3.7a | 2310001H12Rik | Unknown |
| F-box only protein 32 | Fbxo32 | Protein modification |
| v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | Erbb3 | Apoptosis |
| Expressed sequence AI661017 | AI661017 | Unknown |
| cDNA sequence BC024760 | BC024760 | Unknown |
| Kelch-like24 | klhl24 | Signaling |
| Interleukin 16 | Il16 | Signaling |
| BCL2-like 11 (apoptosis facilitator) | Bim | Apoptosis |
| Gene name . | Abbreviation . | Process . |
|---|---|---|
| Genes significantly up-regulated | ||
| Cystathionase (cystathionine gamma-lyase) | Cth | Amino acid/nucleotide synthesis |
| Dynactin 5 | Dctn5 | Transport |
| Glycogen synthase 3, brain | Gys3 | Glucose uptake/ metabolism |
| Squalene epoxidase | Sqle | Lipogenesis |
| Tspy-like 4 | Tspyl4 | Chromatin assembly |
| 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1 | Hmgcs1 | Lipogenesis |
| Low-density lipoprotein receptor | Ldlr | Lipogenesis |
| Solute carrier family 2 (facilitated glucose transporter), member 1 | Slc2a1 | Nutrient uptake |
| Stanniocalcin 2 | Stc2 | Signaling |
| Translocase of inner mitochondrial membrane 8 homolog a (yeast) | Timm8a | Glucose uptake/metabolism |
| Genes significantly down-regulated | ||
| Yippee-like3 | Ypel3 | Apoptosis |
| Transcription factor 2 | Tcf2 | Transcription/ RNA processing |
| Hypothetical protein R07G3.7a | 2310001H12Rik | Unknown |
| F-box only protein 32 | Fbxo32 | Protein modification |
| v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | Erbb3 | Apoptosis |
| Expressed sequence AI661017 | AI661017 | Unknown |
| cDNA sequence BC024760 | BC024760 | Unknown |
| Kelch-like24 | klhl24 | Signaling |
| Interleukin 16 | Il16 | Signaling |
| BCL2-like 11 (apoptosis facilitator) | Bim | Apoptosis |
IGF-1 down-regulates Bim expression in the murine 5T33MMvv cells through the PI-3K and p42/44 MAPK pathways. (A) Microarray analysis of the murine 5T33MMvv cells showing relative down-regulation of Bim mRNA expression after treatment with IGF-1 for 12 hours. The mean value ± SD of 3 independent experiments is given (*P < .001). The Bim gene is represented on the mouse 430A microarrays from the Affymetrix chip by 5 different probe sets. (B) Western blot analysis showing down-regulation of Bim at protein level in the 5T33MMvv cells. Cells deprived of serum for 1 hour were treated with IGF-1 for 24 hours. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. The numbers below refer to the optical density of the BimEL, BimL, and BimS bands after treatment with IGF-1 as measured with the NIH1.62 image program. The variation between experiments was below 10%. (C-D) IGF-1 inhibits Bim transcription through the PI3-K pathway in the 5T33MMvv cells. (C) Western blot analysis showing activation of the PI-3K and p42/44 MAPK pathway after IGF-1 stimulation. Cells were starved 1 hour before IGF-1 treatment for the indicated time periods. The expression of p-ERK1/2, tot-ERK1/2, p-Akt, tot-Akt, and pFoxO3a were analyzed by Western blot. (D) Cells were starved 1 hour, treated with the PI-3K inhibitor LY294002 (1 hour of preincubation; 10μM) or not and stimulated or not with IGF-1 for 10 minutes, and the levels of p-Akt, tot-Akt, p-FoxO3a, and tot-FoxO3a were analyzed by Western blot. To confirm equal loading, levels of actin were determined. (E-F) IGF-1 induces proteasomal degradation of the BimEL isoform by the p42/44 MAPK pathway. (E) Cells were deprived of serum for 1 hour before stimulation for the indicated time periods, and the levels of phosphorylation and expression of BimEL were analyzed by Western blot. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. (F) Cells were starved for 1 hour, pretreated or not for 1 hour with the specific proteasome inhibitor MG132 (1μM) and treated with IGF-1 for the indicated time periods. Next, the levels of Bim were analyzed by Western blot. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. For all Western blot analyses, 1 experiment representative of 3 is shown.
IGF-1 down-regulates Bim expression in the murine 5T33MMvv cells through the PI-3K and p42/44 MAPK pathways. (A) Microarray analysis of the murine 5T33MMvv cells showing relative down-regulation of Bim mRNA expression after treatment with IGF-1 for 12 hours. The mean value ± SD of 3 independent experiments is given (*P < .001). The Bim gene is represented on the mouse 430A microarrays from the Affymetrix chip by 5 different probe sets. (B) Western blot analysis showing down-regulation of Bim at protein level in the 5T33MMvv cells. Cells deprived of serum for 1 hour were treated with IGF-1 for 24 hours. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. The numbers below refer to the optical density of the BimEL, BimL, and BimS bands after treatment with IGF-1 as measured with the NIH1.62 image program. The variation between experiments was below 10%. (C-D) IGF-1 inhibits Bim transcription through the PI3-K pathway in the 5T33MMvv cells. (C) Western blot analysis showing activation of the PI-3K and p42/44 MAPK pathway after IGF-1 stimulation. Cells were starved 1 hour before IGF-1 treatment for the indicated time periods. The expression of p-ERK1/2, tot-ERK1/2, p-Akt, tot-Akt, and pFoxO3a were analyzed by Western blot. (D) Cells were starved 1 hour, treated with the PI-3K inhibitor LY294002 (1 hour of preincubation; 10μM) or not and stimulated or not with IGF-1 for 10 minutes, and the levels of p-Akt, tot-Akt, p-FoxO3a, and tot-FoxO3a were analyzed by Western blot. To confirm equal loading, levels of actin were determined. (E-F) IGF-1 induces proteasomal degradation of the BimEL isoform by the p42/44 MAPK pathway. (E) Cells were deprived of serum for 1 hour before stimulation for the indicated time periods, and the levels of phosphorylation and expression of BimEL were analyzed by Western blot. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. (F) Cells were starved for 1 hour, pretreated or not for 1 hour with the specific proteasome inhibitor MG132 (1μM) and treated with IGF-1 for the indicated time periods. Next, the levels of Bim were analyzed by Western blot. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. For all Western blot analyses, 1 experiment representative of 3 is shown.
Involvement of the PI-3K and MAPK pathways in the IGF-1–mediated down-regulation of Bim expression in the 5T33MMvv cells
Next, we tried to identify the signaling pathways that regulate the Bim expression in the 5T33MMvv cells. Phosphorylation of Akt and the downstream transcriptions factor FoxO3a have been reported to repress transcription of Bim.20 Moreover, activation of the p44/p42 MAPK pathway has been proven to phosphorylate the BimEL isoform, leading to its proteasomal degradation.21 As shown in Figure 1C and reported earlier, IGF-1 activates the MAPK and PI-3K kinase pathways in the 5T33MMvv myeloma cells.6 Here, we demonstrated phosphorylation of FoxO3a on treatment with IGF-1 (Figure 1C), and pretreatment with the PI-3K inhibitor LY294002 was found to block this IGF-1–induced phosphorylation of Akt and FoxO3a (Figure 1D). Furthermore, a mobility shift of BimEL because of phosphorylation (pBimEL) could be observed 30 minutes after administration of IGF-1 (Figure 1E). In cells preincubated with the proteasome inhibitor MG132, the degradation of pBimEL was delayed, resulting in accumulation of Bim (Figure 1F). However, already after 6 hours of treatment, MG132 induced cell death in these primary cells as evidenced by a 7-amino actinomycin D (7′AAD)/annexin V staining and the Cell Titer Glo Luminescent assay (data not shown). Accumulation of Bim preceding apoptosis has been shown extensively both for MG132 and bortezomib.15 Nevertheless, our results indicate that cell phosphorylation of BimEL by p44/p42 MAPK targets it for proteasomal degradation in the 5T33MMvv.
IGF-1 down-regulates Bim expression in human myeloma cells through the PI-3K and MAPK pathways
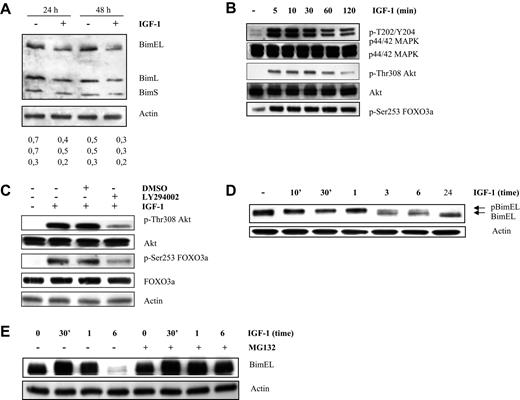
To confirm our data, 3 human myeloma cell lines were tested: Karpas707, OPM-2, and LP-1. The first 2 lines showed strong expression of Bim and expression was clearly down-regulated after 24 hours of IGF-1 treatment (Figures 2A, 3A). In contrast, the LP-1 cells were negative for Bim (data not shown). Next, the PI-3K and MAPK pathways were studied in the Karpas707 and OPM-2 cell lines. For both human cells lines, we demonstrated phosphorylation of Akt and FoxO3a on treatment with IGF-1 (Figures 2B, 3B) and pretreatment with LY294002 blocked this effect (Figures 2C, 3B). Thus, the results indicate that in MM cells the PI-3K pathway regulates Bim expression. In response to IGF-1 stimulation, p44/p42 MAPK was also found to be phosphorylated after 5 minutes in the Karpas707 (Figure 2B) cells. In contrast, phosphorylation of p44/p42 MAPK could be observed in the OPM-2 cells after 6 hours of stimulation (Figure 3B-C). Consequently, a clear shift in mobility of BimEL could only be observed in the Karpas707 cells (Figure 2D). Furthermore, preincubation with MG132 abolished the degradation of pBimEL up to 6 hours upon treatment with IGF-1 in the Karpas707 cells (Figure 2E). In accordance with the murine primary cells, MG132-induced cell death was again observed as evidenced by a 7′AAD/AnnexinV staining and Cell Titer Glo Luminescent assay (data not shown). Although no clear mobility shift of BimEL was shown for the OPM-2 cells (Figure 3D), we did find some accumulation of BimEL in cells pretreated with MG132 in the 6-hour stimulation point (Figure 3E). These results can be explained by the delay in phosphorylation of p44/p42 MAPK. Taken together, our results indicate that phosphorylation of BimEL by p44/p42 MAPK results in proteasomal degradation in MM cells.
IGF-1 down-regulates Bim expression in the human Karpas707 cells through the PI-3K and p42/44 MAPK pathways. (A) Western blot analysis showing down-regulation of Bim at protein level in the Karpas707 cells. Cells deprived of serum for 24 hours were treated with IGF-1 for 24 and 48 hours. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. The numbers below refer to the optical density of the BimEL, BimL, and BimS bands after treatment with IGF-1 as measured with the NIH1.62 image program. Variation between experiments was below 10%. (B-C) IGF-1 inhibits Bim transcription through the PI3-K pathway in the Karpas707 cells. (B) Western blot analysis showing activation of the PI-3K and p42/44 MAPK pathways after IGF-1 stimulation. Cells were starved 24 hours before IGF-1 treatment for the indicated time periods. The expression of p-ERK1/2, tot-ERK1/2, p-Akt, tot-Akt, and pFoxO3a were analyzed by Western blot. (C) Cells were starved 24 hours, pretreated with LY294002 (1 hour of preincubation; 10μM) and stimulated with IGF-1 for 10 minutes. Next, the levels of p-Akt, tot-Akt, p-FoxO3a, and tot-FoxO3a were analyzed by Western blot. To confirm equal loading levels of actin were determined. (D-E) IGF-1 induces proteasomal degradation of the BimEL isoform by the p42/44 MAPK pathway. (D) Cells were deprived of serum for 24 hours before stimulation for the indicated time periods, and the levels of phosphorylation and expression of BimEL were analyzed by Western blot. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. (E) Cells were starved for 24 hours, pretreated for 1 hour with MG132 (1μM), and treated with IGF-1 for the indicated time periods. Next, the levels of Bim were analyzed by Western blot. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. For all Western blot analyses, 1 experiment representative of 3 is shown.
IGF-1 down-regulates Bim expression in the human Karpas707 cells through the PI-3K and p42/44 MAPK pathways. (A) Western blot analysis showing down-regulation of Bim at protein level in the Karpas707 cells. Cells deprived of serum for 24 hours were treated with IGF-1 for 24 and 48 hours. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. The numbers below refer to the optical density of the BimEL, BimL, and BimS bands after treatment with IGF-1 as measured with the NIH1.62 image program. Variation between experiments was below 10%. (B-C) IGF-1 inhibits Bim transcription through the PI3-K pathway in the Karpas707 cells. (B) Western blot analysis showing activation of the PI-3K and p42/44 MAPK pathways after IGF-1 stimulation. Cells were starved 24 hours before IGF-1 treatment for the indicated time periods. The expression of p-ERK1/2, tot-ERK1/2, p-Akt, tot-Akt, and pFoxO3a were analyzed by Western blot. (C) Cells were starved 24 hours, pretreated with LY294002 (1 hour of preincubation; 10μM) and stimulated with IGF-1 for 10 minutes. Next, the levels of p-Akt, tot-Akt, p-FoxO3a, and tot-FoxO3a were analyzed by Western blot. To confirm equal loading levels of actin were determined. (D-E) IGF-1 induces proteasomal degradation of the BimEL isoform by the p42/44 MAPK pathway. (D) Cells were deprived of serum for 24 hours before stimulation for the indicated time periods, and the levels of phosphorylation and expression of BimEL were analyzed by Western blot. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. (E) Cells were starved for 24 hours, pretreated for 1 hour with MG132 (1μM), and treated with IGF-1 for the indicated time periods. Next, the levels of Bim were analyzed by Western blot. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. For all Western blot analyses, 1 experiment representative of 3 is shown.
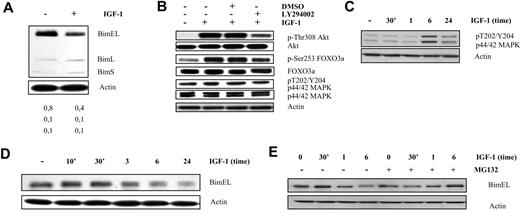
IGF-1 down-regulates Bim expression in the human OPM-2 cells through the p42/44 MAPK and PI-3K pathways. (A) Western blot analysis showing down-regulation of Bim at the protein level in the OPM-2 cells. Cells deprived of serum for 24 hours were treated with IGF-1 for 24 hours. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. The numbers below refer to the optical density of the BimEL, BimL, and BimS bands after treatment with IGF-1 as measured with the NIH1.62 image program. Variation between experiments was below 10%. (B) IGF-1 inhibits Bim transcription through the PI3-K pathway in the OPM-2 cells. Cells were starved 24 hours, pretreated with LY294002 (1 hour of preincubation; 10μM), and stimulated with IGF-1 for 10 minutes. Next, the levels of p-Akt, tot-Akt, p-FoxO3a, tot-FoxO3a, p-ERK1/2, tot-ERK1/2, and actin were analyzed by Western blot. (C) Activation of the p42/44 MAPK pathway on IGF-1 treatment is delayed in the OPM-2 cells. Cells were deprived of serum for 24 hours before stimulation for the indicated time periods, and the levels of p-ERK1/2 and actin were analyzed by Western blot. (D-E) IGF-1 induces proteasomal degradation of the BimEL isoform by the p42/44 MAPK pathway. (D) OPM-2 cells were deprived of serum for 24 hours before stimulation for the indicated time periods, and the levels of phosphorylation and expression of BimEL were analyzed by Western blot. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. (E) Cells were starved for 24 hour, pretreated for 1 hour with MG132 (1μM), and treated with IGF-1 for the indicated time periods. Next, the levels of Bim were analyzed by Western blot. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. For all Western blot analyses, 1 experiment representative of 3 is shown.
IGF-1 down-regulates Bim expression in the human OPM-2 cells through the p42/44 MAPK and PI-3K pathways. (A) Western blot analysis showing down-regulation of Bim at the protein level in the OPM-2 cells. Cells deprived of serum for 24 hours were treated with IGF-1 for 24 hours. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. The numbers below refer to the optical density of the BimEL, BimL, and BimS bands after treatment with IGF-1 as measured with the NIH1.62 image program. Variation between experiments was below 10%. (B) IGF-1 inhibits Bim transcription through the PI3-K pathway in the OPM-2 cells. Cells were starved 24 hours, pretreated with LY294002 (1 hour of preincubation; 10μM), and stimulated with IGF-1 for 10 minutes. Next, the levels of p-Akt, tot-Akt, p-FoxO3a, tot-FoxO3a, p-ERK1/2, tot-ERK1/2, and actin were analyzed by Western blot. (C) Activation of the p42/44 MAPK pathway on IGF-1 treatment is delayed in the OPM-2 cells. Cells were deprived of serum for 24 hours before stimulation for the indicated time periods, and the levels of p-ERK1/2 and actin were analyzed by Western blot. (D-E) IGF-1 induces proteasomal degradation of the BimEL isoform by the p42/44 MAPK pathway. (D) OPM-2 cells were deprived of serum for 24 hours before stimulation for the indicated time periods, and the levels of phosphorylation and expression of BimEL were analyzed by Western blot. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. (E) Cells were starved for 24 hour, pretreated for 1 hour with MG132 (1μM), and treated with IGF-1 for the indicated time periods. Next, the levels of Bim were analyzed by Western blot. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. For all Western blot analyses, 1 experiment representative of 3 is shown.
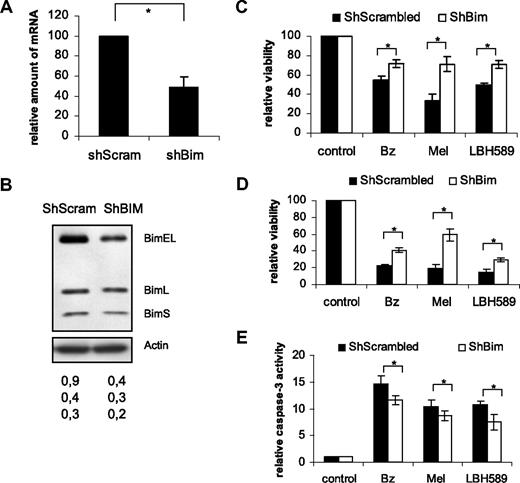
Silencing of Bim protects Karpas707 cells from drug-induced cell death
To address the relevance of Bim down-regulation in MM, cells were stably transduced with a lentiviral vector containing an RNA interference (RNAi) cassette against the Bim gene and the effect on drug-induced cell death was studied. Silencing of Bim was evaluated by quantitative RT-PCR and Western blotting comparing samples transduced with a ShScrambled (control) and a ShBim-bearing lentiviral vector. As shown in Figure 4A and B, Bim expression was partially silenced (∼ 55% on the mRNA level). Cells were exposed to bortezomib, melphalan, or LBH589 for 24 and 48 hours after which time cell viability and caspase-3 activity (only for 24 hours) were evaluated. Silencing of Bim partially protected cells from bortezomib-, melphalan-, and LBH589-induced cell death (Figure 4C-E). Collectively, these findings strongly indicate that Bim plays an important role in MM cell survival and drug resistance.
Bim silencing protects Karpas707 cells from drug-induced apoptosis. (A-B) Silencing of Bim in the Karpas707 cells. (A) Comparison of the Bim expression of the ShScrambled (control) and ShBim Karpas707 cell lines with the use of quantitative RT-PCR analysis. The mean value ± SD of 3 independent experiments is given (*P < .05). (B) Comparison of the Bim expression of the ShScrambled (lane 1) and ShBim (lane 2) Karpas707 cell lines by Western blot analysis. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. One experiment representative of 3 is shown. The numbers below refer to the optical density of the BimEL, BimL, and BimS bands as measured with the NIH1.62 image program. Variation between experiments was below 10%. (C-E) Cells were treated with 10nM bortezomib, 15μM melphalan, and 15nM LBH589 for (C) 24 and (D) 48 hours. Viability was measured by the Celltiter-Glo luminescent cell viability assay. Caspase-3 activity after 24 hours of treatment (E) was measured with the Caspase-Glo 3/7 assay. Experiments were performed 4 times, and the results are given as the percentage of control (untreated) viability or caspase-3 activity (mean value ± SD; *P < .05).
Bim silencing protects Karpas707 cells from drug-induced apoptosis. (A-B) Silencing of Bim in the Karpas707 cells. (A) Comparison of the Bim expression of the ShScrambled (control) and ShBim Karpas707 cell lines with the use of quantitative RT-PCR analysis. The mean value ± SD of 3 independent experiments is given (*P < .05). (B) Comparison of the Bim expression of the ShScrambled (lane 1) and ShBim (lane 2) Karpas707 cell lines by Western blot analysis. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. One experiment representative of 3 is shown. The numbers below refer to the optical density of the BimEL, BimL, and BimS bands as measured with the NIH1.62 image program. Variation between experiments was below 10%. (C-E) Cells were treated with 10nM bortezomib, 15μM melphalan, and 15nM LBH589 for (C) 24 and (D) 48 hours. Viability was measured by the Celltiter-Glo luminescent cell viability assay. Caspase-3 activity after 24 hours of treatment (E) was measured with the Caspase-Glo 3/7 assay. Experiments were performed 4 times, and the results are given as the percentage of control (untreated) viability or caspase-3 activity (mean value ± SD; *P < .05).
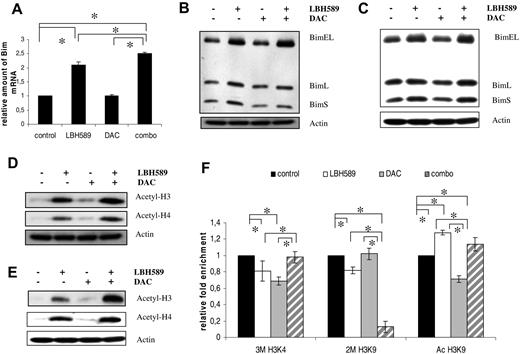
DAC/LBH589 treatment of the human myeloma cells
In MM, HDACi's have extensively been shown to regulate Bim expression. However, the mechanisms by which demethylation agents and/or HDACi affect Bim expression still remain to be elucidated.36 Cells were treated with either the demethylation agent DAC for 72 hours, the HDACi LBH589 for 24 hours, or a combination of both. Treatment of Karpas707 and OPM-2 cells with LBH589 resulted in a clear up-regulation in the expression of BimEL both at the mRNA and protein levels (Figure 5A-C). In contrast, treatment with DAC had no effect. However, DAC significantly increased the effects of LBH589. To verify whether the effect of LBH589 is mediated through hyperacetylation of histones, we studied the histone H3 and H4 acetylation. Cells treated with LBH589 for 24 hours exhibited a strong increase in acetylation over the untreated control cells (Figure 5D-E). Moreover, cotreatment exhibited a clear increase in acetylation over the LBH589-treated cells. Finally, the LP-1 cells still remained negative for Bim expression even after treatment with LBH589 and/or DAC, indicating that the absence of Bim is not due to epigenetic silencing in this cell line.
LBH589 treatment enhances Bim expression of the human Karpas707 and OPM-2 myeloma cells. (A) Bim mRNA expression in Karpas707 cells after treatment with LBH589, DAC, and LBH589/DAC. Cells were treated with 15nM LBH589 for 24 hours, 500nM DAC for 72 hours, and a combination of both. The Assays on Demand detected an mRNA sequence identifying only the BimEL isoform. The mean value ± SD of 3 independent experiments is given (*P < .05). (B-C) Bim expression in the Karpas707 (B) and OPM-2 (C) cells after treatment with LBH589, DAC, and LBH589/DAC as shown by Western blot analysis. Cells were treated with 15nM LBH589 for 24 hours, 500nM DAC for 72 hours, and a combination of both. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. One experiment representative of 3 is shown. (D-E) Acetylation status of histone 3 (H3) and H4 in the Karpas707 (D) and OPM-2 (E) cells after LBH589, DAC, and combination treatment. Equivalent amounts of lysates were immunoblotted with anti–acetyl H3, anti–acetyl H4, and anti-actin to confirm equal loading. One experiment representative of 3 is shown. (F) ChIP analysis of the Bim promoter in the Karpas707 cells after treatment with LBH589 and/or DAC. Chromatin DNA was immunoprecipitated with antibodies for normal IgG (negative control), anti–acetyl-histone H3 lysine 9 (Ac H3K9), anti–trimethyl-histone H3 lysine 4 (3M H3K4), anti–dimethyl-histone H3 lysine 9 (2M H3K9), and anti–histone H3 (positive control). A DNA fragment corresponding to the Bim promoter region was amplified by quantitative RT-PCR. Each ChIP and RT-PCR were repeated, respectively, 3 and 2 times (*P < .05). SDs refer to the 3 independent experiments.
LBH589 treatment enhances Bim expression of the human Karpas707 and OPM-2 myeloma cells. (A) Bim mRNA expression in Karpas707 cells after treatment with LBH589, DAC, and LBH589/DAC. Cells were treated with 15nM LBH589 for 24 hours, 500nM DAC for 72 hours, and a combination of both. The Assays on Demand detected an mRNA sequence identifying only the BimEL isoform. The mean value ± SD of 3 independent experiments is given (*P < .05). (B-C) Bim expression in the Karpas707 (B) and OPM-2 (C) cells after treatment with LBH589, DAC, and LBH589/DAC as shown by Western blot analysis. Cells were treated with 15nM LBH589 for 24 hours, 500nM DAC for 72 hours, and a combination of both. Equivalent amounts of lysates were immunoblotted with anti-Bim and anti-actin to confirm equal loading. One experiment representative of 3 is shown. (D-E) Acetylation status of histone 3 (H3) and H4 in the Karpas707 (D) and OPM-2 (E) cells after LBH589, DAC, and combination treatment. Equivalent amounts of lysates were immunoblotted with anti–acetyl H3, anti–acetyl H4, and anti-actin to confirm equal loading. One experiment representative of 3 is shown. (F) ChIP analysis of the Bim promoter in the Karpas707 cells after treatment with LBH589 and/or DAC. Chromatin DNA was immunoprecipitated with antibodies for normal IgG (negative control), anti–acetyl-histone H3 lysine 9 (Ac H3K9), anti–trimethyl-histone H3 lysine 4 (3M H3K4), anti–dimethyl-histone H3 lysine 9 (2M H3K9), and anti–histone H3 (positive control). A DNA fragment corresponding to the Bim promoter region was amplified by quantitative RT-PCR. Each ChIP and RT-PCR were repeated, respectively, 3 and 2 times (*P < .05). SDs refer to the 3 independent experiments.
Epigenetic regulation of the Bim promoter in the Karpas707 cells
To study the effect of DAC and/or LBH589 treatment on the methylation status of the Bim promoter, pyrosequencing analysis was performed on the Karpas707 cells. The Bim promoter region was found to be hypomethylated both before and after treatment with DAC and/or LBH589 (Table 2). On the basis of our data, we hypothesized that posttranslational histone modifications might be involved in the epigenetic regulation of Bim. To confirm this, ChIP analysis of the Bim promoter was performed (Figure 5F). Treatment with DAC resulted in a reduced acetylation of histone H3 tail Lysine 9 (Ac H3K9) and trimethylation of histone H3 tail Lysine 4 (3M H3K4) levels, whereas the dimethylation of histone H3 tail lysine 9 (2M H3K9) remained unchanged. For LBH589 treatment, an increased H3K9 acetylation and a reduction of the 2M H3K9 and 3M H3K4 levels were observed. Finally, cotreatment resulted in a drastic and significant reduction of the 2M H3K9 methylation levels accompanied by an increase in H3K9 acetylation.
Methylation status of the Bim promoter after treatment with DAC and/or LBH589
| Mean methylation % calculated over all CpG sites (31 CpGs) . | . |
|---|---|
| Control | 2.07 |
| DAC | 0.59 |
| LBH589 | 1.94 |
| DAC + LBH589 | 1.02 |
| Mean methylation % calculated over all CpG sites (31 CpGs) . | . |
|---|---|
| Control | 2.07 |
| DAC | 0.59 |
| LBH589 | 1.94 |
| DAC + LBH589 | 1.02 |
Methylation percentage below 5 is considered to be noise.
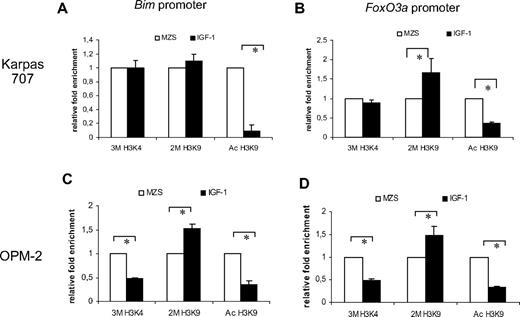
Effect of IGF-1 treatment on DNA methylation and histone modifications of the Bim/FoxO3a promoter
To investigate whether IGF-1 and epigenetic modifications act as independent regulators of Bim expression, Karpas707 cells were subjected to 4 days of IGF-1 treatment, and the DNA methylation status of the Bim promoter was assayed by pyrosequencing. Because these results did not show any relevant changes in DNA methylation of the promoter (data not shown), we performed ChIP analysis of the Bim and FoxO3a promoter. For the Karpas707 cells, the levels of Ac H3K9 were significantly reduced for both promoters (Figure 6A-B). Moreover, IGF-1 treatment resulted in a modest increase of 2M H3K9 for the Bim promoter (Figure 6A) and a strong increase for the FoxO3a promoter (Figure 6B). For the OPM-2 cells, both the levels of Ac H3K9 and 3M H3K4 were significantly reduced for both promoters (Figure 6C-D). In addition, a strong increase of 2M H3K9 was found for both promoters.
ChIP analysis of the Bim and FoxO3a promoter. ChIP analysis of the Bim promoter (A-C) and FoxO3a promoter (B-D) in the Karpas707 (A-B) and OPM-2 (C-D) cells after treatment with IGF-1 for 4 days. Chromatin DNA was immunoprecipitated with antibodies for normal IgG (negative control), anti–acetyl-histone H3 lysine 9 (Ac H3K9), anti–trimethyl-histone H3 lysine 4 (3M H3K4), anti–dimethyl-histone H3 lysine 9 (2M H3K9), and anti–histone H3 (positive control). A DNA fragment corresponding to the Bim or FoxO3a promoter region was amplified by quantitative RT-PCR. Each ChIP and RT-PCR were repeated, respectively, 3 and 2 times (*P < .05). SDs refer to the 3 independent experiments.
ChIP analysis of the Bim and FoxO3a promoter. ChIP analysis of the Bim promoter (A-C) and FoxO3a promoter (B-D) in the Karpas707 (A-B) and OPM-2 (C-D) cells after treatment with IGF-1 for 4 days. Chromatin DNA was immunoprecipitated with antibodies for normal IgG (negative control), anti–acetyl-histone H3 lysine 9 (Ac H3K9), anti–trimethyl-histone H3 lysine 4 (3M H3K4), anti–dimethyl-histone H3 lysine 9 (2M H3K9), and anti–histone H3 (positive control). A DNA fragment corresponding to the Bim or FoxO3a promoter region was amplified by quantitative RT-PCR. Each ChIP and RT-PCR were repeated, respectively, 3 and 2 times (*P < .05). SDs refer to the 3 independent experiments.
Discussion
Despite recent advances in the transplantation protocols, combinations of chemotherapy and the introduction of novel agents, MM remains an incurable malignancy. Importantly, the BM microenvironment provides a sanctuary for subpopulations of MM cells to evade or circumvent drug-induced cell death. This form of de novo drug resistance is mediated by (1) a paracrine mechanism because of soluble cytokine and/or growth factors and (2) adhesion to BM stromal cells/fibronectin, termed cell adhesion–mediated drug resistance (or CAM-DR).2,25,37 CAM-DR is accompanied with an up-regulation of genes involved in the cholesterol biosynthesis and the down-regulation of the proapoptotic protein Bcl2-like11 (Bim).
Only very recently, IGF-1 was identified as a major growth factor for myeloma cell lines, and aberrant IGF-1R expression was shown in a subset of patients and correlated with adverse prognosis.5 These data underline the importance of IGF-1 in MM progression and provide the rationale for IGF-1–targeted therapy in a subset of patients expressing IGF-1R. In an attempt to better understand IGF-1–mediated survival and growth of the MM cells, we investigated the effect of IGF-1 on the gene expression in the murine 5T33MM model. Microarray data comparing untreated 5T33MM cells with IGF-1–stimulated cells identified a similar gene expression pattern as observed for CAM-DR. Because target genes for CAM-DR and IGF-1 are comparable (Bim and genes involved in cholesterol biosynthesis), we hypothesized from these data that the interplay between IGF-1 (and growth factors in general) and β1 integrin–mediated adhesion might induce de novo drug resistance.
Because apoptotic pathways are crucial for the survival of tumor cells and drug-induced cell death, the down-regulation of Bim was further analyzed. We confirmed in the 5T33MMvv cells the IGF-1–induced down-regulation of Bim on the mRNA level. Moreover, Western blot analysis showed in both the murine 5T33MMvv cells and the 2 human MM cell lines the down-regulation of Bim upon IGF-1 treatment. Next, we tried to elucidate the mechanism(s) behind this altered expression. Several reports have identified the regulation of Bim through the Akt and MAPK pathways.20,21 We confirmed activation of both pathways on IGF-1 treatment both in the murine as well as the human MM cell lines, although there was a delay in the activation of the MAPK pathway in the OPM-2 cells. The increased MAPK activity resulted in the phosphorylation of BimEL and its subsequent proteasomal degradation, whereas the increased Akt activity resulted in the phosphorylation of the transcription factor FoxO3a. This phosphorylated form is excluded from the nucleus, thereby abolishing FoxO3a-mediated transcription of genes such as Bim.38 IGF-1– and IL-6–induced BimEL phosphorylation has been shown earlier in the human U266 and the RPMI8226 cell line.39 Moreover, recently Bim was reported to play a critical role in leukemia cell death triggered by concomitant inhibition of the PI-3K and MAPK pathways, and indications were found that Bim expression is negatively regulated by MAPK activation.40 Collectively, all these data strongly indicate that, in MM cells, IGF-1 negatively regulates Bim expression by the MAPK and PI-3K pathways.
To examine the functional implications of the down-regulation of Bim by IGF-1 stimulation, Karpas707 cells were transduced with a lentiviral vector bearing a shRNA sequence against Bim. The reduction in Bim expression was confirmed both on transcriptional and translational levels. Because Bim is a stress sensor, we expected that cells with a decreased expression of Bim would show increased viability when exposed to drugs. Indeed, partial silencing protected cells from cell death induced by bortezomib, melphalan, and LBH589. Reduced Bim levels have also been reported to protect MM cells and/or leukemia cells from cell death induced by arsenic trioxide, PD184352/ perifosine, and SBHA/ABT737.40–42 Recently, an imbalance between survivin and Bim was shown to mediate the MM tumor growth and to correlate with poor prognosis in patients with MM.43 This further underlines the importance of Bim in MM disease progression.
In a second part, we investigated the influence of epigenetic phenomena on the expression of Bim. Two of the best documented interacting epigenetic modifications are (1) DNA methylation and (2) posttranslational (non)histone modifications (PTMs).44,45 Both the interaction between these 2 and the dynamic interplay between the PTMs have a profound effect on the transcriptional regulation (cfr histone code or PTM code hypothesis).36,45 Hypermethylation of CpG islands within promoter regions of genes and/or deacetylation of the histones are common features of cancer that result in transcriptional silencing of (tumor suppressor) genes.44,46 Importantly, transcriptional gene silencing is reversible by epigenetic therapies with HDACi's and DNA-methyltransferase inhibitors. In MM, hypermethylation of genes, including p15, p16, DAP-kinase, BAD, BAK, BAX, and BIK, has been reported.47,48 Furthermore, HDACi's have been shown to decrease survival of human MM cells in vitro by affecting genes involved in the cell cycle and cell death pathways and by potentiating other proapoptotic agents.49,50 We treated OPM-2 and Karpas707 cells with a demethylase agent (DAC) or a HDACi (LBH589) and showed a clear up-regulation in the Bim mRNA expression after treatment with LBH589 alone. In contrast, DAC treatment did not alter Bim expression levels, but, when combined with LBH589, it resulted in an increased effect. Because the Bim promoter is prone to DNA hypermethylation in other cancers,24 we subjected (treated) Karpas707 cells to bisulfite treatment and pyrosequencing. Our results showed that the CpG sites within the Bim promoter have a very low methylation status, and this was not significantly altered by treatment with DAC and/or LBH589. These results were not very surprising because DAC treatment did not lead to an increased Bim expression.
Treatment of the Karpas707 and OPM-2 cells with LBH589 (alone or in combination with DAC) also increased the global levels of acetylated histones 3 and 4. This resulted in a more open and permissive chromatin state.46 Because these data gave only information about the total acetylation levels, we wanted to know if this was also true at the Bim promoter locus. Indeed, ChIP analysis for the Karpas707 cells confirmed that, within the Bim promoter chromatin, Ac H3K9 levels were increased on treatment with LBH589 (alone or in combination with DAC). However, because Ac H3K9 acetylation levels were lower in the combination treatment versus LBH589 alone, these findings alone cannot explain the up-regulation in the Bim expression. Therefore, we analyzed the methylation status of the histone H3 tail lysines that are known to elicit permissive (3M H3K4) or repressive (2M H3K9) chromatin states. The most favorable configuration for Bim expression would show high Ac H3K9 and 3M H3K4 marks (both permissive marks) and low 2M H3K9 levels (repressive mark).45,51 Combination of both inhibitors induced a favorable signature and therefore lead to the highest level of Bim reactivation. Cells treated with DAC showed totally the opposite (low Ac H3K9 and 3M H3K4), whereas LBH589-treated cells were intermediate. From all the above-mentioned findings, we conclude that the expression of the Bim gene is also regulated by epigenetic mechanisms in MM cells and more specifically by the interplay of acetylation and/or methylation of lysines on the H3 histone tails. In contrast, the LP-1 cells were shown to be negative for Bim expression, even after treatment with LBH589 and/or DAC, indicating that the absence of Bim expression is not due to epigenetic silencing. We found the lack of Bim expression to be the result of homozygous deletion of the Bim gene (data not shown). These deletions of Bim have also been reported in some B-cell non-Hodgkin lymphoma subtypes.24 Collectively, these data indicate that in MM cells silencing of Bim, either through epigenetic or genetic alterations, results in tumor progression.
In a final series of experiments, we investigated the possible role of IGF-1 in altering the epigenetic status of a promoter. Therefore, we compared Karpas707 cells grown without serum or treated with IGF-1. Again, DNA methylation analysis did not show any hypermethylation of the CpG sites within the Bim promoter (data not shown); therefore, we turned to ChIP analysis. Compared with untreated cells, IGF-1–treated cells showed a reduced Ac H3K9 level and an increased 2M H3K9 level. Moreover, in the OPM-2 cells, the 3M H3K4 levels were also found significantly reduced upon IGF-1 treatment. This is a typical signature of repressive chromatin45,51 and we concluded that IGF-1 not only influences Bim expression by the classical pathways but also through changed histone marks in the promoter region. Furthermore, we investigated if IGF-1 also epigenetically regulated the FoxO3A promoter. Indeed, IGF-1–treated cells were found to have high 2M H3K9 levels and showed reduced Ac H3K9 and 3M H3K4 at the FoxO3A promoter locus. The reduction in 3M H3K4 was again more pronounced in the OPM-2 cells. These data are in line with the observed repression of the expression of FoxO genes on treatment with IGF-1.52 Although DNA methylation maintains long-term silencing, PTMs (and thus histone modifications) are dynamic modifications responding to the microenvironment according to the needs of the cell.36,53 Thus, down-regulation by promoter histone modifications is a reversible and more immature state of silencing, which can be modulated or selected for in relation to the microenvironment.54 This further supports our observation that histone modifications, rather than DNA methylation, are the epigenetic modifications involved in the IGF-1–mediated silencing of Bim.
Collectively, we investigated the role of IGF-1 in the regulation of Bim expression in MM. In both murine and human cells, we demonstrated the direct regulation of Bim by IGF-1 by the activation of the Akt and the MAPK pathway. Moreover, we described the direct regulation of Bim by epigenetics in MM and are the first to identify the mechanism behind the histone modifications involved in the transcriptional (re)activation of the Bim promoter. Further, we demonstrate a role for IGF-1 in the epigenetic regulation of the Bim promoter. Therefore, we added an important factor to the list of mechanisms by which Bim is regulated. These findings not only deliver a more profound insight into the regulation of Bim but also show the potential of combining novel compounds that target this class of proapoptotic genes such as Bim (eg, BH3 mimetics) with compounds targeting the IGF-1 receptor.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank L. VanLommel, C. Heirman, E. Vaeremans, A. Willems, and especially C. Seynaeve for expert technical assistance. Furthermore, we thank Prof K. Thielemans (Vrije Universiteit Brussel) for providing both cloning and lentiviral facilities.
The work was supported by the International Myeloma Foundation, the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO-Vl), the Belgische Federatie tegen Kanker, Vlaamse Liga tegen Kanker–Stichting Emmanuel Van der Schueren, Multiple Myeloma Research Foundation, the European Stem Cell Network (EUFP6 MSCNET), the Onderzoeksraad Vrije Universiteit Brussel (GOA48, OZR-VUB), Swedish Cancer Society, and Swedish Research Council. E.M. and E.V.V. are postdoctoral fellows of FWO-Vl.
Authorship
Contribution: E.D.B. and T.J.B. designed the work, performed experiments, interpreted the data, and wrote the manuscript; F.S. designed and performed experiments, interpreted the data, and revised the manuscript; E.V.V. and E.M. revised the manuscript; L.T. managed and interpreted data; P.A. and H.J.-W. contributed vital reagents and revised the manuscript; and K.V. designed experiments and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Dr Karin Vanderkerken, Vrije Universiteit Brussel (VUB), Department of Hematology and Immunology, Laarbeeklaan 103, B-1090 Brussels, Belgium; e-mail: karin.vanderkerken@vub.ac.be.
References
Author notes
E.D.B. and T.J.B. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal