In this issue of Blood, Stam and colleagues use gene expression profiling to provide new insights into the diverse types of ALL in infants.1
Acute lymphoblastic leukemia (ALL) diagnosed in infants has attracted a very high level of basic and clinical research, with 4800 citations currently in PubMed. There are several reasons for this high level of interest, including prenatal origins, mixed-lineage phenotypes, and clear demonstration of transcription factor dysregulation, especially involving MLL fusion genes.2,3
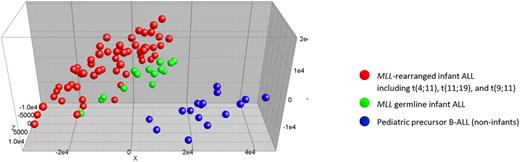
Unsupervised principal component analysis (PCA) of gene-expression profile shows that infants with germline/wild-type MLL (green dots) clustered closely to MLL-rearranged infant cases (red dots). Noninfant B precursor ALL cases (blue dots) were separable from both types of infant ALL. See the complete figure by Stam et al beginning on page 2835.
Unsupervised principal component analysis (PCA) of gene-expression profile shows that infants with germline/wild-type MLL (green dots) clustered closely to MLL-rearranged infant cases (red dots). Noninfant B precursor ALL cases (blue dots) were separable from both types of infant ALL. See the complete figure by Stam et al beginning on page 2835.
Although infant ALL is relatively uncommon, it still has a grim prognosis. The study by Stam et al evaluated a large number of infants (73) from the INTERINFANT 99 study4 with gene expression profiling and MLL gene rearrangement status. The investigators provide convincing evidence that MLL-rearranged infants, MLL germline infants, and MLL germline noninfant children can be distinguished based on gene expression profiling. Principal component analysis (PCA) showed that the MLL germline infants clustered closely to (but could be separated from) MLL-rearranged infant cases (see figure). The close clustering of infant ALL regardless of MLL status shows gene expression similarities that are likely to be important to the biology of these leukemias.
Previous studies of MLL-rearranged ALL have emphasized the expression of high levels of HOXA genes, for example, HOXA9.5 This study showed that not all cases express HOXA genes. Interestingly, although the sample size is small, the study suggests that infants with HOXA-negative MLL-rearranged ALL may have a very high incidence of relapse.
Where does this new information lead us? First, the clustering of all infant ALL separate from noninfant ALL demonstrates that “infant origin” is a biologic variable in itself, suggesting that leukemia in infants originates in different cells than in noninfants. The diversity within infant ALL is largely attributable to MLL rearrangements and within the MLL-rearranged cases, with different fusion partners likely to lead to different biologic characteristics. A number of studies using genetically engineered mice support this hypothesis.6,7 The possibility that human MLL-AF4 leukemia has a different cellular origin than other MLL fusion leukemia is further strengthened by a recent study showing the presence of the MLL fusion gene in mesenchymal stromal cells from MLL-AF4 but not other MLL fusion gene combinations.8 The gene-profiling data from the current and related studies can now be used to study the “cell of origin” issue in detail, with the caveat that the gene expression profile of the bulk leukemia population may differ from that of the cell in which the initial genetic “hit” (eg, a translocation-induced MLL fusion) occurred.
Second, this new information may give leads to help tailor therapies to improve outcomes in this group of patients where the outcomes are not currently satisfactory. If other groups confirm that expression profiles within the MLL fusion group predicts prognosis (eg, HOX cluster genes in this study) or more generally that other genes are uniquely important to the poor outcomes in infants regardless of MLL status, better targeted therapies should result. We certainly are in the very early stages of this journey.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal