Endothelial cell activation and dysfunction underlie many vascular disorders, including atherosclerosis, tumor growth, and sepsis. Endothelial cell activation, in turn, is mediated primarily at the level of gene transcription. Here, we show that in response to several activation agonists, including vascular endothelial growth factor (VEGF), tumor necrosis factor-α, and thrombin, endothelial cells demonstrate rapid and profound induction of the early growth response (Egr) genes egr-1 and egr-3. In VEGF-treated endothelial cells, induction of Egr-3 was far greater and more prolonged compared with Egr-1. VEGF-mediated stimulation of Egr-3 involved the inducible binding of NFATc, serum response factor, and CREB to their respective consensus motifs in the upstream promoter region of Egr-3. Knockdown of Egr-3 markedly impaired VEGF-mediated proliferation, migration, and tube formation of endothelial cells and blocked VEGF-induced monocyte adhesion. Egr-3 knockdown abrogated VEGF-mediated vascular outgrowth from ex vivo aortic rings and attenuated Matrigel plug vascularization and melanoma tumor growth in vivo. Together, these findings suggest that Egr-3 is a critical determinant of VEGF signaling in activated endothelial cells. Thus, Egr-3 represents a potential therapeutic target in VEGF-mediated vasculopathic diseases.
Introduction
The endothelium is a highly malleable cell layer that constantly senses and responds to changes in the extracellular environment. Many extracellular mediators modulate gene transcription in endothelial cells, resulting in such phenotypic changes as cell migration, cell proliferation, angiogenesis, leukocyte adhesion, and hypercoagulability.1 Tight control of these processes is essential for maintaining homeostasis. Endothelial cell activation, if excessive, oversustained, or spatially and temporally misplaced, may lead to vasculopathic disease such as pathologic angiogenesis, inflammation, and atherosclerosis. Thus, an understanding of the molecular pathways leading to endothelial activation may provide novel insights into therapeutic targets.
Among the nuclear regulators of endothelial cell activation is the early growth response (Egr) family of transcription factors (Egr-1 to Egr-4). These proteins have a highly conserved DNA-binding domain comprising 3 C2H2 zinc finger motifs that recognize a 9-bp DNA consensus element (GCG(G/T)GGGCG). Activation of target gene transcription by Egr family members requires their de novo protein synthesis. Egr-1, the founding member of this family, initially was identified as an immediate early gene in growth factor–treated cells.2,–4 The Egr-3 gene was cloned from a serum-activated cDNA library5,6 and was originally described as a T-cell receptor–induced cyclosporine A sensitive factor responsible for the up-regulation of FasL.7 Both Egr-1 and Egr-3 are rapidly induced by extracellular stimuli, and both have been implicated in the proliferation and differentiation of several different cell types, including endothelial cells. Egr-1–null mice display female infertility, whereas Egr-3–null mice demonstrate abnormal neuronal and T-cell development.6,8,9
Egr proteins exert overlapping yet distinct functions. Isoform-specific functions may reflect differences in their interactions with coactivators. For example, Egr-1 but not Egr-2/-3 cooperates with NFATc1 in regulating T-cell development.10 Isoform-specific function also is regulated at the level of feedback inhibition by the NAB family of corepressors (NAB-1 and NAB-2).11,12 For example, Egr-4 alone is insensitive to inhibition by NAB proteins.13 Interestingly, in some cases, NAB-2 may function as a coactivator of Egr-1–mediated gene transcription.14
Egr-1 has been implicated in several vascular disease states, including ischemia/reperfusion lung injury,15 atherosclerosis,16,–18 and fibroblast growth factor-2 (FGF-2)–dependent angiogenesis/tumor growth.19 Although Egr-3 has been studied primarily in the context of lymphocyte and neuromuscular development, recent evidence points to a role for Egr-3 in transducing signals in endothelial cells. For example, we and others20,21 have shown that Egr-3 is one of the most highly inducible genes in vascular endothelial cell growth factor (VEGF)–treated endothelial cells. Moreover, Egr-3 knockdown attenuated VEGF- and FGF-2–induced growth of cultured endothelial cells.22
Here, we show that activation of primary human endothelial cells with VEGF, thrombin, or tumor necrosis factor-α (TNF-α) results in the rapid up-regulation of both Egr-1 and -3. VEGF-mediated induction of Egr-3 is greater and more prolonged compared with Egr-1; involves the inducible binding of NFAT, serum response factor (SRF), and CREB to the Egr-3 proximal promoter; and regulates cell growth, migration, neoangiogenesis, hemostatic balance, and leukocyte adhesion. These findings provide new insights into the molecular basis of endothelial cell activation and suggest that Egr-3 may be therapeutically targeted in states of pathologic angiogenesis and inflammation.
Methods
Cell culture
Human umbilical vein endothelial cells (HUVECs), human coronary artery endothelial cells (HCAECs), human pulmonary artery endothelial cells (HPAECs), human dermal microvascular endothelial cells (HDMVECs), and human skin fibroblasts were purchased from Clonetics. All primary vascular endothelial cells were cultured in EGM-2 MV complete medium (Clonetics). Fibroblasts were culture in Dulbecco modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum (FBS). Human embryonic kidney (HEK)–293 (ATCC CRL-1573), mouse pancreatic endothelial cells (MS-1; ATCC CRL-2279), mouse Lewis lung carcinoma cells (ATCC CRL-1642), and B16-F10 cells (ATCC CRL-6475) were cultured in Dulbecco modified Eagle medium supplemented with 10% FBS. Human U937 cells (JCRB-9021) were grown in RPMI-1640 medium plus 10% FBS.
ChIP assays
Chromatin immunoprecipitation (ChIP) assays were performed as previously described.23,24 In brief, sonicated chromatin was immunoprecipitated by the use of antibodies against NFATc1 (Affinity BioReagents), NFATc2 (BD Biosciences), SRF (Santa Cruz Biotechnology), and CREB (Cell Signaling Technology) or normal rabbit/mouse immunoglobulin G (IgG; Sigma-Aldrich), and the precipitates were collected on protein A/G-Sepharose beads (GE Healthcare). Real-time polymerase chain reactions (PCRs) were performed with the primer pairs as shown in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Cell migration assays
Endothelial cells were treated with siRNAs for 24 hours and then serum-starved and labeled with PKH2 dye (Sigma-Aldrich). Migration assays were carried out by the use of the BD Biocoat angiogenesis system (BD Biosciences). Fluorescently labeled cells were seeded on the upper chamber (105 cells/250 mL of EBM-2) and incubated with 750 mL of EBM-2 with or without 50 ng/mL VEGF in the lower chamber. After 24 hours, migrated cells were visualized under the fluorescent microscope (Leica [DMIRB]) and quantified by the use of a fluorescence detect-cell image analyzer (Kurabo).
Matrigel plug assay
Growth factor-reduced Matrigel (BD Biosciences) containing 50 ng of VEGF and either 109 plaque-forming units (pfu) of Ad-miControl or Ad-miEgr-3 was injected subcutaneously into the flanks of C57BL/6 mice. After 14 days, Matrigel plugs were removed for histologic sections. Alternatively, mice were injected intravenously with 100 μL of 1% Evans blue dye. After 40 minutes, mice were perfused with phosphate-buffered saline containing 2mM EDTA (ethylenediaminetetraacetic acid). Matrigel plugs were removed and incubated with formamide for 2 days to elute Evans blue dye, which was then measured with the use of a spectrophotometer (620 nm). All animal studies involving mice were approved by the Institutional Animal Care and Use Committee at the University of Tokyo.
Aortic ring assay
C57BL/6J mice were injected intravenously with 1.5 × 109 pfu AdmiControl or Ad-miEgr-3. After 4 days, the descending thoracic aorta was isolated, cut into approximately 1-mm segments, and embedded in Matrigel (BD Biosciences). Aortic rings were incubated with MCDB131 medium (Sigma-Aldrich) plus VEGF or vehicle control in the presence of 108 pfu Ad-miControl or Ad-miEgr-3. Culture medium was exchanged every 4 days. Then, 2 weeks later, vessel outgrowth was observed with phase-contrast microscopy. Total tube length was calculated by the use of the image analyzer software from Kurabo.
Monocyte adhesion assay
HUVECs were transfected with siRNAs and seeded on 24-well tissue-culture plates. Cells were grown to confluence, incubated with EBM-2 containing 0.5% FBS for 18 hours, and then treated in the presence or absence of 50 ng/mL VEGF for 6 hours. Cells were then incubated with control IgG or neutralizing antibodies against vascular cell adhesion molecule-1 (VCAM-1) or E-selectin for 30 minutes and overlaid with 3 × 105 PKH-26 (Sigma-Aldrich)–labeled U937 cells (per well). At 90 minutes later, cells were washed with Hanks buffered saline (Invitrogen) and examined with fluorescent microscopy (Leica [DMIRB]). The adhesion intensities were measured with Metamorph (Molecular Devices Corp) and a cell image analyzer (Kurabo).
Solid tumor model
We mixed 106 logarithmically growing B16-F10 melanoma cells with 100 μL of Matrigel (BD Biosciences) and implanted subcutaneously into the flank of C57/BL6 mice. When the tumor reached 10 mm3 in volume, it was injected with 5 × 109 pfu of Ad-miControl or Ad-miEgr-3. Tumor volume (mm3) was measured by the use of calipers. Additional experimental procedures were shown in the supplemental Methods.
Results
Activation agonists induce Egr-1 and Egr-3 mRNA and protein expression in primary human endothelial cells
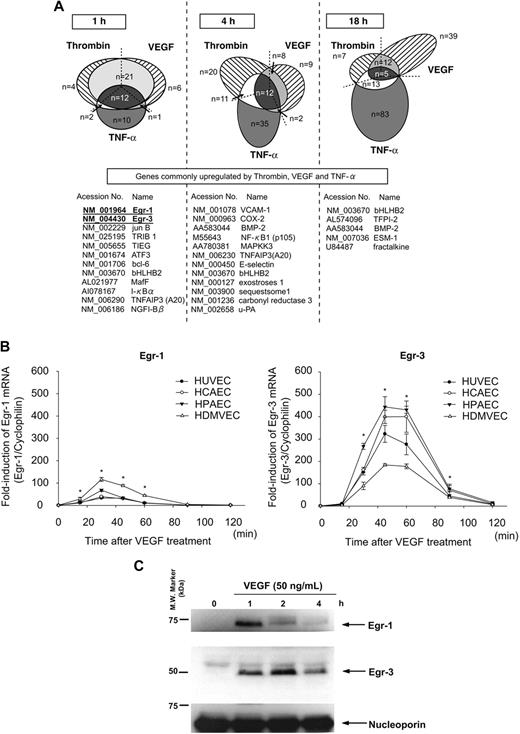
We have previously shown that different activation agonists result in overlapping yet distinct patterns of gene expression in endothelial cells.21,25 To more comprehensively define these patterns, we used DNA microarrays to assay transcriptional profiles in HUVECs treated in the absence or presence of VEGF, thrombin, or TNF-α for 1, 4, and 18 hours. As shown in Figure 1A, the majority of thrombin-responsive genes were up-regulated at 4 hours, whereas VEGF- and TNF-α–responsive genes were most highly represented at 18 hours. The 3 agonists induced the expression of a common set of genes at each time point (Figure 1A). At 1 hour, all such genes were transcription factors/cofactors. The most highly induced of these early-response genes were Egr-1 and Egr-3 (>10-fold).
Activation agonists result in rapid, high-level induction of Egr-3 in primary human endothelial cells. (A) DNA microarrays of HUVECs treated with 50 ng/mL VEGF, 2 U/mL thrombin, or 10 ng/mL TNF-α for 1, 4 or 18 hours. Shown is Venn diagram of agonist-induced genes. (B) Real-time PCR time-course analysis of Egr-1 and -3 mRNA expression in VEGF-treated HUVECs, HCAECs, HPAECs, and HDMVECs. *P < .001 compared with 0 hours (no treatment) in HUVECs, HCAECs, HPAECs, and HDMVECs. (C) Time-dependent induction of Egr-1 and -3 protein expression in VEGF-stimulated HUVECs. Western blot analysis was performed by the use of antibodies against Egr-1 or -3. Antinucleoporin antibody was used as an internal loading control. The data are representative of 3 independent experiments.
Activation agonists result in rapid, high-level induction of Egr-3 in primary human endothelial cells. (A) DNA microarrays of HUVECs treated with 50 ng/mL VEGF, 2 U/mL thrombin, or 10 ng/mL TNF-α for 1, 4 or 18 hours. Shown is Venn diagram of agonist-induced genes. (B) Real-time PCR time-course analysis of Egr-1 and -3 mRNA expression in VEGF-treated HUVECs, HCAECs, HPAECs, and HDMVECs. *P < .001 compared with 0 hours (no treatment) in HUVECs, HCAECs, HPAECs, and HDMVECs. (C) Time-dependent induction of Egr-1 and -3 protein expression in VEGF-stimulated HUVECs. Western blot analysis was performed by the use of antibodies against Egr-1 or -3. Antinucleoporin antibody was used as an internal loading control. The data are representative of 3 independent experiments.
Quantitative real-time PCR was used to assay time-dependent mRNA expression of Egr-1 and -3 in VEGF-treated endothelial cells. In HUVECs, Egr-1 was maximally induced (32-fold, relative to cyclophilin mRNA) at 30 minutes, whereas Egr-3 mRNA expression peaked at 45 minutes (314-fold; Figure 1B). Similar results were obtained with HCAEC, HPAEC, and HDMVEC (Figure 1B). Western blot analyses demonstrated time-dependent induction of Egr-1 and -3 protein expression in VEGF-treated HUVECs, with maximal levels occurring at 1 and 2 hours, respectively (Figure 1C). Compared with Egr-1, expression of Egr-3 was more sustained, returning to baseline levels only after 12 hours (data not shown). Thus, VEGF results in early transient induction of Egr-1 and more pronounced, delayed, and sustained induction of Egr-3 in primary human endothelial cells.
VEGF-mediated induction of Egr-1 and Egr-3 in primary human endothelial cells involves overlapping but distinct signaling pathways
We next wished to determine whether the distinct time course of Egr-1 and -3 induction in VEGF-treated endothelial cells reflected different underlying signal transduction pathways. To that end, real-time PCR was carried out in control and VEGF-treated HUVECs preincubated in the absence or presence of signaling inhibitors. VEGF-mediated expression of Egr-1 and -3 was blocked by neutralizing antibody against VEGFR2/KDR (Table 1). VEGF stimulation of Egr-1 and -3 was similarly attenuated by inhibitors of MEK1/2 (PD98059), c-jun N-terminal kinase (SP600125), and Ca2+ influx (BAPTA-AM) but was unaffected by inhibitors of p38 mitogen-activated protein kinase (SB203580) and de novo protein synthesis (cycloheximide). Classical protein kinase C inhibitors (GF109203x and Gö6976) had a more pronounced effect on VEGF-mediated induction of Egr-3 compared with Egr-1. Finally, VEGF stimulation of Egr-3 but not Egr-1 was significantly blunted by inhibitors of phosphoinositide-3 kinase (LY294002), calcineurin/NFATc (cyclosporine A), protein kinase C-δ (rottlerin), and protein kinase A (H-85; Table 1). Thus, VEGF-mediated expression of Egr-1 and -3 in primary human endothelial cells is governed by overlapping yet distinct signal transduction pathways.
Comparison of the VEGF-mediated Egr-1 and Egr-3 expression signals
| Name . | Target . | egr-1 mRNA reduction rate ± SD (%) . | egr-3 mRNA reduction rate ± SD (%) . |
|---|---|---|---|
| DMSO | 0 | 0 | |
| KDR nAb. | KDR | −81 ± 6.4* | −87 ± 5.7* |
| LY294002 | PI3K | (superinduced by 60-fold) | −73 ± 2.4* |
| PD98059 | MEK1/2 | −68 ± 2.3* | −78 ± 4.4* |
| SB203580 | p38 | (superinduced by 10-fold) | N.E. |
| Cyclosporine A | Calcineurin | N.E. | −75 ± 3.9* |
| BAPTA-AM | Ca2+ | −87 ± 3.9 | −99 ± 0.1* |
| GF109203X | PKCα, βI, βI, σ, γ, ϵ | −44 ± 1.9† | −99 ± 0.1* |
| Gö6976 | PKCα, βI | −40 ± 5.4† | −99 ± 0.1* |
| Rottlerin | PKCσ, θ | (superinduced by 10-fold) | −59 ± 3.2* |
| SP600125 | JNK | −95 ± 2.1* | −99 ± 0.1* |
| H-85 | PKA | (superinduced by 6-fold) | −73 ± 9.7* |
| Cycloheximide | Protein biosynthesis | (superinduced by 120-fold) | (superinduced by 10-fold) |
| Name . | Target . | egr-1 mRNA reduction rate ± SD (%) . | egr-3 mRNA reduction rate ± SD (%) . |
|---|---|---|---|
| DMSO | 0 | 0 | |
| KDR nAb. | KDR | −81 ± 6.4* | −87 ± 5.7* |
| LY294002 | PI3K | (superinduced by 60-fold) | −73 ± 2.4* |
| PD98059 | MEK1/2 | −68 ± 2.3* | −78 ± 4.4* |
| SB203580 | p38 | (superinduced by 10-fold) | N.E. |
| Cyclosporine A | Calcineurin | N.E. | −75 ± 3.9* |
| BAPTA-AM | Ca2+ | −87 ± 3.9 | −99 ± 0.1* |
| GF109203X | PKCα, βI, βI, σ, γ, ϵ | −44 ± 1.9† | −99 ± 0.1* |
| Gö6976 | PKCα, βI | −40 ± 5.4† | −99 ± 0.1* |
| Rottlerin | PKCσ, θ | (superinduced by 10-fold) | −59 ± 3.2* |
| SP600125 | JNK | −95 ± 2.1* | −99 ± 0.1* |
| H-85 | PKA | (superinduced by 6-fold) | −73 ± 9.7* |
| Cycloheximide | Protein biosynthesis | (superinduced by 120-fold) | (superinduced by 10-fold) |
DMCO indicates dimethyl sulfoxide; JNK, c-jun N-terminal kinase; KDR, kinase domain receptor; MEK1/2, MAP kinase/ERK kinase; N.E., no effect; PI3K, phosphoinositide-3 kinase; PKA, protein kinase A; and PKC, protein kinase C.
Indicates significant reduction.
indicates mild reduction.
VEGF induction of Egr-3 in primary human endothelial cells is mediated by NFATc, SRF, and CREB
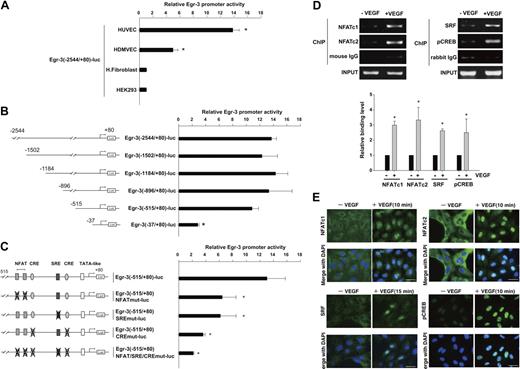
To evaluate the transcriptional mechanisms underlying VEGF-mediated induction of Egr-3, we isolated the human Egr-3 promoter and subcloned a fragment spanning the region between −2544 and +80 into the luciferase vector pGL3. The resulting plasmid (Egr-3 (−2544/+80)-luc) was transiently transfected into primary endothelial cells (HUVECs and HDMVECs) or nonendothelial cells (human skin fibroblasts and HEK-293). The cells were serum-starved and treated in the absence or presence of 50 ng/mL VEGF for 4 hours. As shown in Figure 2A, VEGF resulted in 13.8-fold and 5.1-fold induction of induction of promoter activity in HUVECs and HDMVECs, respectively. In contrast, VEGF failed to activate the promoter in fibroblasts or HEK-293 cells. HUVECs were transiently transfected with a series of progressive 5′ deletions of the Egr-3 promoter. Promoters containing 1502-, 1184-, 896-, and 515-bp 5′-flanking sequences retained full VEGF responsiveness (Figure 2B), whereas further deletion to 37 bp resulted in a significant loss of VEGF induction.
NFAT, SRF, and CREB bind to and transactivate the Egr-3 promoter in primary human endothelial cells. (A) Egr-3 (−2544/+80)-luc was transiently transfected into HUVECs, HDMVECs, human skin fibroblasts (H.Fibroblast), or HEK-293 cells. Cells were treated in the absence or presence of 50 ng/mL VEGF for 4 hours and assayed for reporter gene activity. The results show the mean ± SD of luciferase light units in VEGF-treated cells relative to untreated cells, obtained in triplicate from 3 independent experiments. *P < .001 compared with untreated cells. (B) 5′-deletion analysis of Egr-3 promoter activity in control versus VEGF-treated HUVECs. Successive deletions of the 5′-flanking region of Egr-3 were coupled to luciferase in pGL3, and the resulting constructs were transiently transfected into HUVECs. Cells were treated in the absence or presence of 50 ng/mL VEGF for 4 hours and assayed for reporter gene activity. The results show the mean ± SD of luciferase light units in VEGF-treated cells relative to untreated cells obtained in triplicate from 3 independent experiments. *P < .01 compared with Egr-3 (−2544/+80)-luc. (C) Analysis of mutant Egr-3 promoter activity in control versus VEGF-treated HUVECs. HUVECs were transiently transfected with either wild-type Egr-3 (−515/+80)-luc or Egr-3 (−515/+80)-luc containing point mutations of NFAT, SRF, and/or CRE motifs; treated in the absence or presence of 50 ng/mL VEGF for 4 hours; and assayed for reporter gene activity. The results show the mean ± SD of luciferase light units in VEGF-treated cells relative to untreated cells obtained in triplicate from 3 independent experiments. *P < .01 compared with Egr-3 (−515/+80)-luc. (D) ChIP assays of HUVECs treated in the absence or presence of 50 ng/mL VEGF for 30 minutes. Formalin-fixed chromatin was immunoprecipitated with monoclonal mouse antibodies to NFATc1 or NFATc2 or with mouse control immunoglobulin (IgG; left). Alternatively, formalin-fixed chromatin was immunoprecipitated with rabbit polyclonal antibodies against SRF or phospho-CREB or with rabbit control IgG (right). Precipitated chromatin was PCR amplified (30-35 cycles) and subjected to agarose gel electrophoresis (top). Binding was quantified by the use of real-time PCR (bottom). The results show the mean ± SE of binding level relative to control (without VEGF) obtained from at least 3 independent experiments. *P < .01 compared with control. (E) Immunofluorescent studies of NFAT, SRF, and phospho-CREB (pCREB) in control and VEGF-treated HUVECs. Serum-starved cells were incubated in the presence or absence of 50 ng/mL VEGF for the times indicated. The cells were then fixed and incubated with antibodies against NFATc1, NFATc2, SRF, or phospho-CREB, followed by Alexa 488–conjugated second antibody (green). The nuclei were stained with DAPI (blue). Merged images are shown in the bottom row. White bar indicates 20 μm.
NFAT, SRF, and CREB bind to and transactivate the Egr-3 promoter in primary human endothelial cells. (A) Egr-3 (−2544/+80)-luc was transiently transfected into HUVECs, HDMVECs, human skin fibroblasts (H.Fibroblast), or HEK-293 cells. Cells were treated in the absence or presence of 50 ng/mL VEGF for 4 hours and assayed for reporter gene activity. The results show the mean ± SD of luciferase light units in VEGF-treated cells relative to untreated cells, obtained in triplicate from 3 independent experiments. *P < .001 compared with untreated cells. (B) 5′-deletion analysis of Egr-3 promoter activity in control versus VEGF-treated HUVECs. Successive deletions of the 5′-flanking region of Egr-3 were coupled to luciferase in pGL3, and the resulting constructs were transiently transfected into HUVECs. Cells were treated in the absence or presence of 50 ng/mL VEGF for 4 hours and assayed for reporter gene activity. The results show the mean ± SD of luciferase light units in VEGF-treated cells relative to untreated cells obtained in triplicate from 3 independent experiments. *P < .01 compared with Egr-3 (−2544/+80)-luc. (C) Analysis of mutant Egr-3 promoter activity in control versus VEGF-treated HUVECs. HUVECs were transiently transfected with either wild-type Egr-3 (−515/+80)-luc or Egr-3 (−515/+80)-luc containing point mutations of NFAT, SRF, and/or CRE motifs; treated in the absence or presence of 50 ng/mL VEGF for 4 hours; and assayed for reporter gene activity. The results show the mean ± SD of luciferase light units in VEGF-treated cells relative to untreated cells obtained in triplicate from 3 independent experiments. *P < .01 compared with Egr-3 (−515/+80)-luc. (D) ChIP assays of HUVECs treated in the absence or presence of 50 ng/mL VEGF for 30 minutes. Formalin-fixed chromatin was immunoprecipitated with monoclonal mouse antibodies to NFATc1 or NFATc2 or with mouse control immunoglobulin (IgG; left). Alternatively, formalin-fixed chromatin was immunoprecipitated with rabbit polyclonal antibodies against SRF or phospho-CREB or with rabbit control IgG (right). Precipitated chromatin was PCR amplified (30-35 cycles) and subjected to agarose gel electrophoresis (top). Binding was quantified by the use of real-time PCR (bottom). The results show the mean ± SE of binding level relative to control (without VEGF) obtained from at least 3 independent experiments. *P < .01 compared with control. (E) Immunofluorescent studies of NFAT, SRF, and phospho-CREB (pCREB) in control and VEGF-treated HUVECs. Serum-starved cells were incubated in the presence or absence of 50 ng/mL VEGF for the times indicated. The cells were then fixed and incubated with antibodies against NFATc1, NFATc2, SRF, or phospho-CREB, followed by Alexa 488–conjugated second antibody (green). The nuclei were stained with DAPI (blue). Merged images are shown in the bottom row. White bar indicates 20 μm.
The latter findings suggest that the VEGF-responsive element(s) of the Egr-3 promoter is (are) located between −515 and −37. This region contains a tandem NFAT/NF-κB–like site as well as consensus motifs for CRE and SRE (Figure 2C; supplemental Figure 1).26 To determine whether one or more of these sites plays a functional role in transducing the VEGF signal, we generated Egr-3-luc constructs containing internal deletions or point mutations of these sequences. An internal deletion of a region spanning the tandem NFAT/NF-κB motifs (between −133 and −101) resulted in a 77% reduction in VEGF response (supplemental Figure 1). Consistent with these findings, point mutations of the tandem sites (Egr-3 [−515/+80]-NFATmut-luc) inhibited VEGF induction by greater than 50% (Figure 2C). An internal deletion of the region containing both the SRE and CRE sites (between −102 and −37) resulted in 80% reduction in VEGF response (supplemental Figure 1). A single point mutation of the SRE (Egr-3 (−515/+80)-SREmut-luc) inhibited VEGF induction by 50% (Figure 2C). Combined mutation of the 2 CRE sites (Egr-3 (−515/+80)-CREmut-luc) yielded a 70% decrease in VEGF response (Figure 2C). Finally, disruption of all sites (NFAT/NF-κB, CRE, and SRE) virtually abrogated VEGF-mediated activation of the Egr-3 promoter (Figure 2C; supplemental Figure 1). Thus, the NFAT/NF-κB, SRE, and CRE elements each play a critical role in mediating VEGF stimulation of Egr-3 promoter activity.
To delineate the DNA-binding proteins involved in this response, we carried out electrophoretic mobility shift analysis by the use of nuclear extracts from VEGF-treated HUVECs and radiolabeled probes spanning the SRE, CRE, or NFAT/NF-κB motifs. The SRE probe demonstrated a slowly migrating DNA-protein complex (supplemental Figure 2A lane 2). The complex was abrogated by unlabeled wild-type but not mutant self-competitor (supplemental Figure 2A lanes 3-4). Preincubation of the reaction mixture with antibodies against SRF but not control IgG resulted in a supershift of the DNA-protein complex (supplemental Figure 2A lanes 5-6). Similarly, radiolabeled probes containing the CRE (supplemental Figure 2B) or NFAT/NF-κB motifs (supplemental Figure 2C) also demonstrated slowly migrating DNA-protein complexes, which were inhibited by unlabeled wild-type but not mutant competitor (supplemental Figure 2B-C).
To assess the role for NFATc, CREB, and SRF in mediating VEGF induction of Egr-3 in its native chromatin context, we used ChIP assays. As shown in Figure 2D, VEGF treatment increased binding of NFATc1 (2.9-fold), NFATc2 (3.2-fold), SRF (2.7-fold), and pCREB (2.5-fold) but not control IgG to the proximal Egr-3 promoter. As a negative control, VEGF failed to alter binding of these transcription factors to the endogenous MyoD promoter (data not shown). Together, these findings suggest that VEGF induces binding of NFATc1, NFATc2, SRF, and CREB to the endogenous Egr-3 promoter endothelial cells.
To provide additional evidence for the role of these transcription factors in mediating the effect of VEGF on Egr-3 expression, we used immunohistochemistry to localize the proteins of interest in control- and VEGF-treated HUVEC. As shown in Figure 2E, VEGF resulted in marked nuclear localization of NFATc1, NFATc2, and phospho-CREB at 10 minutes and SRF at 15 minutes.
Finally, to test whether NFATc, SRF, and CREB can transactivate the Egr-3 promoter, cotransfection assays were carried by the use of wild-type or mutant Egr-3 luciferase constructs and expression vectors for these transcription factors. Each factor (NFATc, SRF, and CREB) was capable of transactivating the Egr-3 promoter, an effect that was abrogated by mutation of their respective DNA binding sites (supplemental Figure 3). Collectively, promoter-luciferase, electrophoretic mobility shifts, ChIP, and immunohistochemistry assay results strongly implicate a role for NFATc1, NFATc2, SRF, and phospho-CREB in mediating VEGF induction of Egr-3 expression in endothelial cells.
NAB-1 is the predominant NAB isoform in primary human endothelial cells and inhibits Egr-1 but not Egr-3 activity
Svaren et al12 have implicated a role for NAB-1 and NAB-2 in inhibiting Egr-1 activity. In real-time PCR, HUVECs expressed much greater levels of NAB-1 compared with NAB-2 (supplemental Figure 4A). To determine whether NAB-1 or NAB-2 repress Egr-1 and/or -3 activity, HUVECs were cotransfected with 1 of 2 Egr-responsive promoters (tissue factor or Flt-1) and expression vectors for Egr-1, Egr-3, NAB-1, and/or NAB-2. Egr-1–mediated transactivation of TF and Flt-1 was abrogated both by NAB-1 and NAB-2 (supplemental Figure 4B). In contrast, Egr-3–mediated transactivation of these 2 promoters was inhibited by NAB-2 alone (supplemental Figure 4B). Cotransfection with plasmids expressing Egr-1, Egr-3, NAB-1, and NAB-2 resulted in comparable expression of their respective genes, as measured by real-time PCR and Western blots (data not shown). Thus, NAB-1 is the predominant NAB isoform in HUVECs and selectively inhibits Egr-1 activity.
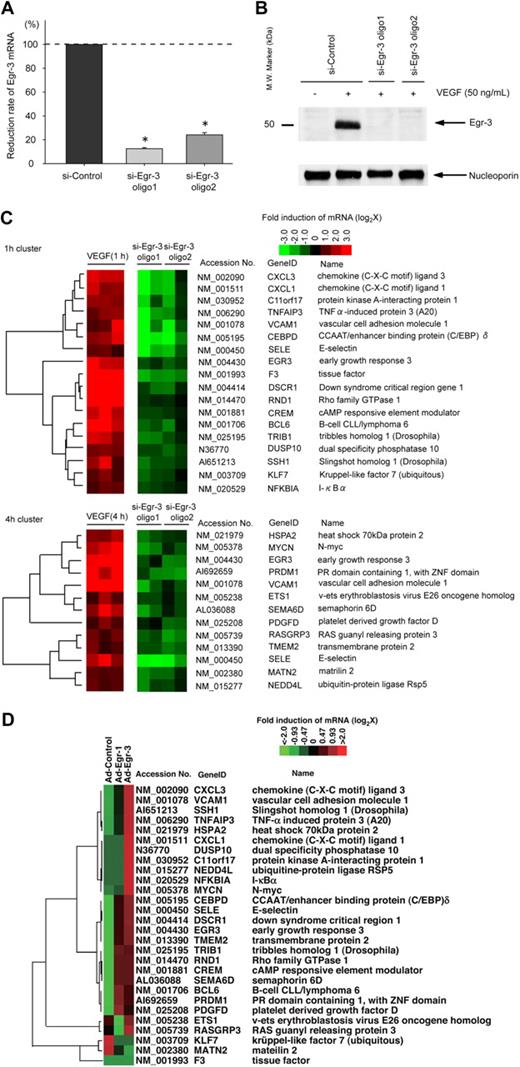
Egr-3 knockdown/overexpression in human primary endothelial cells results in altered VEGF-mediated transcriptional profiles
We next wished to analyze the functional relevance of VEGF-inducible Egr-3 in endothelial cells. To that end, we used siRNA to knock down expression of Egr-3. Transient transfection of HUVECs with 2 independent siRNAs against Egr-3 (si-Egr-3 oligo 1 and oligo 2) resulted in significant inhibition of VEGF-responsive Egr-3 mRNA expression (90% and 72% reduction, respectively; Figure 3A) and virtual abrogation of inducible Egr-3 protein expression (Figure 3B). As evidence of specificity, si-Egr-3 failed to inhibit VEGF induction of Egr-1 (supplemental Figure 5). DNA microarrays were carried out in control and Egr-3–deficient HUVECs treated in the absence or presence of 50 ng/mL VEGF. The data were filtered for those genes that were induced greater than 2-fold by VEGF and whose induction was inhibited at least 50% by the 2 independent Egr-3 siRNAs compared with control siRNA (Figure 3C). Among the genes represented were those associated with migration/angiogenesis (eg, CXCL1, SSH1, and ETS-1), coagulation (eg, tissue factor), and inflammation/leukocyte trafficking (eg, VCAM-1 and E-selectin). These changes were validated by the use of quantitative real-time PCR (supplemental Figure 6). These changes were validated by the use of quantitative real-time PCR (supplemental Figure 6). These microarray data have been deposited in the Gene Expression Omnibus (National Center for Biotechnology Information) and are accessible through GEO Series accession number GSE18913.
Egr-3 is required for VEGF-mediated induction of proangiogenic and proinflammatory genes in primary human endothelial cells. (A) Quantitative real-time PCR analysis of Egr-3 mRNA expression in HUVECs transfected with control siRNA (si-Control) or 2 independent siRNAs against Egr-3 (si-Egr-3 oligo1 and -2) and then treated in the presence of 50 ng/mL VEGF for 1 hour. Egr-3 expression is normalized to Cyclophilin A mRNA levels. The results show the mean ± SD of expression relatives relative to si-Control obtained from at least 5 independent experiments. *P < .001 compared with si-Control. (B) Western blot analysis of Egr-3 protein in HUVECs transfected with control siRNA or 2 independent siRNAs against Egr-3 and treated in the absence of presence of 50 ng/mL VEGF for 1 hour. Nucleoporin was used as a loading control. The data are representative of 4 independent experiments. (C) DNA microarrays of HUVECs transfected with si-Control, si-Egr-3 oligo1, or si-Egr-3 oligo2 and treated with 50 ng/mL VEGF for 1 hour. Shown are heat maps of genes whose VEGF response was most profoundly inhibited by si-Egr-3. Transcriptome data were derived from triplicate arrays of VEGF-treated si-Control–transfected cells and duplicate arrays of each of the VEGF-treated si-Egr-3–transfected cells. (D) DNA microarrays of HUVECs transduced with Ad-control or Ad-expressing Egr-1 (Ad-Egr-1) or Egr-3 (Ad-Egr-3; multiplicity of infection = 5). Cells were harvested for RNA at 24 hours after infection. Shown is heat map of siEgr-3–mediated down-regulated genes in Ad-Egr vs Ad-control–transduced cells.
Egr-3 is required for VEGF-mediated induction of proangiogenic and proinflammatory genes in primary human endothelial cells. (A) Quantitative real-time PCR analysis of Egr-3 mRNA expression in HUVECs transfected with control siRNA (si-Control) or 2 independent siRNAs against Egr-3 (si-Egr-3 oligo1 and -2) and then treated in the presence of 50 ng/mL VEGF for 1 hour. Egr-3 expression is normalized to Cyclophilin A mRNA levels. The results show the mean ± SD of expression relatives relative to si-Control obtained from at least 5 independent experiments. *P < .001 compared with si-Control. (B) Western blot analysis of Egr-3 protein in HUVECs transfected with control siRNA or 2 independent siRNAs against Egr-3 and treated in the absence of presence of 50 ng/mL VEGF for 1 hour. Nucleoporin was used as a loading control. The data are representative of 4 independent experiments. (C) DNA microarrays of HUVECs transfected with si-Control, si-Egr-3 oligo1, or si-Egr-3 oligo2 and treated with 50 ng/mL VEGF for 1 hour. Shown are heat maps of genes whose VEGF response was most profoundly inhibited by si-Egr-3. Transcriptome data were derived from triplicate arrays of VEGF-treated si-Control–transfected cells and duplicate arrays of each of the VEGF-treated si-Egr-3–transfected cells. (D) DNA microarrays of HUVECs transduced with Ad-control or Ad-expressing Egr-1 (Ad-Egr-1) or Egr-3 (Ad-Egr-3; multiplicity of infection = 5). Cells were harvested for RNA at 24 hours after infection. Shown is heat map of siEgr-3–mediated down-regulated genes in Ad-Egr vs Ad-control–transduced cells.
We next wished to determine whether genes that were affected by Egr-3 knockdown are reciprocally regulated by Egr-3 overexpression. To that end, HUVECs were transduced with doxycycline-regulated adenovirus expressing no cDNA or Egr-3 (Ad-Egr-3). At 2 days after infection, HUVECs were treated in the absence or presence of 1 μg/mL doxycycline for 2 days.
In DNA microarrays, conditional doxycycline-mediated overexpression of Egr-3 resulted in the induction of 24 of the 28 genes that were down-regulated by si-Egr-3 (Figure 3D). Compared with conditional overexpression of Egr-1, Egr-3 induced significantly greater levels of VCAM-1, SSH-1, and CXCL1 (Figure 3D). A comprehensive analysis of the microarray data from Egr-1 versus Egr-3 overexpressing cells revealed a preferential role for Egr-3 in inducing genes related to cell growth and hemostasis (supplemental Figure 7; whole gene lists are shown in supplemental Table 2).
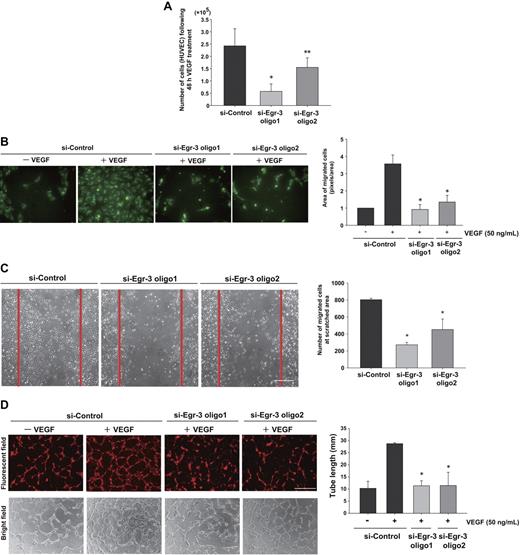
Egr-3 knockdown attenuates VEGF-mediated growth, migration, and tube formation of human primary endothelial cells
On the basis of the Egl-3–mediated transcriptional profiles, we hypothesized that Egr-3 plays a critical role in mediating the phenotypic response of endothelial cells to VEGF, including migration/angiogenesis. To test this hypothesis, HUVECs were transfected with si-Control or si-Egr-3, serum-starved, and treated with or without 50 ng/mL VEGF. Cells were then enumerated over time. VEGF treatment of si-Control–transfected cells resulted in a 2.4-fold increase in cell number after 48 hours (Figure 4A). This effect was inhibited 36% to 75% by Egr-3 knockdown (Figure 4A). There was no evidence of increased apoptosis in si-Egr-3–transfected HUVECs (data not shown).
Egr-3 is required for VEGF-mediated induction of proliferation, migration, and tube formation of primary human endothelial cells. (A) Proliferation assay. HUVECs (105) were transfected with si-Control, si-Egr-3 oligo1, or si-Egr-3 oligo2. Cells were serum-starved for 18 hours, incubated in the presence of 50 ng/mL VEGF for 48 hours, and subsequently enumerated. The results show the mean ± SD derived from 6 independent experiments. *P < .01; **P < .04 compared with si-Control. (B) Migration assay. HUVECs were transfected with si-Control, si-Egr-3 oligo1, or si-Egr-3 oligo2; labeled with PKH2; serum-starved; and plated in upper layer of a Transwell. A total of 50 ng of VEGF (or vehicle) was added to the lower chamber. After 24 hours' incubation, migrated cells were detected by the use of a fluorescent microscope. The number of migrated cells (green) was quantified with image analysis software. Means ± SD were derived from 3 independent experiments, each performed in triplicate. *P < .01 compared with si-Control plus VEGF. (C) Scratch wound assay. Confluent HUVECs transfected with si-Control, si-Egr-3 oligo1, or si-Egr-3 oligo2 were scratched with the use of a 1-mm fine tip. After 24 hours of VEGF treatment, the number of cells migrating into the scratched area was counted. Red lines correspond to the borders of the scratched area. The graph shows mean ± SD of migrated cells derived from 4 independent experiments, each performed in triplicate. *P < .01, compared with si-Control. (D) Tube-formation assay. HUVECs were transfected with si-Control si-Egr-3 oligo1 or si-Egr-3 oligo2, labeled with PKH26, serum-starved, and grown on a collagen gel in the presence or absence of 50 ng/mL VEGF. Cells were observed under fluorescence (top) or bright field (bottom). White bar indicates 100 μm. The mean ± SD of total tube length was calculated with image analyzer from 3 independent experiments performed in triplicate (bottom bar graph). *P < .02 compared the activity from si-Control plus VEGF.
Egr-3 is required for VEGF-mediated induction of proliferation, migration, and tube formation of primary human endothelial cells. (A) Proliferation assay. HUVECs (105) were transfected with si-Control, si-Egr-3 oligo1, or si-Egr-3 oligo2. Cells were serum-starved for 18 hours, incubated in the presence of 50 ng/mL VEGF for 48 hours, and subsequently enumerated. The results show the mean ± SD derived from 6 independent experiments. *P < .01; **P < .04 compared with si-Control. (B) Migration assay. HUVECs were transfected with si-Control, si-Egr-3 oligo1, or si-Egr-3 oligo2; labeled with PKH2; serum-starved; and plated in upper layer of a Transwell. A total of 50 ng of VEGF (or vehicle) was added to the lower chamber. After 24 hours' incubation, migrated cells were detected by the use of a fluorescent microscope. The number of migrated cells (green) was quantified with image analysis software. Means ± SD were derived from 3 independent experiments, each performed in triplicate. *P < .01 compared with si-Control plus VEGF. (C) Scratch wound assay. Confluent HUVECs transfected with si-Control, si-Egr-3 oligo1, or si-Egr-3 oligo2 were scratched with the use of a 1-mm fine tip. After 24 hours of VEGF treatment, the number of cells migrating into the scratched area was counted. Red lines correspond to the borders of the scratched area. The graph shows mean ± SD of migrated cells derived from 4 independent experiments, each performed in triplicate. *P < .01, compared with si-Control. (D) Tube-formation assay. HUVECs were transfected with si-Control si-Egr-3 oligo1 or si-Egr-3 oligo2, labeled with PKH26, serum-starved, and grown on a collagen gel in the presence or absence of 50 ng/mL VEGF. Cells were observed under fluorescence (top) or bright field (bottom). White bar indicates 100 μm. The mean ± SD of total tube length was calculated with image analyzer from 3 independent experiments performed in triplicate (bottom bar graph). *P < .02 compared the activity from si-Control plus VEGF.
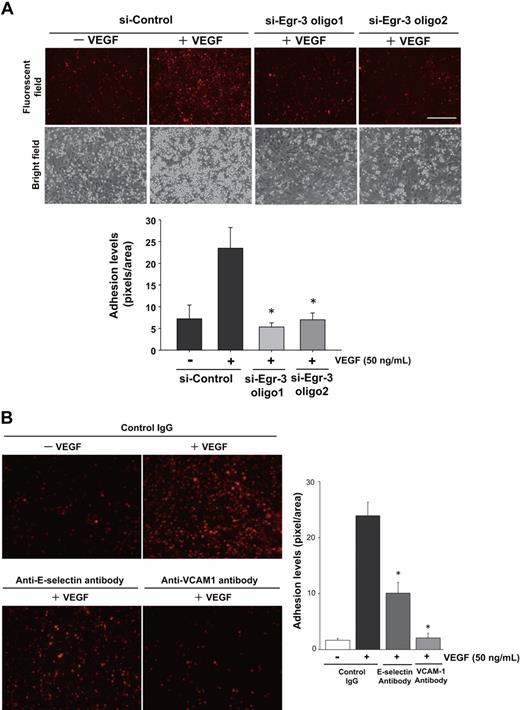
In modified Boyden chamber assays, VEGF resulted in a 3.6-fold increase in migration of si-Control–transfected HUVEC (Figure 4B). In contrast, VEGF failed to induce the migration of Egr-3 siRNA-treated HUVECs (Figure 4B). Similarly, Egr-3 knockdown inhibited cell migration in a 24-hour cell scratch assay (Figure 4C). VEGF treatment of si-Control–transfected HUVECs induced the formation of tube-like structures in 3-dimensional collagen gels (Figure 4D). This effect was markedly inhibited by each of the 2 siRNAs against Egr-3 (96% or 92% of basal level, respectively; Figure 4D). Collectively, these findings suggest that Egr-3 may play a role in mediating the effect of VEGF on new blood vessel formation.
Egr-3 knockdown attenuates VEGF-mediated monocyte adhesion and tissue factor activity in cultured endothelial cells
The DNA microarrays shown in Figure 3 support a role for Egr-3 in mediating VEGF induction of cell adhesion molecules. To determine whether this effect is functionally relevant, we carried out cell adhesion assays using U937 monocytic cells and HUVECs. VEGF treatment of si-Control–transfected HUVECs resulted in 4.1-fold increased monocyte adhesion. In contrast, VEGF had no effect on U937 cell adhesion to Egr-3–deficient HUVECs (Figure 5A). VEGF-mediated induction of E-selectin and VCAM-1 expression was strongly attenuated by Egr-3 knockdown (Figure 3C; supplemental Figure 6). VEGF-mediated U937 monocyte adhesion was inhibited 64% and 95% in the presence of neutralizing antibody against E-selectin and VCAM-1, respectively (Figure 5B). Egr-3 was also found to mediate VEGF induction of tissue factor (Figure 3C; supplemental Figure 6). Consistent with these findings, Egr-3 knockdown blocked VEGF-mediated tissue factor–dependent activation of factor X (data not shown). Together, these findings suggest that Egr-3 plays a critical role in VEGF-mediated E-selectin/VCAM-1–dependent leukocyte adhesion and tissue factor activity in endothelial cells.
Egr-3 is required for VEGF-mediated induction of leukocyte adhesion to primary human endothelial cells. HUVECs were transfected with si-Control, si-Egr-3 oligo1, or si-Egr-3 oligo2 (A) or preincubated with control IgG or antibodies against E-selectin, VCAM-1 (B) and treated with or without 50 ng/mL VEGF for 6 hours. PKH26-stained U937 monocytes were plated on top of HUVEC monolayers and incubated for 90 minutes. After washing, adhered U937 cells were observed under fluorescent and phase-contrast microscopy. Adhesion levels were quantified on the basis of fluorescent intensity by the use of image analysis software. The mean ± SD was derived from 3 arbitral optical images with 5 independent experiments (bar graph). *P < .01 compared with si-Control plus VEGF.
Egr-3 is required for VEGF-mediated induction of leukocyte adhesion to primary human endothelial cells. HUVECs were transfected with si-Control, si-Egr-3 oligo1, or si-Egr-3 oligo2 (A) or preincubated with control IgG or antibodies against E-selectin, VCAM-1 (B) and treated with or without 50 ng/mL VEGF for 6 hours. PKH26-stained U937 monocytes were plated on top of HUVEC monolayers and incubated for 90 minutes. After washing, adhered U937 cells were observed under fluorescent and phase-contrast microscopy. Adhesion levels were quantified on the basis of fluorescent intensity by the use of image analysis software. The mean ± SD was derived from 3 arbitral optical images with 5 independent experiments (bar graph). *P < .01 compared with si-Control plus VEGF.
Egr-3 knockdown impairs VEGF-dependent vascular sprouting in aortic ring assays and neovascularization of in vivo Matrigel plugs
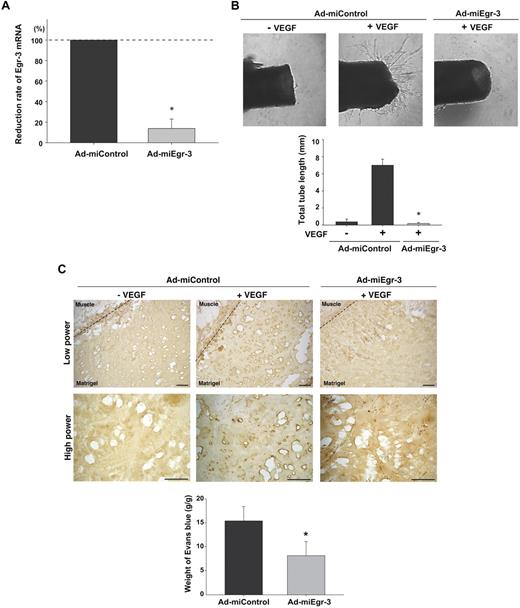
Having established a critical role for Egr-3 in VEGF-mediated proliferation and migration of cultured endothelial cells, we wished to evaluate the functional relevance of these findings in vivo. First, we generated adenovirus expressing microRNA against murine Egr-3 (Ad-miEgr-3). Infection of mouse pancreatic microvascular cells (MS-1) with Ad-miEgr-3 resulted in greater than 80% knockdown of VEGF-inducible Egr-3 mRNA expression (Figure 6A). Adenoviral gene delivery was then used to knock down Egr-3 expression in an aortic ring assay. To that end, mice were systemically administered Ad-miControl or Ad-miEgr-3. Aortas were removed 3 days later and plated in Matrigel in the presence or absence of VEGF plus Ad-miControl or Ad-miEgr-3. As shown in Figure 6B, VEGF induced sprouting of new vessels from Ad-miControl–treated aortas (18-fold increase in total tube length). In contrast, VEGF failed to simulate sprouting from Ad-miEgr-3–treated aortas (Figure 6B).
Egr-3 plays a role in VEGF-mediated neovascularization of ex vivo aortic explants and in vivo Matrigel plugs in mice. (A) Quantitative real-time PCR analysis of Egr-3 mRNA expression in VEGF-treated murine MS-1 cells transduced with adenovirus expressing miControl (Ad-miControl) or miRNA against Egr-3 (Ad-miEgr-3). Egr-3 expression is normalized to Cyclophilin A mRNA levels. The results show the mean ± SD of expression levels relative to miControl obtained from 3 independent experiments. *P < .001 compared with Ad-miControl. (B) C57/BL6 mice were injected intravenously with 109 pfu Ad-miControl or Ad-miEgr-3. At 3 days later, a short segment of the aorta was removed, embedded in Matrigel, and incubated with MCDB131 medium containing 109 pfu of Ad-miControl or Ad-miEgr-3. Outgrowth of neovessels from the aorta was observed under phase-contrast microscopy, and tube length was calculated by the use of a cell image analyzer. The mean ± SD were derived from 4 independent experiments. *P < .001 compared with Ad-miControl plus VEGF. (C) Matrigel containing 109 pfu Ad-miControl or Ad-miEgr-3 was injected subcutaneously into the flank of C57/BL6 mice. Fourteen days later, Matrigel plugs were removed, and sections were immunostained with anti–PECAM-1 antibody. Broken line indicates the boundary between flank muscle and explanted Matrigel. Bar indicates 50 μm. The data are representative of 6 independent experiments. To quantify neoangiogenesis, 1% Evans blue was injected intravenously into mice. At 10 minutes later, Matrigel plugs were removed and incubated in formamide. The amount of Evans blue dye was quantified by OD620 and normalized to weight of Matrigel. The mean ± SD were derived from 6 Matrigel plugs in each condition. *P < .01 compared with Ad-miControl.
Egr-3 plays a role in VEGF-mediated neovascularization of ex vivo aortic explants and in vivo Matrigel plugs in mice. (A) Quantitative real-time PCR analysis of Egr-3 mRNA expression in VEGF-treated murine MS-1 cells transduced with adenovirus expressing miControl (Ad-miControl) or miRNA against Egr-3 (Ad-miEgr-3). Egr-3 expression is normalized to Cyclophilin A mRNA levels. The results show the mean ± SD of expression levels relative to miControl obtained from 3 independent experiments. *P < .001 compared with Ad-miControl. (B) C57/BL6 mice were injected intravenously with 109 pfu Ad-miControl or Ad-miEgr-3. At 3 days later, a short segment of the aorta was removed, embedded in Matrigel, and incubated with MCDB131 medium containing 109 pfu of Ad-miControl or Ad-miEgr-3. Outgrowth of neovessels from the aorta was observed under phase-contrast microscopy, and tube length was calculated by the use of a cell image analyzer. The mean ± SD were derived from 4 independent experiments. *P < .001 compared with Ad-miControl plus VEGF. (C) Matrigel containing 109 pfu Ad-miControl or Ad-miEgr-3 was injected subcutaneously into the flank of C57/BL6 mice. Fourteen days later, Matrigel plugs were removed, and sections were immunostained with anti–PECAM-1 antibody. Broken line indicates the boundary between flank muscle and explanted Matrigel. Bar indicates 50 μm. The data are representative of 6 independent experiments. To quantify neoangiogenesis, 1% Evans blue was injected intravenously into mice. At 10 minutes later, Matrigel plugs were removed and incubated in formamide. The amount of Evans blue dye was quantified by OD620 and normalized to weight of Matrigel. The mean ± SD were derived from 6 Matrigel plugs in each condition. *P < .01 compared with Ad-miControl.
To further address the functional role of Egr-3 in mediating angiogenesis, we used an in vivo Matrigel plug assay. In these experiments, Matrigel impregnated with or without VEGF, Ad-miControl, or Ad-miEgr-3 was implanted subcutaneously into the flank of C57BL/6 mice. Fourteen days later, the Matrigel plugs were harvested, processed for tissues sectioning, and stained for endothelial-restricted PECAM-1 as a marker for blood vessels. In the presence of Ad-miControl, VEGF induced significant new blood vessel formation (Figure 6C middle, shown with dark brown). In contrast, Ad-miEgr-3 treatment markedly attenuated VEGF-mediated neovascularization (Figure 6C left). Evans blue was used to quantify the extent of neoangiogenesis. Matrigel that contained no virus or Ad-miControl yielded comparable levels of Evans blue (data not shown), whereas Matrigel containing Ad-miEgr-3 demonstrated a marked (56%) reduction in Evans blue content (Figure 6C bottom bar graph).
Egr-3 knockdown impairs tumor growth in mice
On the basis of the findings of Egr-3–regulated endothelial cell activation, we hypothesized that Egr-3–dependent angiogenesis may be important for tumor growth. Indeed, immunohistochemical studies of B16-F10 melanoma and Lewis lung carcinoma xenografts revealed colocalization of Egr-3 and PECAM-1 in tumor microvascular endothelium (Figure 7A arrows). B16-F10 melanoma xenografts (10 mm3 in volume) were injected with Ad-miControl or Ad-miEgr-3 (n = 6, each condition), and measured daily for volume. Compared with control, Ad-miEgr-3 treatment resulted in greater than 50% reduction of tumor size beginning 3 days after the injection and lasting until the end of the experiment (day 14; Figure 7B). Moreover, Egr-3 was associated with significantly reduced (88%) blood vessel density (Figure 7C).
Egr-3 contributes to B16-melanoma tumor progression in mice. (A) B16-F10 melanoma cells and Lewis lung carcinoma (LLC) cells were injected subcutaneously into C57/BL6j mice. After 14 days, solid tumor xenografts (1 mm3) were removed, cryosectioned, and immunostained with anti–Egr-3 antibody (left) or anti–PECAM-1 antibody (middle). Merged images with DAPI are shown on the right. White bar indicates 50 μm. (B) Gross pathology of B16-F10 melanoma from Ad-miControl– or Ad-miEgr-3–injected groups immediately after resection (left). Graph shows tumor volume in Ad-miControl–injected (●) or Ad-miEgr-3–injected (○) mice. Arrow indicates the day of adenovirus injection. Data represent mean ± SEM n = 6. *P < .01 compared with Ad-control and Ad-miEgr3. (C) Representative cryosections of B16-F10 melanoma stained with anti–PECAM-1 antibody (left). Vascular density was calculated on the basis of the number of PECAM-1–positive cells. The mean ± SD were derived from 3 optical images of 3 separate xenografts in each condition (right). *P < .001 compared with Ad-miControl. (D) Representative cryosections of B16-F10 melanoma stained with anti-CD45 antibody. Inflammatory leukocytes infiltration levels were calculated on the basis of numbers of CD45-positive cells per optical field (×200, Leica [DMLB]). The mean ± SD were derived from 6 optical images of 6 independent xenografts in each condition (bottom). *P < .001 compared with Ad-miControl.
Egr-3 contributes to B16-melanoma tumor progression in mice. (A) B16-F10 melanoma cells and Lewis lung carcinoma (LLC) cells were injected subcutaneously into C57/BL6j mice. After 14 days, solid tumor xenografts (1 mm3) were removed, cryosectioned, and immunostained with anti–Egr-3 antibody (left) or anti–PECAM-1 antibody (middle). Merged images with DAPI are shown on the right. White bar indicates 50 μm. (B) Gross pathology of B16-F10 melanoma from Ad-miControl– or Ad-miEgr-3–injected groups immediately after resection (left). Graph shows tumor volume in Ad-miControl–injected (●) or Ad-miEgr-3–injected (○) mice. Arrow indicates the day of adenovirus injection. Data represent mean ± SEM n = 6. *P < .01 compared with Ad-control and Ad-miEgr3. (C) Representative cryosections of B16-F10 melanoma stained with anti–PECAM-1 antibody (left). Vascular density was calculated on the basis of the number of PECAM-1–positive cells. The mean ± SD were derived from 3 optical images of 3 separate xenografts in each condition (right). *P < .001 compared with Ad-miControl. (D) Representative cryosections of B16-F10 melanoma stained with anti-CD45 antibody. Inflammatory leukocytes infiltration levels were calculated on the basis of numbers of CD45-positive cells per optical field (×200, Leica [DMLB]). The mean ± SD were derived from 6 optical images of 6 independent xenografts in each condition (bottom). *P < .001 compared with Ad-miControl.
To determine whether the attenuation of Egr-3 would suppress infiltration of tumors with inflammatory leukocytes, the xenografts were digested, sectioned, and stained with anti-CD45 antibody. As shown in Figure 7D, Ad-miControl–treated xenografts demonstrated large numbers of CD45-positive cells. Ad-miEgr-3 treatment resulted in greater than 85% reduction of CD45-positive cells per area. The latter effects were accompanied by a parallel reduction in VCAM-1 and E-selectin levels (supplemental Figure 8 shows VCAM-1). There was no difference in body weight between the 2 groups (data not shown). Together, these findings suggest that Egr-3 attenuates inflammation, neovascularization, and secondary tumor growth in mice.
Discussion
Activation of endothelial cells by extracellular stimuli is a key mechanism underlying the development of vascular disease. In the present study, we used DNA microarrays to analyze the global gene expression profiles of agonist-treated primary human endothelial cells. We have shown that VEGF, thrombin, and TNF-α result in rapid, high-level induction of the early-immediate transcription factors Egr-1 and Egr-3. VEGF-mediated up-regulation of Egr-3 was greater and more sustained compared with Egr-1. Egr-3, in turn was shown to mediate many downstream functions of VEGF, including proinflammatory activity, cell growth and migration in vitro, and neovascularization of Matrigel and tumor growth in vivo.
Previous studies of the Egr-1 promoter have implicated an important functional role for 2 CRE motifs and 5 SRE sites in mediating inducible gene expression.27,–29 In contrast to Egr-1, remarkably little is known about the transcriptional regulation of Egr-3. In T cells, cyclosporine was shown to inhibit Egr-3 induction, suggesting a positive effect of NFAT transcription factors.7,30 However, cyclosporine A did not influence the induction of Egr-3 in primary human fibroblasts.30 In endothelial cells, our data implicate a role for NFAT in mediating the positive effect of VEGF on Egr-3 but not Egr-1 expression. Specifically, we have shown that 2 NFAT/NF-κB–like elements in the upstream promoter of Egr-3 are necessary for transcriptional activation, that NFATc1 and NFATc2 each bind to this region, and that NFATc-mediated transactivation of the promoter is dependent on intact NFAT/NF-κB–like elements.
Our data are the first to demonstrate a role for SRF and CREB in mediating inducible expression of Egr-3. Unlike Egr-1, Egr-3 contains only a single SRE consensus motif. We have shown that VEGF induces SRF binding to this element. Moreover, mutation of the SRE abrogated SRF-mediated transactivation of Egr-3. Similar to Egr-1, Egr-3 has 2 CRE motifs. VEGF promoted the binding of phospho-CREB to the CRE motif and mutation of CRE abrogated CREB-mediated Egr-3 promoter activation. Thus, although certain transcriptional control mechanisms are shared between Egr-1 and Egr-3 (eg, they both have functional CRE and SRE sites), others are fundamentally different (eg, the presence or absence of NFAT, and the number of SRE motifs). Further studies are required to determine how these convergent and divergent pathways ultimately govern Egr-dependent endothelial activation.
There is increasing evidence that negative feedback pathways play a key role in modulating the temporal course of endothelial cell activation. For example, we have previously demonstrated that VEGF activation of the calcineurin-NFATc pathway induces expression of DSCR-1s, which in turn negatively feeds back to inhibit NFATc activity.21,24 Our findings suggest that Egr-3 contributes to the up-regulation of DSCR-1 expression in VEGF-treated endothelial cells and that DSCR-1 in turn inhibits NFAT-dependent activation of Egr-3 (Figure 3C-D).
Previous studies have shown that the corepressors NAB-1 and NAB-2 inhibit Egr-1 activity.11,12,31 Consistent with these findings, we have demonstrated that NAB-1 and NAB-2 inhibit Egr-1 activity in endothelial cells. In contrast, NAB-2 alone inhibited Egr-3 activity. Given that HUVECs constitutively express NAB-1≫NAB-2, it seems likely that NAB serves as an immediate negative feedback inhibitor of Egr-1. In contrast, overexpression of Egr-3 was shown to induce NAB-2 (and NAB-1) levels, suggesting that NAB-2 may result in delayed feedback inhibition of Egr-3. These observations may explain why VEGF induction of Egr-3 is more profound and sustained compared with Egr-1. Collectively, our data point to dual feedback control of Egr-3 whereby VEGF induces Egr-3 whereas at the same time triggering expression of NAB-2 and DSCR-1. NAB-2 and DSCR-1, in turn, feed back to inhibit Egr-3 activity and expression, respectively.
In a previous study, DNAzyme-mediated inhibition of Egr-1 blocked serum-mediated proliferation, migration, and tube formation of endothelial cells in vitro and repressed neovascularization in corneas and tumors.19 Here, we have shown that Egr-3 plays a critical role in VEGF-mediated endothelial cell growth, migration, and tube formation in vitro and angiogenesis in vivo. These findings suggest that both Egr-1 and Egr-3 may contribute to new blood vessel formation. Mice that are null for Egr-1 or Egr-3 develop normally.6,8,9 Thus, it is possible that these transcription factors compensate for one another during development of the vascular system. Future studies are required to determine whether Egr-3 is required for pathologic angiogenesis in adult mice.
In addition to promoting angiogenesis, VEGF induces a proinflammatory phenotype. For example, VEGF increases permeability, promotes a procoagulant state, and induces the expression of cell adhesion molecules (eg, VCAM-1, E-selectin) and chemokines (eg, CXCL1, interleukin-8).21 Elevated levels of VEGF play a pathogenic role in sepsis.32 Our present findings suggest that Egr-3 mediates many of the effects of VEGF on inflammation. siRNA knockdown of Egr-3 significantly inhibited the effect of VEGF on the expression of inflammatory mediators. Moreover, Egr-3 knockdown abolished VEGF-mediated monocyte adhesion. We recently reported that DSCR-1 inhibits VEGF-calcineurin-NFATc induction of inflammatory gene expression and monocyte adhesion,21,24 in essence phenocopying the effect of Egr-3 knockdown. These data suggest that many of the proinflammatory effects of VEGF-calcineurin-NFATc signaling are mediated by Egr-3.
During the course of our studies, Liu et al20,22 reported that VEGF induces Egr-3 expression and transcriptional activity in HUVECs. In the latter study, Egr-3 siRNA inhibited VEGF- and FGF-2–mediated cell proliferation and tube formation in cultured endothelial cells.22 Our data add to these findings in several important ways. First, we compare the dynamics of Egr-3 expression with that of Egr-1. Second, we address the role of Egr-3 multiple types of endothelial cells, including those from veins, arteries, and capillaries. Third, we delineate for the first time the transcriptional mechanisms underlying agonist induction of Egr-3 in endothelial cells. Fourth, we have used DNA microarrays to systematically identify Egr-3–dependent VEGF target genes in endothelial cells. Finally, and most importantly, we provide the first evidence for a role of Egr-3 in mediating angiogenesis in vivo.
An important goal in vascular biology is to understand the molecular mechanisms by which the microenvironment regulates vascular function in space and time. In this study, we have uncovered a key role for Egr-3 in VEGF-mediated signaling, gene transcription, and cell phenotype. On the basis of this knowledge, Egr-3 may represent a novel target in vascular diseases.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Mai Miura, Mika Kobayashi, and Akashi Izumi (RCAST, The University of Tokyo) for technical assistance.
This study was supported by the fund for Science and Technology from Ministry of Education, Culture, Sports, Science, and Technology (Japan; T.M.) and supported in part by Takeda Science Foundation (Japan; T.M.). This study was also supported in part by National Institues of Health grants HL082927 and HL076540 (W.C.A.).
National Institutes of Health
Authorship
Contribution: J.-i.S. performed research and analyzed data; T.H. and T.K. contributed vital new reagents; W.C.A. designed research and wrote the paper; and T.M. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Takashi Minami, PhD, Research Center for Advanced Science and Technology, University of Tokyo, 4-6-1 Komaba, Meguro Tokyo, 153-8904, Japan; e-mail: minami@med.rcast.u-tokyo.ac.jp; or William C. Aird, MD, Beth Israel Deaconess Medical Center, 330 Brookline Ave, Boston MA 02215; e-mail: waird@bidmc.harvard.edu.

![Figure 7. Egr-3 contributes to B16-melanoma tumor progression in mice. (A) B16-F10 melanoma cells and Lewis lung carcinoma (LLC) cells were injected subcutaneously into C57/BL6j mice. After 14 days, solid tumor xenografts (1 mm3) were removed, cryosectioned, and immunostained with anti–Egr-3 antibody (left) or anti–PECAM-1 antibody (middle). Merged images with DAPI are shown on the right. White bar indicates 50 μm. (B) Gross pathology of B16-F10 melanoma from Ad-miControl– or Ad-miEgr-3–injected groups immediately after resection (left). Graph shows tumor volume in Ad-miControl–injected (●) or Ad-miEgr-3–injected (○) mice. Arrow indicates the day of adenovirus injection. Data represent mean ± SEM n = 6. *P < .01 compared with Ad-control and Ad-miEgr3. (C) Representative cryosections of B16-F10 melanoma stained with anti–PECAM-1 antibody (left). Vascular density was calculated on the basis of the number of PECAM-1–positive cells. The mean ± SD were derived from 3 optical images of 3 separate xenografts in each condition (right). *P < .001 compared with Ad-miControl. (D) Representative cryosections of B16-F10 melanoma stained with anti-CD45 antibody. Inflammatory leukocytes infiltration levels were calculated on the basis of numbers of CD45-positive cells per optical field (×200, Leica [DMLB]). The mean ± SD were derived from 6 optical images of 6 independent xenografts in each condition (bottom). *P < .001 compared with Ad-miControl.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/12/10.1182_blood-2009-07-233478/5/m_zh89990947780007.jpeg?Expires=1767762072&Signature=yCxmYhwxz6c3vVEv8mvD6YtDOKVnbkxfFkV2XBaR-ipf9f32SjDLXRSy3vnVGinqXiN7j48u2IHMC51kGrHApaMfa-U8TX1HiQLexbrnf6IGzNRXE25ZCzgMQIoUGwYPJmQlI--8AxqebHQ4-HLY1cXfF5hb0Bflwb8dnv8BuUxJ9SYHT~1WA13Y2R4a6x8D2kL2usHJpRcd7bKJbyeLFfxq6pDvlahpDP4DoJr3SGalVtFLvbGiICeBL6WQ33Ft2rofA3zWWagIZ~3pcujfPKUfmZ2Qabi-HTGtCIoZsSiXUwQv2WfIFj~WqotSiZVKlj47Dt5V0VIpZ3bUsxrJYA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal