Abstract
The molecular and genetic factors induced by human T-lymphotropic virus type-1 (HTLV-1) that initiate adult T-cell leukemia/lymphoma (ATLL) remain unclear, in part from the lack of an animal model that accurately recapitulates leukemogenesis. HTLV-1–infected humanized nonobese diabetic severe combined immunodeficiency (HU-NOD/SCID) mice were generated by inoculation of NOD/SCID mice with CD34+ hematopoietic progenitor and stem cells (CD34+ HP/HSCs) infected ex vivo with HTLV-1. HTLV-1-HU-NOD/SCID mice exclusively developed CD4+ T-cell lymphomas with characteristics similar to ATLL and elevated proliferation of infected human stem cells (CD34+CD38−) in the bone marrow were observed in mice developing malignancies. Purified CD34+ HP/HSCs from HTLV-1–infected patient peripheral blood mononuclear cells revealed proviral integrations suggesting viral infection of human bone marrow–derived stem cells. NOD/SCID mice reconstituted with CD34+ HP/HSCs transduced with a lentivirus vector expressing the HTLV-1 oncoprotein (Tax1) also developed CD4+ lymphomas. The recapitulation of a CD4+ T-cell lymphoma in HU-NOD/SCID mice suggests that HSCs provide a viral reservoir in vivo and act as cellular targets for cell transformation in humans. This animal model of ATLL will provide an important tool for the identification of molecular and cellular events that control the initiation and progression of the lymphoma and potential therapeutic targets to block tumor development.
Introduction
Human T-lymphotropic virus type-1 (HTLV-1) has been linked to the development of adult T-cell leukemia/lymphoma (ATLL), a monoclonal malignancy of mature CD4+ T cells, which often express the activation marker CD25. It is estimated that approximately 15 to 20 million patients are infected with HTLV-1 worldwide, and although a majority of infected patients remain clinically asymptomatic, approximately 2% to 6% will develop some form of ATLL.1 Infection rates for HTLV-1 vary between 0.1% and 30% within endemic populations and occur more sporadically among at-risk groups from nonendemic regions, such as urban areas in the United States and Europe.2-4
HTLV-1 typically is transmitted in endemic areas from mother to child through breastfeeding,5 but it also can be transmitted by exposure to infected blood or through the sharing of needles among intravenous drug users.6 Sexual transmission is less efficient but an important mode of transmission.7 The development of ATLL generally manifests in patients after a prolonged latency period (20-40 years).8-10 The chronic-stage ATLL is characterized by leukocytosis, whereas the acute stage is characterized by multicentric lymphomatous masses, lymphoadenopathy, and hepatosplenomegaly.11-13 Diagnostic criteria of ATLL include demonstration of clonally integrated HTLV-1 in neoplastic lymphocytes.14,15
Identification of primary target cells that harbor HTLV-1 infection in infants and characterization of factors that promote virally induced transformation are difficult to identify primarily because of the extremely long clinical latent period to ATLL development and the lack of appropriate animal models that accurately recapitulate leukemogenesis.16 Inoculation of nonobese diabetic severe combined immunodeficiency (NOD/SCID) mice and NOD/SCID IL-2γ chainnull (NSG) mice with human CD34+ hematopoietic progenitor/stem cells (HP/HSCs) results in robust and extensive human hematopoiesis, including the maturation of human monocyte/macrophages and B and T lymphocytes, and sustained maintenance and expansion of human stem cells in the murine bone marrow (BM).17-19
The ability of humanized SCID mice (HU-NOD/SCID and HU-NSG) to support maturation of all human hematopoietic lineages facilitates the evaluation of the effects of viral infection and gene transduction on human hematopoiesis in vivo. We previously demonstrated that HTLV-1 efficiently infects human CD34+ HP/HSCs ex vivo and that integrated proviral sequences persist in cells throughout maturation and differentiation in vitro.20-23 Herein, we report that HTLV-1 infection and Tax1 transduction of CD34+ HP/HSCs results in highly reproducible induction of CD4+ human T-cell lymphoma with clinical and pathologic features of ATLL in HU-SCID mice. In parallel, we provide data that CD34+ HP/HSCs from HTLV-1–infected patients contain proviral integrations, suggesting a role of human BM-derived stem cells in leukemogenesis. This animal model of ATLL will be useful to determine early factors that initiate or block the development of HTLV-1–associated lymphoma.
Methods
Ex vivo infection of CD34+ cells with HTLV-1 and generation of HU-NOD/SCID mice
Fetal liver (FL)–derived CD34+ HP/HSCs were freshly isolated without cryopreservation and infected with HTLV-1 or transduced with Tax1 or Tax1-lentiviral vectors (LV) as described previously.22-24 CD34+ HPCs (5 × 106) were injected into sublethally irradiated (350 rads) 4- to 6-week-old NOD/SCIDpkrdc mice (The Jackson Laboratory) intravenously through the tail vein, as previously described25-27 to generate HTLV-1-HU-NOD/SCID or Tax1-HU-NOD/SCID mice. HTLV-1–infected or mock CD34+ HPCs (2 × 105) were injected into sublethally irradiated (25 rads) 1- to 2-day-old NSG mice (The Jackson Laboratory) intrahepatically to generate HTLV-1-HU-NSG and mock HU-NSG mice as described previously.18 Mice were housed under specific pathogen–free conditions. Mice were killed after the administration of isoflurane and tissue collected under aseptic condition for flow cytometric, polymerase chain reaction (PCR), and histopathologic analysis.
Flow cytometry
The flow cytometric analysis was performed as described previously.20,27 In brief, cells from various murine organs were resuspended in phosphate-buffered saline with 2% human serum (Jackson ImmunoResearch Laboratories) and 2% mouse serum (Invitrogen). For surface antigen detection, the cells were stained with a combination of 5 μL each of murine anti–human CD4-fluorescein isothiocyanate, CD3-PE, CD8-Pe-CY7, CD19-APC-CY7, CD45-APC, CD25-APC, CD34-PE,CD38-APC, and CD14-PE-Cy7 (BD Biosciences) and rabbit anti–mouse CD45-peridinin chlorophyll protein complex (BD Biosciences) antibody at 4°C for 20 minutes. For detection of intracellular HTLV-1 p19gag, the cells were permeabilized/fixed by the use of Cytofix/Cytoperm solution (BD Biosciences) as per manufacturer's protocol and then incubated with mouse anti–HTLV-1 p19gag antibody (ZeptoMetrix; 3 μL/106 cells) for 30 minutes at 4°C, washed twice with phosphate-buffered saline, and then incubated with fluorescein isothiocyanate- or CY5-conjugated goat anti–mouse IgG monoclonal antibody (1 μL/106 cells; Dako) for 30 minutes in dark at 4°C. Then, 1 × 104 viable and/or HTLV-1 p19gag+ cells were acquired with the use of an LSR II flow cytometer (BD Biosciences). Acquired samples were analyzed with the use of the FlowJo 8.3 program (TreeStar Inc).
Histopathologic analysis
Tissue specimens were fixed directly in neutral buffered formalin (Sigma-Aldrich) embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E) solution. In addition, tumor cells derived from NOD/SCID engrafted intraperitoneally with lymphomas from HTLV-1-HU-SCID/NOD and Tax1-HU-SCID/NOD mice were fixed in Cytospun (Sigma-Aldrich) solution and stained with the Giemsa stain. Embedded sections also were stained with anti–human CD45 antibody. All slides were examined under a light microscope, and 40× and 100× photomicrographs were generated with the use of a Nikon camera. Tissue samples were fixed in 10% neutral buffered formalin, conventionally processed, and embedded in paraffin. Three micron sections were obtained and mounted on charged slides. A representative section from each block was stained for H&E. Immunohistochemistry was performed on the BONDX automated stainer (Leica Microsystems). All aspects of the staining, including deparaffinization and epitope retrieval, are included in the protocols. The detection protocol was modified to include a 10-minute incubation of Rodent Block M (Biocare Medical) to reduce any endogenous mouse immunoglobulin activity. Mouse monoclonal anti–human antibodies were obtained (Leica Microsystems) and included CD45 (X16/99), CD20 (MJ1), and CD3 (LN10). Negative controls were performed by elimination of the primary antibody to address nonspecific staining of the detection system. Immunodetection was performed by the use of Bond Polymer Refine Detection (Leica Microsystems) with horseradish peroxidase as the visualization enzyme and DAB as the substrate chromogen. Slides were counterstained with hematoxylin, dehydrated through graded alcohol, cleared, and mounted with synthetic mounting media.
Serial transplantation of tumor cells into NOD/SCID mice
All animal studies were approved by the SUNY Upstate Medical University Committee for the Humane Use of Animals. Cells were harvested from the spleen and mesenteric lymph nodes of the primary HTLV-1–infected mice or from LV-Tax1–transduced mice and resuspended in serum-free Iscove modified Dulbecco medium (Invitrogen). Cells (5 × 106) were then injected intraperitoneally into naive NOD/SCID mice. Mice were killed when they displayed extended abdomens or evidence of ascites, and flow cytometric and histopathologic analyses were performed as described.
Real-time PCR detection of proviral sequences in patient peripheral blood mononuclear cell samples
Peripheral blood from HTLV-1– and HTLV-2–infected donors was collected after informed consent in accordance with the Declaration of Helsinki, under a research protocol approved by the Medical College of Wisconsin or the University of Miami Institutional Review Board. Cryopreserved peripheral blood mononuclear cell specimens were then shipped without unique identifiers to the Upstate Medical University of New York for the proposed molecular analysis. Quantitative PCR was performed with QuantiTect SYBR Green (QIAGEN) and primers specific for the HTLV-1 proviral genes gag (forward: 5′-CCTTCGTAGAACGCCTCAAC-3′ and reverse: 5′-ACAAGCCCGCAACATATCTC-3′), env (forward: 5′-TGGGAGCAAGGAGGATTATG-3′ and reverse: 5′-CGACAAGGGTGATTCCAGTT-3′), and tax (forward: 5′-TGTTTAGAGACTGTGTACAAGGCG-3′ and reverse: 5′-GTTGTATGAGTGATTGGCGGGGTAA-3′). The DNA products in each sample were adjusted for the total input DNA as monitored by the human β-globin gene detected with huBG240 primers (forward: 5′-GCCCTGGGCAGGTTGGTATCAAGGT-3′ and reverse: 5′-TGAGCCTTCACCTTAGGGTTGCCCA-3′). After an initial denaturation of 95°C for 15 minutes, followed by a 50-cycle PCR (94°C for 30 seconds, 60°C for 30 seconds, 72°C for 1 minute, and 80°C for 30 seconds) with detection points at 72°C and 80°C, the reaction was concluded with a 0.5°C melt-curve from 60°C to 95°C with use of the IQ-5 iCycler (Bio-Rad Laboratories). Standard curve analysis was performed with the IQ-5 program, with R2 values of at least 0.990.
T-cell receptor rearrangement analysis
DNA from lymphoma samples were analyzed for combinatorial diversity of the human β-chain of the T-cell receptor (TCR) by multiplex PCR analysis (ImmunID Technologies).
Statistical analysis
Statistical analyses were performed with the Student t test.
Results
Development of CD4+ T-cell lymphoma in HTLV-1-HU-NOD/SCID mice
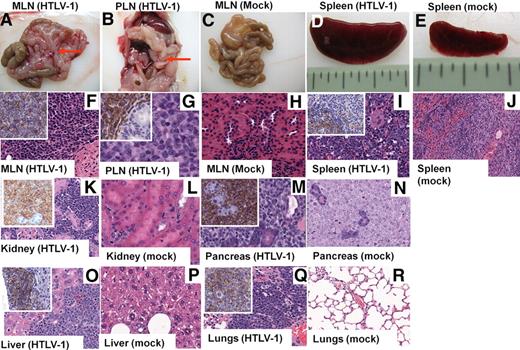
To determine the effects of HTLV-1 infection on hematopoiesis in vivo, human FL-derived CD34+ HP/HSCs were infected with HTLV-1 and then inoculated intravenously into NOD/SCID mice to reconstitute hematopoiesis. A high percentage (43%) of HTLV-1-HU-NOD/SCID mice developed lymphoma after an average latency period of approximately 17 weeks (Table 1). Reconstitution of neonatal NSG mice with HTLV-1–infected CD34+ FL cells similarly resulted in the manifestation of CD4+ T-cell lymphomas. Mice exhibited multicentric lymphoma with masses in the mesenteric lymph nodes (MLNs), liver, and in the spleen, resulting in splenomegaly (Table 1; Figure 1). HU-NOD/SCID mice inoculated with uninfected donor-matched CD34+ cells (“mock”) did not develop malignancies or lesions. Gross and histopathologic examination of affected mice revealed that presence of highly infiltrative lymphomas in the MLNs and spleen of infected mice that expressed human CD45 by immunohistochemistry (Figure 1F, G, and I). Infiltration of human lymphoma cells (CD45+) was also detected in the lungs, liver, kidney, pancreas, small intestine, and the BM in HTLV-1-HU-NOD/SCID mice (Figure 1K-R).
Lymphomagenesis in HU-NOD/SCID and HU-NSG mice infected with HTLV-1
| CD34+ cell donor tissue ID* . | Mouse strain and age of inoculation† . | Virus‡ . | No. of mice with lymphoma§ . | Percentage of lymphoma‖ . | Avg. time to disease, weeks ± SD . | Avg. time to lymphogenesis, weeks¶ . | Malignant phenotype# . | No. lymphomas serially transplanted** . | Phenotype of transplanted lymphomas†† . |
|---|---|---|---|---|---|---|---|---|---|
| 7698 | NOD/SCID | HTLV-1 | 2/9 | 43.20 | 18.3 ± 2.2 | 16.7 | CD4+CD25− | 2 | CD4+CD25− |
| 15267 | NOD/SCID | HTLV-1 | 2/10 | 18.6 ± 1.01 | CD4+CD25− | 2 | CD4+ CD25− | ||
| 7241 | NOD/SCID | HTLV-1 | 3/5 | 20 ± 3.6 | CD4+CD25low | 3 | CD4+CD25low | ||
| 7749 | NOD/SCID | HTLV-1 | 5/8 | 14 ± 2.2 | CD4+CD25low | 4 | CD4+CD25low | ||
| 7762 | NOD/SCID | HTLV-1 | 4/5 | 16 ± 1.5 | CD4+CD25− | 4 | CD4+CD25− | ||
| 1594 | NSG | HTLV-1 | 4/8 | 62.50 | 14 ± 1.52 | 14.66 | CD4+CD25− | 2 | CD4+CD25− |
| 9321 | NSG | HTLV-1 | 2/3 | 12 ± 3.16 | CD4+CD25− | 1 | CD4+CD25− | ||
| 1735 | NSG | HTLV-1 | 4/5 | 18 ± 2.51 | CD4+CD25− | 3 | CD4+CD25− | ||
| 4399 | NOD/SCID | LV-Tax 1(+) | 0/4 | 26.30 | 0 | 11.7 | NA | ||
| 7749 | NOD/SCID | LV-Tax 1(+) | 2/5 | 8 ± 2.4 | CD4+CD25low | 2 | CD4+CD25low | ||
| 7762 | NOD/SCID | LV-Tax 1(+) | 2/5 | 12 ± 4.2 | CD4+CD25− | 2 | CD4+CD25− | ||
| 15267 | NOD/SCID | Mock | 0/6 | 0 | NA | NA | NA | ||
| 7241 | NOD/SCID | Mock | 0/5 | NA | NA | ||||
| 7749 | NOD/SCID | Mock | 0/8 | NA | NA | ||||
| 7762 | NOD/SCID | Mock | 0/6 | NA | NA | ||||
| 1594 | NSG | Mock | 0/5 | NA | NA | ||||
| 9321 | NSG | Mock | 0/5 | NA | NA |
| CD34+ cell donor tissue ID* . | Mouse strain and age of inoculation† . | Virus‡ . | No. of mice with lymphoma§ . | Percentage of lymphoma‖ . | Avg. time to disease, weeks ± SD . | Avg. time to lymphogenesis, weeks¶ . | Malignant phenotype# . | No. lymphomas serially transplanted** . | Phenotype of transplanted lymphomas†† . |
|---|---|---|---|---|---|---|---|---|---|
| 7698 | NOD/SCID | HTLV-1 | 2/9 | 43.20 | 18.3 ± 2.2 | 16.7 | CD4+CD25− | 2 | CD4+CD25− |
| 15267 | NOD/SCID | HTLV-1 | 2/10 | 18.6 ± 1.01 | CD4+CD25− | 2 | CD4+ CD25− | ||
| 7241 | NOD/SCID | HTLV-1 | 3/5 | 20 ± 3.6 | CD4+CD25low | 3 | CD4+CD25low | ||
| 7749 | NOD/SCID | HTLV-1 | 5/8 | 14 ± 2.2 | CD4+CD25low | 4 | CD4+CD25low | ||
| 7762 | NOD/SCID | HTLV-1 | 4/5 | 16 ± 1.5 | CD4+CD25− | 4 | CD4+CD25− | ||
| 1594 | NSG | HTLV-1 | 4/8 | 62.50 | 14 ± 1.52 | 14.66 | CD4+CD25− | 2 | CD4+CD25− |
| 9321 | NSG | HTLV-1 | 2/3 | 12 ± 3.16 | CD4+CD25− | 1 | CD4+CD25− | ||
| 1735 | NSG | HTLV-1 | 4/5 | 18 ± 2.51 | CD4+CD25− | 3 | CD4+CD25− | ||
| 4399 | NOD/SCID | LV-Tax 1(+) | 0/4 | 26.30 | 0 | 11.7 | NA | ||
| 7749 | NOD/SCID | LV-Tax 1(+) | 2/5 | 8 ± 2.4 | CD4+CD25low | 2 | CD4+CD25low | ||
| 7762 | NOD/SCID | LV-Tax 1(+) | 2/5 | 12 ± 4.2 | CD4+CD25− | 2 | CD4+CD25− | ||
| 15267 | NOD/SCID | Mock | 0/6 | 0 | NA | NA | NA | ||
| 7241 | NOD/SCID | Mock | 0/5 | NA | NA | ||||
| 7749 | NOD/SCID | Mock | 0/8 | NA | NA | ||||
| 7762 | NOD/SCID | Mock | 0/6 | NA | NA | ||||
| 1594 | NSG | Mock | 0/5 | NA | NA | ||||
| 9321 | NSG | Mock | 0/5 | NA | NA |
HTLV-1 indicates human T-lymphotropic virus type-1; HU, humanized; LV, lentiviral vector; NA, not applicable; NOD/SCID, nonobese diabetic severe combined immunodeficiency; NSG, NOD/SCID IL-2γ chainnull.
Mice were inoculated with fresh CD34+ cells derived from human fetal liver.
Three- to 5-week-old NOD/SCID mice or 1- to 2-day-old neonate NSG mice were inoculated with either 5 × 106 (for adults) or 5 × 105 (for neonates) fresh CD34+ cells.
CD34+ HP/HSCs were infected with HTLV-1 or transduced with LVs ex vivo before inoculation into NOD/SCID or NSG mice.
Number of mice that displayed physical signs of leukemia/lymphoma, characterized by hunched posture, lethargy, anemia, and ruffled coat.
Percentage of mice developing lymphoma for each group.
Average time to lymphomagenesis in weeks for each group.
Phenotype of predominant human lymphocytic population in the mesenteric lymph nodes.
Number of lymphomas that engrafted after intraperitoneal injection into naive NOD/SCID recipient mice.
Phenotype displayed by lymphoma cells recovered by peritoneal lavage from secondary recipient mice.
Lymphomagenesis in HTLV-1 HU-NOD/SCID mice. Representative histologic analysis of HTLV-1-HU-NOD/SCID mice and mock mice killed 14 to 16 weeks after reconstitution. Development of lymphomas localized to the mesenteric (MLN) and lymph nodes surrounding the pancreas (PLN; red arrows) and spleen (A-B,D) in HTLV-1-HU-NOD/SCID mice in comparison with mock-infected HU-NOD/SCID mice (C,E). H&E staining of MLN (F), pancreatic lymph nodes (G), and infiltrating lymphocytes in spleen (I) of HTLV-1-HU-NOD/SCID mice in contrast to lymph nodes (H) and spleen (J) of mock-infected HU-NOD/SCID mice. H&E staining shows diffuse large scale lymphomas in the kidney, pancreas, liver, and lungs (K,M,O,Q) of HTLV-1-HU-NOD/SCID mice in comparison with kidney, pancreas, liver, and lungs (L,N,P,R) of mock-infected HU-NOD/SCID mice. Insets (K,M,O,Q) show immunohistochemical analysis for human CD45.
Lymphomagenesis in HTLV-1 HU-NOD/SCID mice. Representative histologic analysis of HTLV-1-HU-NOD/SCID mice and mock mice killed 14 to 16 weeks after reconstitution. Development of lymphomas localized to the mesenteric (MLN) and lymph nodes surrounding the pancreas (PLN; red arrows) and spleen (A-B,D) in HTLV-1-HU-NOD/SCID mice in comparison with mock-infected HU-NOD/SCID mice (C,E). H&E staining of MLN (F), pancreatic lymph nodes (G), and infiltrating lymphocytes in spleen (I) of HTLV-1-HU-NOD/SCID mice in contrast to lymph nodes (H) and spleen (J) of mock-infected HU-NOD/SCID mice. H&E staining shows diffuse large scale lymphomas in the kidney, pancreas, liver, and lungs (K,M,O,Q) of HTLV-1-HU-NOD/SCID mice in comparison with kidney, pancreas, liver, and lungs (L,N,P,R) of mock-infected HU-NOD/SCID mice. Insets (K,M,O,Q) show immunohistochemical analysis for human CD45.
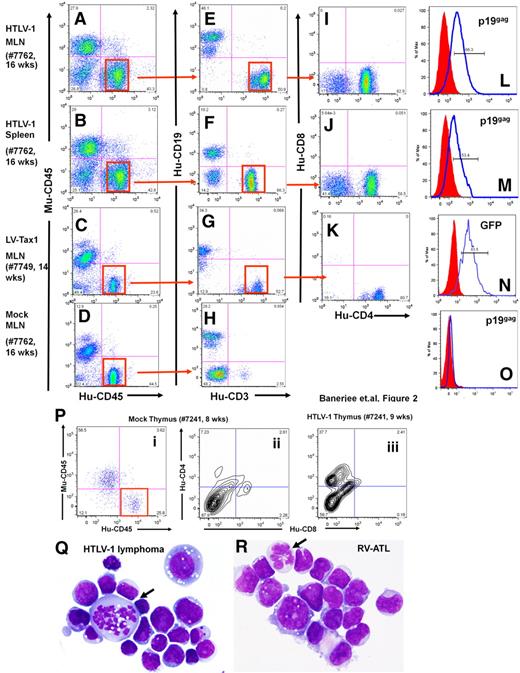
Lymphoma cells from HTLV-1-HU-NOD/SCID mice were CD4+/CD8−, coexpressed HTLV-1 viral p19gag envelope, and occasionally expressed CD25+, demonstrating that a mature T-cell lineage was the predominant phenotype among the infected lymphoma cells derived from these mice (Figure 2).28 Leukemic cells were detected in the peripheral blood, and Giemsa-stained tumor cells displayed an abnormal cellular morphology (Figure 2Q-R). HTLV-1–infected mice demonstrated CD4+/CD8− cells in the MLN and spleen, in contrast to the predominantly CD19+ B cells in the spleens of mock-infected HU-NOD/SCID mice as previously reported (Figure 2E-H).25,26 Cells recovered from the murine thymus of HTLV-1–infected mice demonstrated the presence of CD4+/CD8− human T cells in contrast to mock mice, which predominantly displayed CD4−/CD8− cells (Figure 2P).
Phenotype analysis of lymphomas from HTLV-1-HU-NOD/SCID and Tax1-HU-NOD/SCID mice. Phenotype analysis of lymphocytes from the MLN (A,E,I,L) and the spleen (B,F,J,M) of HTLV-1-HU-NOD/SCID mouse (#7762, 16 weeks after injection) and MLN of Tax1-HU-NOD/SCID (#7749, 14 weeks after injection) mouse (C,G,K,N). Human CD3+ cells are gated and analyzed in panels I, J, and K. CD4+/CD8− are the predominant populations in HTLV-1-HU-NOD/SCID mice MLN (I) and spleen (J) and Tax1-HU-NOD/SCID mice MLN (K). Human lymphocytes in the mesenteric lymph nodes (D) of mock-infected donor-tissue matched HU-NOD/SCID (#7762, 16 weeks after injection) mice are predominantly of CD19+ (B-cell) lineage (H). HTLV-1 p19gag expression in CD4+ T cells from the MLN (L) and spleen (M) of the HTLV-1-HU-NOD/SCID mouse. Expression of GFP in the CD4+ T cells from (N) the MLN of the Tax1-HU-NOD/SCID mouse and (O) in cells from the MLN of mock-infected mouse. (P) Proliferation of CD4+/CD8− subpopulation among gated human lymphocytes in the thymus of the HTLV-1-HU-NOD/SCID mouse (#7241, 9 weeks after injection; iii) in comparison with mock HU-NOD/SCID mouse (#7241, 8 weeks after injection; ii). Comparative Giemsa staining of lymphoma cells derived from HTLV-1-HU-NOD/SCID mouse (Q) and RV-ATL tumor cell line (R).28
Phenotype analysis of lymphomas from HTLV-1-HU-NOD/SCID and Tax1-HU-NOD/SCID mice. Phenotype analysis of lymphocytes from the MLN (A,E,I,L) and the spleen (B,F,J,M) of HTLV-1-HU-NOD/SCID mouse (#7762, 16 weeks after injection) and MLN of Tax1-HU-NOD/SCID (#7749, 14 weeks after injection) mouse (C,G,K,N). Human CD3+ cells are gated and analyzed in panels I, J, and K. CD4+/CD8− are the predominant populations in HTLV-1-HU-NOD/SCID mice MLN (I) and spleen (J) and Tax1-HU-NOD/SCID mice MLN (K). Human lymphocytes in the mesenteric lymph nodes (D) of mock-infected donor-tissue matched HU-NOD/SCID (#7762, 16 weeks after injection) mice are predominantly of CD19+ (B-cell) lineage (H). HTLV-1 p19gag expression in CD4+ T cells from the MLN (L) and spleen (M) of the HTLV-1-HU-NOD/SCID mouse. Expression of GFP in the CD4+ T cells from (N) the MLN of the Tax1-HU-NOD/SCID mouse and (O) in cells from the MLN of mock-infected mouse. (P) Proliferation of CD4+/CD8− subpopulation among gated human lymphocytes in the thymus of the HTLV-1-HU-NOD/SCID mouse (#7241, 9 weeks after injection; iii) in comparison with mock HU-NOD/SCID mouse (#7241, 8 weeks after injection; ii). Comparative Giemsa staining of lymphoma cells derived from HTLV-1-HU-NOD/SCID mouse (Q) and RV-ATL tumor cell line (R).28
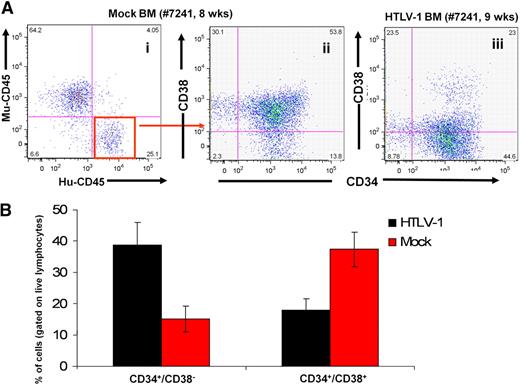
Collectively, cells coexpressing CD45+/CD4+/p19gag+ were the predominant human cell population in the BM, thymus, spleen, and the MLN of HTLV-1-HU-NOD/SCID mice. Most HTLV-1-HU-NOD/SCID mice, which did not develop disease, failed to demonstrate engraftment of CD34+ cells, suggesting that the incidence of lymphomagenesis by HTLV-1 may be significantly greater than calculated. The slightly elevated incidence of lymphomagenesis in NSG mice most likely reflects the greater engraftment rate of CD34+ HP/HSCs in this mouse strain in contrast to NOD/SCID mice (P.B. and G.F., unpublished observations, December 2009). This finding demonstrates that HTLV-1 infection of CD34+ HP/HSCs induces the development of a mature CD4+ T-cell lymphoma within a relatively short time frame (12-20 weeks after reconstitution) with histopathologic, immunologic, and clinical characteristic features of ATLL in HU-NOD/SCID and HU-NSG mice. Furthermore, hyperproliferation of HTLV-1–infected stem cells in the BM suggests that infection of stem cells may be a predeterminant of lymphomagenesis (Figure 3). Intrahepatic injection of neonatal NSG mice with HTLV-1–infected CD34+ HP/HSCs also induced lymphoma and infected mice demonstrated a dramatic shift toward CD4+/CD8− T-cell maturation in comparison with mock-infected HU-NSG mice reconstituted with tissue-matched CD34+ cells (Figures 4–5).
Hyperproliferation of infected HSCs in the BM of HTLV-1-HU-NOD/SCID mice. (A) Predominance of CD34+/CD38− human stem cell subpopulation among human lymphocytes in the BM of the HTLV-1-HU-NOD/SCID mouse (iii) in comparison with mock-infected HU-NOD/SCID mouse (ii) when gated on human CD45 subpopulation (i). (B) Cumulative quantization of CD34+/CD38− and CD34+/CD38+ subpopulations in BM of HTLV-1 infected (n = 7) and mock-infected HU-NOD/SCID mice (n = 8).
Hyperproliferation of infected HSCs in the BM of HTLV-1-HU-NOD/SCID mice. (A) Predominance of CD34+/CD38− human stem cell subpopulation among human lymphocytes in the BM of the HTLV-1-HU-NOD/SCID mouse (iii) in comparison with mock-infected HU-NOD/SCID mouse (ii) when gated on human CD45 subpopulation (i). (B) Cumulative quantization of CD34+/CD38− and CD34+/CD38+ subpopulations in BM of HTLV-1 infected (n = 7) and mock-infected HU-NOD/SCID mice (n = 8).
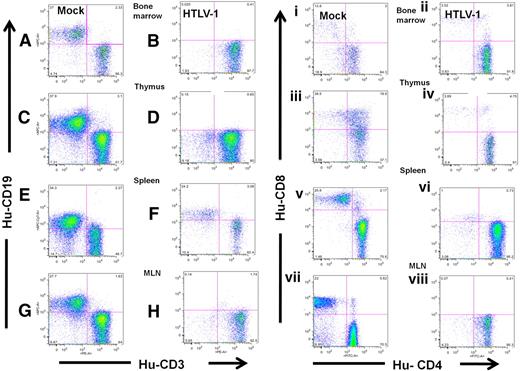
HTLV-1 infection skews hematopoiesis to the T-cell lineage in HU-NSG mice. Expression of CD19 and CD3 on lymphocytes (gated on human CD45) from the BM (A-B), thymus (C-D), spleen (E-F), and MLN (G-H) of HTLV-1-HU-NSG mouse (14 weeks after injection) in comparison with mock-infected HU-NSG mouse reconstituted with tissue-matched CD34+ donor cells. Human CD3+ cells were then gated and subsequently analyzed for CD4 and CD8 expression (i-viii). CD4+/CD8− single-positive T cells are the predominant population in HTLV-1-HU-NSG mice BM (ii), thymus (iv), spleen (vi), and MLN (viii) in comparison with mock-infected HU-NSG mice. Human lymphocytes in mock-infected donor tissue–matched HU-NSG mice (14 weeks after injection) mice show broader and more diverse hematopoietic lineage development, including the presence of B cells (CD19+), mature single CD8+ T cells, and immature CD4+/CD8+ T cells (i,iii,v,vii).
HTLV-1 infection skews hematopoiesis to the T-cell lineage in HU-NSG mice. Expression of CD19 and CD3 on lymphocytes (gated on human CD45) from the BM (A-B), thymus (C-D), spleen (E-F), and MLN (G-H) of HTLV-1-HU-NSG mouse (14 weeks after injection) in comparison with mock-infected HU-NSG mouse reconstituted with tissue-matched CD34+ donor cells. Human CD3+ cells were then gated and subsequently analyzed for CD4 and CD8 expression (i-viii). CD4+/CD8− single-positive T cells are the predominant population in HTLV-1-HU-NSG mice BM (ii), thymus (iv), spleen (vi), and MLN (viii) in comparison with mock-infected HU-NSG mice. Human lymphocytes in mock-infected donor tissue–matched HU-NSG mice (14 weeks after injection) mice show broader and more diverse hematopoietic lineage development, including the presence of B cells (CD19+), mature single CD8+ T cells, and immature CD4+/CD8+ T cells (i,iii,v,vii).
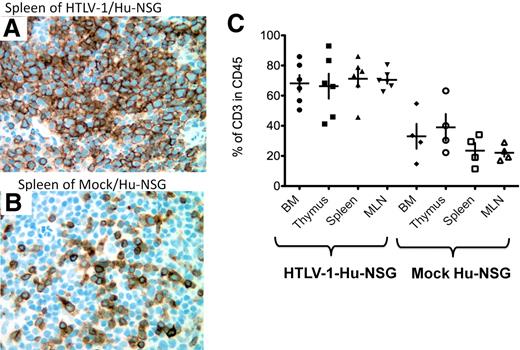
Hyperproliferation of CD3+ T cells in HTLV-1/HU-NSG mice. Representative histologic analysis of the spleen of HTLV-1/HU-NSG mice and mock HU-NSG mice killed 14 weeks after reconstitution. (A-B) Immunohistochemical analysis of human CD3 expression in the spleen of HTLV-1-HU-NSG mice in comparison with mock-infected HU-NSG mice (×40 magnification). (C) Quantification of hyperproliferation of CD3+ T cells in the BM, thymus, spleen, and MLN of HTLV-1-HU-NSG mice in comparison with mock mice reconstituted with the same donor CD34+ cell preparation (HTLV-1, n = 6; mock, n = 4).
Hyperproliferation of CD3+ T cells in HTLV-1/HU-NSG mice. Representative histologic analysis of the spleen of HTLV-1/HU-NSG mice and mock HU-NSG mice killed 14 weeks after reconstitution. (A-B) Immunohistochemical analysis of human CD3 expression in the spleen of HTLV-1-HU-NSG mice in comparison with mock-infected HU-NSG mice (×40 magnification). (C) Quantification of hyperproliferation of CD3+ T cells in the BM, thymus, spleen, and MLN of HTLV-1-HU-NSG mice in comparison with mock mice reconstituted with the same donor CD34+ cell preparation (HTLV-1, n = 6; mock, n = 4).
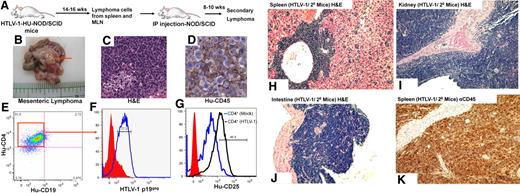
To determine whether the lymphoproliferations from the HTLV-1-HU-NOD/SCID were fully transformed and tumorigenic, cells from the MLN and the spleen were pooled and inoculated intraperitoneally into naive NOD/SCID mice. Secondary recipient mice developed CD4+ lymphomas in the peritoneal cavity and in the MLN, and engrafted tumor cells expressed HTLV-1 p19gag (Figure 6). HTLV-1–induced lymphomas showed primarily oligoclonal patterns of T-cell proliferation as determined by TCR rearrangement patterns (Table 2). Serially transplanted lymphomas developed into monoclonal proliferations in secondary recipient mice for 81% of the HTLV-1–induced lymphomas (21/26 lymphoma cell lines), and these secondary lymphomas demonstrated infiltration into the spleen, kidney, and small intestine in these mice (Figure 6H-K).
Serial transplantation and engraftment of lymphomas cells from HTLV-1-HU-NOD/SCID mice. (A) Pooled cells from the spleen and MLNs of HTLV-1-HU-NOD/SCID mouse (#7749) were serially transplanted intraperitoneally into naive NOD/SCID mice. Cells from lymphomas in the mesenteric lymph nodes (B), in the secondary recipient mice (#7749-2) as visualized by H&E (C), and immunohistochemistry staining for human CD45 (Hu-CD45; D). Phenotypic analysis of lymphoma cells in the MLN of secondary recipients demonstrate CD4+ staining (E), coexpression of HTLV-1 gag antigen p19gag (F), and expression of CD25 (G). H&E staining demonstrating diffuse large-scale lymphomas in the spleen (H), kidney (I), and small intestine (J) of secondary recipient NOD/SCID mice. (K) Immunohistochemical analysis for human CD45 in the spleen of secondary recipient mice that developed lymphoma.
Serial transplantation and engraftment of lymphomas cells from HTLV-1-HU-NOD/SCID mice. (A) Pooled cells from the spleen and MLNs of HTLV-1-HU-NOD/SCID mouse (#7749) were serially transplanted intraperitoneally into naive NOD/SCID mice. Cells from lymphomas in the mesenteric lymph nodes (B), in the secondary recipient mice (#7749-2) as visualized by H&E (C), and immunohistochemistry staining for human CD45 (Hu-CD45; D). Phenotypic analysis of lymphoma cells in the MLN of secondary recipients demonstrate CD4+ staining (E), coexpression of HTLV-1 gag antigen p19gag (F), and expression of CD25 (G). H&E staining demonstrating diffuse large-scale lymphomas in the spleen (H), kidney (I), and small intestine (J) of secondary recipient NOD/SCID mice. (K) Immunohistochemical analysis for human CD45 in the spleen of secondary recipient mice that developed lymphoma.
Clonal analysis of lymphomas by T-cell receptor rearrangement patterns
| . | Mouse strain* . | Mouse no. . | Virus . | Clonality† . | Major rearrangements . |
|---|---|---|---|---|---|
| Tissue ID‡ | |||||
| 7762 | NOD/SCID | 1 | HTLV-1 | Oligoclonal | BV15-BJ1.6, BV27-BJ1.1-1.2, BV12-BJ1.1-1.2 |
| 7762 | NOD/SCID | 1 | HTLV-1 | Oligoclonal | ND |
| 7762 | NOD/SCID | 5 | HTLV-1 | Oligoclonal | ND |
| 7241 | NOD/SCID | 2 | HTLV-1 | Oligoclonal | ND |
| 7241 | NOD/SCID | 1 | HTLV-1 | Oligoclonal | ND |
| 1594 | NSG | 4 | HTLV-1 | Monoclonal | BV05-BV2.6 |
| 1594 | NSG | 5 | HTLV-1 | Oligoclonal | BV05-J1.5, BV11-BJ1.5, BV24-BJ2.3 |
| 9321 | NSG | 2 | HTLV-1 | Oligoclonal | ND |
| 7749 | NOD/SCID | 3 | LV-Tax1 | Monoclonal | BV12-BJ1.1-1.2 |
| 7749 | NOD/SCID | 4 | LV-Tax1 | Monoclonal | BV12-BJ1.1-1.3 |
| Controls | |||||
| Hu-NSG (spleen)§ | NSG | Mock | Polyclonal | ||
| RV-ATL‖ | NA | Monoclonal | BV29-BJ2.2 | ||
| SLB-1 cell line¶ | NA | Monoclonal | BV15-BJ1.6, BV27-BJ1.1-1.2 |
| . | Mouse strain* . | Mouse no. . | Virus . | Clonality† . | Major rearrangements . |
|---|---|---|---|---|---|
| Tissue ID‡ | |||||
| 7762 | NOD/SCID | 1 | HTLV-1 | Oligoclonal | BV15-BJ1.6, BV27-BJ1.1-1.2, BV12-BJ1.1-1.2 |
| 7762 | NOD/SCID | 1 | HTLV-1 | Oligoclonal | ND |
| 7762 | NOD/SCID | 5 | HTLV-1 | Oligoclonal | ND |
| 7241 | NOD/SCID | 2 | HTLV-1 | Oligoclonal | ND |
| 7241 | NOD/SCID | 1 | HTLV-1 | Oligoclonal | ND |
| 1594 | NSG | 4 | HTLV-1 | Monoclonal | BV05-BV2.6 |
| 1594 | NSG | 5 | HTLV-1 | Oligoclonal | BV05-J1.5, BV11-BJ1.5, BV24-BJ2.3 |
| 9321 | NSG | 2 | HTLV-1 | Oligoclonal | ND |
| 7749 | NOD/SCID | 3 | LV-Tax1 | Monoclonal | BV12-BJ1.1-1.2 |
| 7749 | NOD/SCID | 4 | LV-Tax1 | Monoclonal | BV12-BJ1.1-1.3 |
| Controls | |||||
| Hu-NSG (spleen)§ | NSG | Mock | Polyclonal | ||
| RV-ATL‖ | NA | Monoclonal | BV29-BJ2.2 | ||
| SLB-1 cell line¶ | NA | Monoclonal | BV15-BJ1.6, BV27-BJ1.1-1.2 |
HPCs indicate hematopoietic progenitor cells; HTLV-1, human T-lymphotropic virus type-1; LV, lentiviral vector; NA, not applicable; ND, no data available; NOD/SCID, nonobese diabetic severe combined immunodeficiency; and NSG, NOD/SCID IL-2γ chainnull.
NOD/SCID mice were inoculated intravenously either with HTLV-1–infected or LV-Tax–transduced CD34+ HPCs, as described in Table 1. NSG mice were inoculated with HTLV-1–infected CD34+ HPCs at 1-2 days of age, intrahepatically.
DNA from lymphomas from HTLV-1-HU-NOD/SCID, HTLV-1-HU-NSG, and LV-Tax1-HU-NOD/SCID mice were analyzed for combinatorial diversity of the human T-cell receptor β-chain. Samples demonstrating 1-2 predominant rearrangements are designated as monoclonal, 3-50 rearrangements as oligoclonal, and > 50 rearrangements as polyclonal.
ID for corresponding fetal liver donor tissue as the source for CD34+ HPCs as described in Table 1.
Spleen of a HU-NSG mice reconstituted with CD34+ HPCs derived from donor matched fetal liver tissue.
ATL cell line serially propagated in NOD/SCID mice.28
HTLV-1 in vitro–transformed cell line.
LVs expressing Tax1 induce T-cell lymphomas
LVs, which coexpress HTLV-1 tax and green fluorescent protein (GFP; pHR'CMV-Tax1/GFP), have previously been described22-24 and were used to transduce CD34+ HP/HSCs ex vivo before inoculation into NOD/SCID mice. Tax1-HU-NOD/SCID mice developed CD4+ malignancies with a slightly accelerated kinetics in comparison with HTLV-1-HU-NOD/SCID mice (Table 1; Figure 2). Some of the primary Tax1-generated lymphomas demonstrated monoclonality by TCR rearrangement patterns (Table 2). Tax1-induced lymphomas expressed GFP and demonstrated tissue distribution patterns similar to HTLV-1–induced lymphomas (Figure 2). HU-NOD/SCID mice reconstituted with CD34+ cells transduced with LVs encoding the Tax1 gene in the anti-sense orientation did not show any pathology (data not shown). This finding demonstrates that constitutive expression of Tax1 in CD34+ cells results in CD4+ lymphomas, which develop more rapidly into monoclonal proliferations, in contrast to HTLV-1–induced lymphomas. In addition, 100% (4/4 lymphoma cell lines) of Tax1-induced lymphomas engrafted and formed malignancies after intraperitoneal inoculation into naive NOD/SCID mice.
HTLV-1 infection induces hyperproliferation of HSCs in BM
We hypothesize that HTLV-1 infects CD34+ HP/HSCs and that the BM milieu allows productive expansion of HTLV-infected HSCs in vivo. HTLV-1-HU-NOD/SCID mice demonstrated 2- to 3-fold greater levels of infected CD34+/CD38− HSCs in the BM in comparison with HU-NOD/SCID mice generated from the same donor CD34+ cells (Figure 3). This finding demonstrates that HTLV-1 infection induces expansion of infected stem cell subpopulations in the BM and that the BM milieu may provide a suitable environment conducive for establishment of an HTLV-1–infected leukemic stem cell (ILSC).
HTLV-1 proviral sequences in CD34+ cells from patients
To determine whether HTLV-1 infection of CD34+ HP/HSCs is biologically relevant and detected in patient-derived CD34+ HP/HSCs, CD34+ and CD34− cells were purified from peripheral blood lymphocytes of human T-lymphotropic virus type I–associated myelopathy/tropical spastic paraparesis and HTLV-1–seropositive patients by fluorescence-activated cell sorting and subjected to real-time PCR by the use of primers specific for HTLV genes (Table 3). HTLV-1 proviral sequences were detected by PCR for all patient samples, and proviral loads were relatively elevated in purified CD34+ cells in comparison with CD34− cells, suggesting that HP/HSCs preferentially harbor HTLV proviral sequences in patients.
HTLV-1 proviral genes detected in CD34+ HP/HSCs from patients
| Patient details . | Viral copies per 106 PBMCs* . | HTLV-1 proviral genes detected in purified CD34+ cells† . | Clinical diagnosis . | |||
|---|---|---|---|---|---|---|
| ID . | Age, y/sex . | Gag . | Env . | Tax . | ||
| Uninfected | 31/male | ND | − | − | − | None |
| 7854/5885 | ND/ND | ND | ++ | − | ++ | HTLV-1 asymptomatic carrier |
| DB-14 (2 samples) | ND/female | 0 | − | − | − | HTLV-1, asymptomatic carrier |
| AT PBLS | ND/female | ND | − | + | ++ | HTLV-1, TSP/HAM |
| MCB | ND/ND | ND | + | + | ++ | TSP/HAM |
| NXB | ND/ND | ND | − | + | ++ | TSP/HAM |
| LXA | ND/ND | ND | + | + | ++ | TSP/HAM |
| WJ (3 samples) | ND/female | 18 112 | + | + | + | HTLV-1, myelopathy |
| 224/GS (2 samples) | 47/female | 205 000 | ++ | ++ | ++ | HTLV-1, uveitis |
| 6 | 45/female | 598 574 | − | ++ | ++ | HIV/HTLV-1, asymptomatic |
| 261 | 60/male | 501 272 | + | ++ | ++ | HIV/HTLV-1, asymptomatic |
| 88 | 72/male | 77 501 | − | ++ | ++ | HIV/HTLV-1, peripheral neuropathy |
| 154 | 38/female | 29 333 | − | ++ | ++ | HIV/HTLV-2, peripheral neuropathy |
| 76 | 45/male | 404 692 | ++ | ++ | ++ | HIV/HTLV-1, gait ataxia |
| 93 | 47/male | 328 507 | ++ | ++ | ND | HIV-1/HTLV-1, stroke |
| Patient details . | Viral copies per 106 PBMCs* . | HTLV-1 proviral genes detected in purified CD34+ cells† . | Clinical diagnosis . | |||
|---|---|---|---|---|---|---|
| ID . | Age, y/sex . | Gag . | Env . | Tax . | ||
| Uninfected | 31/male | ND | − | − | − | None |
| 7854/5885 | ND/ND | ND | ++ | − | ++ | HTLV-1 asymptomatic carrier |
| DB-14 (2 samples) | ND/female | 0 | − | − | − | HTLV-1, asymptomatic carrier |
| AT PBLS | ND/female | ND | − | + | ++ | HTLV-1, TSP/HAM |
| MCB | ND/ND | ND | + | + | ++ | TSP/HAM |
| NXB | ND/ND | ND | − | + | ++ | TSP/HAM |
| LXA | ND/ND | ND | + | + | ++ | TSP/HAM |
| WJ (3 samples) | ND/female | 18 112 | + | + | + | HTLV-1, myelopathy |
| 224/GS (2 samples) | 47/female | 205 000 | ++ | ++ | ++ | HTLV-1, uveitis |
| 6 | 45/female | 598 574 | − | ++ | ++ | HIV/HTLV-1, asymptomatic |
| 261 | 60/male | 501 272 | + | ++ | ++ | HIV/HTLV-1, asymptomatic |
| 88 | 72/male | 77 501 | − | ++ | ++ | HIV/HTLV-1, peripheral neuropathy |
| 154 | 38/female | 29 333 | − | ++ | ++ | HIV/HTLV-2, peripheral neuropathy |
| 76 | 45/male | 404 692 | ++ | ++ | ++ | HIV/HTLV-1, gait ataxia |
| 93 | 47/male | 328 507 | ++ | ++ | ND | HIV-1/HTLV-1, stroke |
CD34+ cells were purified by fluorescence-activated cell sorting from a total of 105 to 106 PBMCs from patients. The percentage of CD34+ cells ranged from 0.3% to 1.0% of total PBMCs. High-molecular-weight DNA was isolated and analyzed by PCR for individual HTLV genes.
HP/HSCs indicates hematopoietic progenitor/stem cells; HTLV-1, human T-lymphotropic virus type 1; ND, no data available; PBMCs, peripheral blood mononuclear cells; PCR, polymerase chain reaction; TSP/HAM, tropical spastic paraparesis/human T-lymphotropic virus type I–associated myelopathy; −, < 1 viral gene copy detected per 103 human cells; +, 1 viral copy per 1-103 human cells; and ++, > 1 viral copy per 1 human cell.
HTLV virus load per 1 × 106 patient PBMCs.
Mean of HTLV-1 proviral copies calculated per CD34+ cell, purified by fluorescence-activated cell sorting.
Discussion
We describe the development of a novel humanized mouse model that recapitulates ATLL development and speculate that HTLV-1 infection of human CD34+ HP/HSCs contributes to the initiation of lymphomagenesis and the manifestation of this malignant disease. HTLV-1 infection of key target HP/HSCs in infants infected at birth has been proposed to be a predisposing factor in the development of leukemia/lymphoma.14,20,23,29,30 Allogeneic BM transplantation involving HTLV-1–infected stem cell donors has been associated with virus transmission,31 implicating HP/HSCs as harboring viral infection. Reconstitution of hematopoiesis in NOD/SCID mice with the use of HTLV-1–infected or Tax1-transduced human CD34+ HP/HSCs reproducibly recapitulates mature T-cell lymphomas with characteristic genetic, immunologic, histologic, pathologic, and clinical features of ATLL. It is notable that mature CD4+ T-cell lymphomas are established in Tax1-HU-NOD/SCID mice with the use of an LV that is not exclusively targeted to the human T-cell lineage. Recent reports of a Tax-transgenic mouse model demonstrates in vivo transformation of immature and mature T cells32,33 and a role for cancer stem cells in the generation of Tax-induced T-cell malignancy.30 It is noteworthy that HTLV-1 viral gene expression and Tax1 expression in lymphomas induced in HU-SCID mice are not reflective of the silencing of viral gene expression demonstrated by primary ATLL cells from patients. It is also important to note that the lymphomas initiate and progress in the absence of any major immune selective mechanisms in the HU-SCID mouse and that it has not yet been determined whether hallmarks of ATLL, such as hypercalcemia and constitutive NF-κB activation, are recapitulated in the HU-SCID mouse model. Moreover, the short time frame in the manifestation of these lymphomas in this small animal model could suggest that the progression to malignancy in HU-SCID mice may ultimately fail to reflect secondary mutations or immune evasion mechanisms that may be involved in the full progression to ATLL in human patients. Despite these shortcomings, this chimeric human-mouse model will be an important tool to systematically recapitulate ATLL disease and dissect HTLV-1 determinants of pathogenesis and oncogenesis.
Previous attempts to directly infect mature human T cells in the thymus-liver conjoint organ in HU-SCID mice with HTLV-1 failed to induce malignancy.21 This finding suggests that the combination of the appropriate immature hematopoietic target cells in the BM and the immunologically defective environment of the SCID mouse are both required for the manifestation of the ATLL phenotype. Our data demonstrate that infection of CD34+ HP/HSCs is a prerequisite to T-cell lymphoma development in HTLV-1-HU-SCID mice. CD34+ HP/HSCs represent a heterogeneous cell population encompassing early and committed multipotent progenitor cells, noncommitted differentiating cells and stem cells.34 Although Melkus et al35 failed to detect human T-cell development in humanized NOD/SCID mice, our ability to detect human T-cell maturation in NOD/SCID mice receiving freshly isolated FL-derived CD34+ cells is in agreement with other reports19,26 and suggests FL-derived CD34+ HP/HSCs are more pluripotent in robustly reconstituting lymphopoiesis.
HTLV-1 infection of CD34+ HP/HSCs dramatically skews hematopoiesis toward T-cell development, presumably at the expense of other hematopoietic lineages. It has recently been shown that HTLV-1 infection in HP/HSCs results in a perturbation of miRNA expression, resulting in the skewing of hematopoiesis toward T-cell lineages.36 Alternatively, the ability of HTLV-1–transformed T cells to preferentially proliferate in vivo may overshadow normal hematopoietic patterns. Clearly, HTLV-1 infection of HP/HSCs plays a pivotal role in the initiation and accelerated progression of malignancy during the course of HTLV-1 disease, and the immunodeficient environment in the SCID mouse allows for the rapid manifestation of a T-cell malignancy. It is interesting that HTLV-1 infection primarily induced oligoclonal T-cell proliferations in contrast to the predominantly monoclonal T-cell malignancies induced by LV-Tax1 expression. This finding could imply that transformation by HTLV-1 infection in HU-NOD/SCID mice occurs across a broader number of infected CD34+ cells or that numerous infected CD34+ HP/HSCs can progress into a T-cell malignancy. Alternatively, overexpression of Tax1 by the CMV promoter by LV may be a relatively strong selective event driving the expansion of a monoclonal, dominant transformed T-cell clone in comparison with HTLV-1–mediated transformation. The ability of the LV-Tax vector to induce lymphomagenesis relatively rapidly in comparison with HTLV-1 infection also may reflect the different dynamics involving transduction of stem cells with a LV in comparison with infection with HTLV-1. Clearly, evaluation of stages of transformation in vivo and quantitative analyses of Tax1 levels will be critical in deciphering the kinetics and mechanics of in vivo transformation by HTLV-1 infection and Tax1 expression.
HTLV-1 proviral DNA has previously been detected in cord blood samples, generally a rich source of HSCs.37 Our results demonstrate that CD34+ cells from patients consistently harbor HTLV-1 proviral DNA at relatively high levels in comparison with other hematopoietic lineages. It was previously reported that HTLV-1 proviral sequences were not detected in CD34+ BM cells from ATLL patients.38 Although this observation appears to be in discordance with our results, we speculate that ATLL patient BM-derived CD34+ cells are not truly representative of early acute stages of HTLV-1 infection in stem cells in neonates or pediatric cases but, rather, represent an end-stage disease process that may not recapitulate the BM environment of an early-stage infection. Analysis of BM samples for pediatric HTLV-1 infections would be essential in confirming the hypothesis that HTLV-1 infection enters and is sequestered in the CD34+ cells in the BM.
Manifestation of ATLL in patients generally occurs decades after infection, and it is most likely that HTLV-1 latently infects BM stem cells that are sequestered from immunologic surveillance. It is conceivable that initiation of the leukemogenesis in HSCs involves the generation of an ILSC that eventually manifests into the monoclonal ATLL malignancy. Clearly, the expansion of HTLV-1–infected CD34+/CD38− HSCs in the BM of HTLV-1-HU-NOD/SCID mice is consistent with the fact that infection increases the absolute numbers of retrovirally infected stem cells in the milieu of the BM environment, although it should be noted that this has not yet been demonstrated in human pediatric patients. To definitively establish whether HTLV-1 induces generation of ILSCs, we are currently testing whether BM-derived CD34+ cells purified from HTLV-1-HU-NOD/SCID mice are capable of re-establishing ATLL after transplantation.
The HU-NOD/SCID mouse model of ATLL provides a novel tool for characterizing the role of proviral integration sites and somatic mutations associated with induction of lymphoid malignancies and will also allow evaluation of the contribution of the HTLV-1 auxiliary genes in viral pathogenesis. Moreover, this model will allow preclinical therapeutic evaluation studies to be evaluated for a fatal malignancy that lacks effective treatment regimens.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA124595 to G.F. and P01 CA100730 to M.D.L.) and by the Empire State Stem Cell Fund through New York State Department of Health Contract (NYSTEM #C023059 and #N08G-127) to G.F.
This article is dedicated to the memory of William Harrington Jr, MD.
The opinions expressed here are solely those of the authors and do not necessarily reflect those of the Empire State Stem Cell Board, the New York State Department of Health, or the State of New York.
National Institutes of Health
Authorship
Contribution: P.B., A.T., and L.C. performed the majority of experiments involving generation of lentivirus vectors, infection of stem cells, and reconstitution of SCID mice; P.B., L.C., and M.S. performed flow cytometric analyses and PCR analysis of DNA samples recovered from mice; M.D.L. performed histologic analyses; J.C.R. and W.H. analyzed tumor cells for TCR rearrangement patterns; M.A.B. provided peripheral blood lymphocytes patient samples; and G.F. supervised the experiments, interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gerold Feuer, Department of Microbiology and Immunology, SUNY Upstate Medical University, Syracuse, NY 13210; e-mail: feuerg@upstate.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal