Abstract
Acute lymphoblastic leukemia (ALL) in infants (< 1 year) is characterized by a poor prognosis and a high incidence of MLL translocations. Several studies demonstrated the unique gene expression profile associated with MLL-rearranged ALL, but generally small cohorts were analyzed as uniform patient groups regardless of the type of MLL translocation, whereas the analysis of translocation-negative infant ALL remained unacknowledged. Here we generated and analyzed primary infant ALL expression profiles (n = 73) typified by translocations t(4;11), t(11;19), and t(9;11), or the absence of MLL translocations. Our data show that MLL germline infant ALL specifies a gene expression pattern that is different from both MLL-rearranged infant ALL and pediatric precursor B-ALL. Moreover, we demonstrate that, apart from a fundamental signature shared by all MLL-rearranged infant ALL samples, each type of MLL translocation is associated with a translocation-specific gene expression signature. Finally, we show the existence of 2 distinct subgroups among t(4;11)–positive infant ALL cases characterized by the absence or presence of HOXA expression, and that patients lacking HOXA expression are at extreme high risk of disease relapse. These gene expression profiles should provide important novel insights in the complex biology of MLL-rearranged infant ALL and boost our progress in finding novel therapeutic solutions.
Introduction
In recent years, genome-wide assessment of gene activity has proven to be of great value in tumor classification as well as in identifying unique gene expression signatures associated with drug response, prognosis, metastasis, angiogenesis, and tumorigenesis. In pediatric acute lymphoblastic leukemia (ALL), oligonucleotide microarray analyses have been shown to accurately predict 6 major prognostic and genetically distinct patient groups, including specific precursor B-cell lineage subtypes characterized by E2A-PBX1, BCR-ABL, TEL-AML1, and MLL translocations, or hyperdiploidy (> 50 chromosomes), and T-cell lineage ALL (T-ALL).1-3 In addition, our laboratory recently identified a novel subgroup among children with genetically yet unclassified precursor B-ALL.3 In other studies, we demonstrated how gene expression profiling can identify unique gene expression signatures associated with resistance to prednisone, vincristine, L-asparaginase, and daunorubicin in pediatric ALL.4,5 Moreover, these gene expression signatures appeared to be highly predictive for clinical outcome for the patients under investigation as well as in a completely independent patient cohort.4
Among the different genetic subgroups of pediatric ALL, MLL-rearranged ALL represents the most unfavorable type of leukemia and is most frequently diagnosed in infants (ie, children younger than 1 year). In infant ALL, approximately 80% of the cases are typified by leukemia-specific chromosomal translocations involving the Mixed Lineage Leukemia (MLL) gene,6 fusing the N-terminal portion of MLL to the C-terminal region of one of its many translocation partner genes. By far the most frequent MLL translocations found among infant ALL patients are t(4;11), t(11;19), and t(9;11),7,8 giving rise to the fusion proteins MLL-AF4, MLL-ENL, and MLL-AF9, respectively. These chimeric MLL fusion proteins exhibit pronounced transforming capacities9 and independently contribute to an unfavorable prognosis.7,10 To date, event-free survival rates for MLL-rearranged infant ALL range between 20% and 50%, depending on the treatment protocol.7 Approximately 20% of the infant ALL patients carry germline (or wild-type) MLL genes, and nowadays have a far better prognosis with event-free survival chances of 75% to 95%.7,11
Multiple microarray studies demonstrated that MLL translocations specify a distinct gene expression profile that is clearly distinguishable from other ALL subtypes and from acute myeloid leukemia (AML).1-3,12,13 Moreover, Zangrando et al recently reported a gene expression signature commonly shared by MLL-rearranged ALL and AML patients, identifying dysregulated genes specifically associated with the MLL translocation, irrespective of the type of leukemia.14 In most of these studies, however, rather small numbers of MLL-rearranged ALL samples were analyzed as a uniform patient group, regardless of the type of MLL translocation. Nevertheless, MLL-rearranged ALL may well represent heterogeneous biologic entities characterized by a fundamental gene expression profile shared by all patients despite the MLL fusion partner, whereas underlying expression signatures may discriminate between the different types of MLL translocations. To test this, we generated and analyzed gene expression profiles in a relatively large cohort of MLL-rearranged infant ALL samples, and indeed reveal the existence of specific gene expression signatures associated with the different MLL translocations frequently found in infant ALL. Furthermore, we sought to determine whether infant ALL patients carrying germline (or wild-type) MLL genes display gene expression profiles that resemble those of childhood ALL patients older than one year of age (noninfants), or whether these patients form yet another genetically distinct ALL subgroup, and concluded the latter. Finally, we show that, among t(4;11)-positive infant ALL cases, 2 distinct subgroups can be identified based on the absence or presence of HOXA9, HOX10, HOXA7, HOXA5, and HOXA3 expression, and show dramatic differences in relapse-free survival.
Methods
Patient samples
Bone marrow or peripheral blood samples from untreated infants (younger than 1 year) diagnosed with ALL were collected at the Erasmus MC–Sophia Children's Hospital and other institutes participating in the recently published international collaborative INTERFANT-99 treatment protocol.7 Samples from pediatric ALL patients older than 1 year (ie, noninfants) were selected from our cell bank. For all primary patient samples used in this study, approval was obtained from the Erasmus MC Institutional Review Board, and authorization was acquired from the parents or legal guardians of the children via informed consent in accordance with the Declaration of Helsinki. Patient characteristics are listed in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Sample preparation
All samples were processed within 24 hours after sampling as described recently.15 Briefly, mononuclear cells were isolated by density gradient centrifugation using Lymphoprep (Nycomed Pharma), and nonleukemic cells were removed using immunomagnetic beads.16 All leukemia samples used in this study contained more than 90% leukemic cells, as determined morphologically on May-Grünwald-Giemsa (Merck)–stained cytospins.
Gene expression profiles
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions, and quantified on a Nanodrop ND-1000 spectrophotometer (Isogen). The integrity of the extracted RNA was assessed on an Agilent 2100 Bioanalyzer (Agilent). High-quality RNA was reverse transcribed using T7-linked oligo-dT primers, and the obtained cDNA was used as a template to synthesize biotinylated cRNA. Labeled cRNA was then fragmented and hybridized to HU133plus2.0 GeneChips (Affymetrix) according to the manufacturer's guidelines. Raw microarray data for all the patients used in this study are listed in supplemental Table 2. Moreover, the infant ALL gene expression data presented in this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus17 and is accessible via GEO Series accession number GSE19475. The gene expression data for the pediatric precursor B-ALL samples were deposited as GSE13351 as part of a recently published study.3
Quantitative real-time PCR analyses
Total RNA was extracted from a minimum of 5 × 106 leukemic cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The quality of the extracted RNA was assessed on 1% agarose gels. Extracted RNA was reverse transcribed as described before,18 and the obtained cDNA was used to quantify mRNA expression of HOXA9, HOXA7, HOXA5, HOXA10, and HOXA3 relative to the housekeeping gene B2M (encoding β-2-microtubulin), using quantitative real-time polymerase chain reaction (PCR). For this, PCR products were amplified using the DyNAmo SYBR Green qPCR kit (Finnzymes) according to the manufacturer's recommendations, using SYBR Green as a fluorophore to detect transcripts on an ABI Prism 7900 sequence detection system (Applied Biosystems).
Statistical analyses
Raw array data were collectively normalized using variance-stabilizing normalization,19 and differential gene expression was statistically evaluated using linear models for microarray analyses.20,21 Differences in gene expression were deemed significant at P values (adjusted for multiple testing according to the step-up procedure of Benjamini22 ) of less than .01 (ie, false discovery rate [FDR] < 0.01). All statistical analyses were performed in the statistical environment R using Bioconductor packages. Heatmaps were generated in GenePattern,23 and graphical representations of principal component analyses (PCA) were produced using the GeneMath XT 1.6.1 software (Applied Maths).
As a measure of internal validation for the subtype-specific gene expression signatures, the global test24 was applied to evaluate whether gene lists were significantly associated with a certain patient group. In all instances, the global test indicated that the expression of all selected probe sets was significantly associated with the corresponding patient group. To produce informative representations of discriminative probe sets, we chose to visualize the top 50 most significantly overexpressed probe sets for each subgroup in each comparison.
Results
MLL-rearranged infant ALL versus pediatric precursor B-ALL: dataset validation
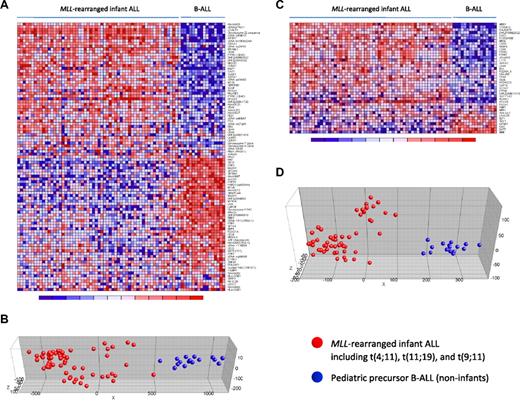
Nowadays, proper validation of gene expression profiling data is achieved either by a double-loop cross-validation procedure in which the sample population is divided into a training and a test set,3 or by confirming differential gene expression in a truly independent patient cohort (eg, Holleman et al4 ). However, infant ALL is a rare malignancy, and collecting an adequate number of samples to apply such validations remains difficult, even in our INTERFANT-99 patient cohort that currently represents the largest collection of infant ALL samples. Therefore, to avoid reduction of the sample size and maintain sufficient statistical power, we here adopted 2 recently published expression signatures that separate MLL-rearranged ALL from other ALL subtypes, based on which we used our samples as an independent patient cohort to validate the integrity of our dataset. The first signature was reported by Armstrong et al,12 and represents 100 probe sets most significantly discerning between MLL-rearranged ALL (n = 17) and conventional precursor B-ALL samples. The second was published by Yeoh et al,2 and composes 40 genes that distinguished pediatric MLL-rearranged ALL (n = 20) from all other known genetic subtypes of childhood ALL, including E2A-PBX1, BCR-ABL, and TEL-AML1 positive or hyperdiploid (> 50 chromosomes) B-ALL, and T-ALL. As both of these studies were performed on Affymetrix HU95A microarrays (containing 12 600 probe sets), we assessed the corresponding probe sets on the HU133plus2.0 arrays (containing 54 675 probe sets) and determined their discriminative capacity on our samples. For the MLL-rearranged ALL signature by Armstrong et al,12 97 probe sets (HU133plus2.0) could be identified to correspond with the 100 probe sets (HU95A) in the original signature. For the signature reported by Yeoh et al,2 all corresponding probe sets were found. Both signatures clearly separated our MLL-rearranged infant ALL patients (n = 59), consisting of t(4;11) (n = 29), t(11;19) (n = 22), and t(9;11)-positive (n = 8) cases, from our pediatric precursor B-ALL (n = 16) samples (Figure 1). To exclude influences from subtype-specific gene expression signatures underlying pediatric ALL, we intentionally selected B-ALL samples from children older than one year of age that could not be assigned to any of the major genetic ALL subtypes. Of the 97 probe sets corresponding to the MLL-rearranged ALL signature by Armstrong et al,12 80 probe sets (∼ 82%) were significantly differentially expressed (FDR < 0.01) between our MLL-rearranged infant ALL and B-ALL samples. For the signature by Yeoh et al,2 32 of the 40 probe sets (80%) were expressed differentially (FDR < 0.01). Probe set identifications and descriptions, gene names, log-fold changes, and P values are listed in supplemental Tables 3 and 4, respectively.
MLL-rearranged infant ALL versus pediatric precursor B-ALL (HU95A): dataset validation. Heatmaps separating our MLL-rearranged infant ALL (n = 59) from pediatric precursor B-ALL (n = 16) samples based on the MLL-rearranged ALL specific gene expression signatures (obtained on HU95A microarrays) published by Armstrong et al12 (A) and Yeoh et al2 (C). Columns represent patient samples, and rows represent the gene names corresponding to the probe sets. Normalized gene expression is depicted in red (high expression) or blue (low expression). (B,D) PCA for both signatures, respectively. Red dots indicate MLL-rearranged infant ALL samples (including t(4;11) (n = 29), t(11;19) (n = 22), and t(9;11)-positive (n = 8) cases), and blue dots represent pediatric precursor B-ALL cases (n = 16). Patient characteristics and detailed gene descriptions are listed in supplemental Tables 1, 3, and 4.
MLL-rearranged infant ALL versus pediatric precursor B-ALL (HU95A): dataset validation. Heatmaps separating our MLL-rearranged infant ALL (n = 59) from pediatric precursor B-ALL (n = 16) samples based on the MLL-rearranged ALL specific gene expression signatures (obtained on HU95A microarrays) published by Armstrong et al12 (A) and Yeoh et al2 (C). Columns represent patient samples, and rows represent the gene names corresponding to the probe sets. Normalized gene expression is depicted in red (high expression) or blue (low expression). (B,D) PCA for both signatures, respectively. Red dots indicate MLL-rearranged infant ALL samples (including t(4;11) (n = 29), t(11;19) (n = 22), and t(9;11)-positive (n = 8) cases), and blue dots represent pediatric precursor B-ALL cases (n = 16). Patient characteristics and detailed gene descriptions are listed in supplemental Tables 1, 3, and 4.
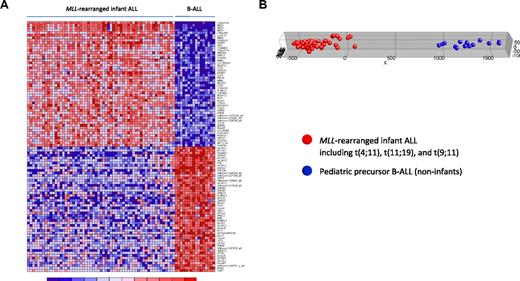
Given the superior number of probe sets on the HU133plus2.0 GeneChips used in the present study over the formerly used first-generation HU95A microarrays, we further explored whether this advantage results in a more pronounced class distinction than reported earlier. Comparing our gene expression profiles of MLL-rearranged infant ALL (n = 59) with those from pediatric B-ALL patients (n = 16), we found 14 246 of the 54 675 probe sets (∼ 26%) to be differentially expressed (FDR < 0.01), of which 6990 were up-regulated in MLL-rearranged infant ALL. Figure 2A shows a heatmap visualization of 100 probe sets most significantly up (n = 50) and down-regulated (n = 50) in MLL-rearranged infant ALL compared with pediatric precursor B-ALL. Probe set identifications and descriptions, gene names, log-fold changes, and P values are listed in supplemental Table 5. PCA revealed that using high-resolution HU133plus2.0 microarrays, by estimate covering the entire human genome, additional genes can be found that more clearly distinguish between MLL-rearranged ALL and conventional B-ALL than the signatures reported before (Figure 2B). For examples, probe sets corresponding to RLP38 (ribosomal protein L38), KCNK12 (potassium channel subfamily K member 12), and MDS027 (also known as HSPC300; hematopoietic stem/progenitor cell protein 300) are not present on HU95 microarrays but did appear among the 50 most significantly up-regulated genes in MLL-rearranged infant ALL samples in our HU133plus2.0-based data (Figure 2A). Of particular interest is the high-level expression of HSPC300, which was recently hypothesized to be associated with the metastatic potential of lung squamous cell carcinoma.25 As such, high level HSPC300 expression may well contribute to the aggressive nature of MLL-rearranged ALL and exemplifies how our HU133plus2.0-based gene expression profiles may further extend our insights in the biology of this malignancy. Given the vast amount of probe sets significantly up- or down-regulated in MLL-rearranged infant ALL, we used high-level HSPC300 expression merely as an example of presumably many genes that have not been associated with MLL-rearranged ALL before.
MLL-rearranged infant ALL versus pediatric precursor B-ALL (HU133plus2.0). (A) Heatmap showing the separation of MLL-rearranged infant ALL (n = 59) from pediatric precursor B-ALL (n = 16) samples based on the 100 probe sets most significantly discriminative between both patient groups as attained in our analyses using HU133plus2.0 GeneChips. Columns represent patient samples, and rows represent the gene names corresponding to the probe sets. Normalized gene expression is depicted in red (high expression) or blue (low expression). The top 50 probe sets are relatively overexpressed and the bottom 50 probe sets relatively underexpressed in MLL-rearranged infant ALL (which include t(4;11) (n = 29), t(11;19) (n = 22), and t(9;11)-positive (n = 8) cases). (B) Graphic representation of PCA based on this gene expression signature, separating the MLL-rearranged infant ALL (red dots) from pediatric precursor B-ALL (blue dots) samples.
MLL-rearranged infant ALL versus pediatric precursor B-ALL (HU133plus2.0). (A) Heatmap showing the separation of MLL-rearranged infant ALL (n = 59) from pediatric precursor B-ALL (n = 16) samples based on the 100 probe sets most significantly discriminative between both patient groups as attained in our analyses using HU133plus2.0 GeneChips. Columns represent patient samples, and rows represent the gene names corresponding to the probe sets. Normalized gene expression is depicted in red (high expression) or blue (low expression). The top 50 probe sets are relatively overexpressed and the bottom 50 probe sets relatively underexpressed in MLL-rearranged infant ALL (which include t(4;11) (n = 29), t(11;19) (n = 22), and t(9;11)-positive (n = 8) cases). (B) Graphic representation of PCA based on this gene expression signature, separating the MLL-rearranged infant ALL (red dots) from pediatric precursor B-ALL (blue dots) samples.
MLL germline infant ALL represents a unique subtype of childhood ALL
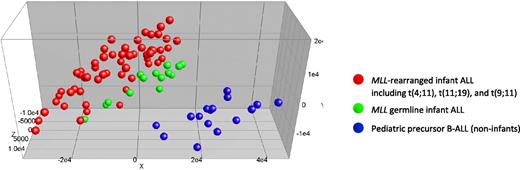
Next we asked whether infant ALL patients bearing germline MLL genes simply represent pediatric ALL patients of very young age (ie, < 1 year) or whether these patients compose an isolated ALL subgroup different from other known ALL subtypes. Therefore, we compared gene expression profiles of MLL germline infant ALL samples (n = 14) to those of the MLL-rearranged infant ALL (n = 59) and the pediatric precursor B-ALL samples (n = 16), lacking known genetic abnormalities. Initially, we performed a PCA, using all 54 675 probe sets present on the HU133plus2.0 GeneChip, without any selection. This unsupervised analysis roughly separated the germline MLL infant ALL samples from both the MLL-rearranged infant ALL and pediatric precursor B-ALL samples (Figure 3). Remarkably, the MLL germline infant ALL samples as a group clustered tightly to, but separately from, the MLL-rearranged infant ALL samples, and clearly away from the pediatric precursor B-ALL samples. Thus, apart from the presence of MLL translocations, young age (< 1 year), characteristically shared by all infants either carrying rearranged or germline MLL genes, also influenced this clustering (Figure 3).
Unsupervised clustering analysis of MLL-rearranged infant ALL, MLL germline infant ALL, and pediatric precursor B-ALL. Completely unsupervised clustering analysis (PCA) of MLL-rearranged infant ALL (n = 59; red dots), MLL germline (wild-type MLL) infant ALL (n = 14; green dots), and pediatric precursor B-ALL (n = 16; blue dots) samples, using all 54 675 probe sets present on the HU133plus2.0 GeneChip.
Unsupervised clustering analysis of MLL-rearranged infant ALL, MLL germline infant ALL, and pediatric precursor B-ALL. Completely unsupervised clustering analysis (PCA) of MLL-rearranged infant ALL (n = 59; red dots), MLL germline (wild-type MLL) infant ALL (n = 14; green dots), and pediatric precursor B-ALL (n = 16; blue dots) samples, using all 54 675 probe sets present on the HU133plus2.0 GeneChip.
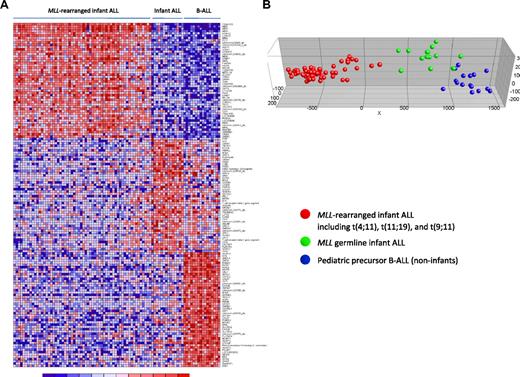
Subsequently, to explore whether specific expression profiles could define these 3 patient groups more accurately, the 50 most significantly up-regulated probe sets for each group (compared with the other 2 groups combined) were selected. Differential expression of these most discriminative probe sets is visualized in a heatmap (Figure 4A). Probe set identifications and descriptions, gene names, log-fold changes, and P values are listed in supplemental Table 6. As expected and consistent with our unsupervised analysis (Figure 3), PCA showed that these 150 probe sets (almost) completely separated the MLL germline infant ALL samples from both the MLL-rearranged infant ALL and the pediatric precursor B-ALL samples (Figure 4B).
Supervised clustering analysis of MLL-rearranged infant ALL, MLL germline infant ALL, and pediatric precursor B-ALL. (A) Heatmap visualizing differential gene expression separating MLL germline infant ALL (n = 14), from MLL-rearranged infant ALL (n = 59) and pediatric precursor B-ALL (n = 16) samples, based on the 50 most significantly up-regulated probe sets for each patient group (compared with the other patient groups combined). Columns represent patient samples, and rows represent the gene names corresponding to the probe sets. Normalized gene expression is depicted in red (high expression) or blue (low expression). (B) Graphical representation of the supervised clustering of the samples based on this expression signature. Red dots indicate MLL-rearranged infant ALL; green dots, MLL germline infant ALL; and blue dots, the pediatric precursor B-ALL samples.
Supervised clustering analysis of MLL-rearranged infant ALL, MLL germline infant ALL, and pediatric precursor B-ALL. (A) Heatmap visualizing differential gene expression separating MLL germline infant ALL (n = 14), from MLL-rearranged infant ALL (n = 59) and pediatric precursor B-ALL (n = 16) samples, based on the 50 most significantly up-regulated probe sets for each patient group (compared with the other patient groups combined). Columns represent patient samples, and rows represent the gene names corresponding to the probe sets. Normalized gene expression is depicted in red (high expression) or blue (low expression). (B) Graphical representation of the supervised clustering of the samples based on this expression signature. Red dots indicate MLL-rearranged infant ALL; green dots, MLL germline infant ALL; and blue dots, the pediatric precursor B-ALL samples.
MLL translocation–specific gene expression profiles among MLL-rearranged infant ALL patients
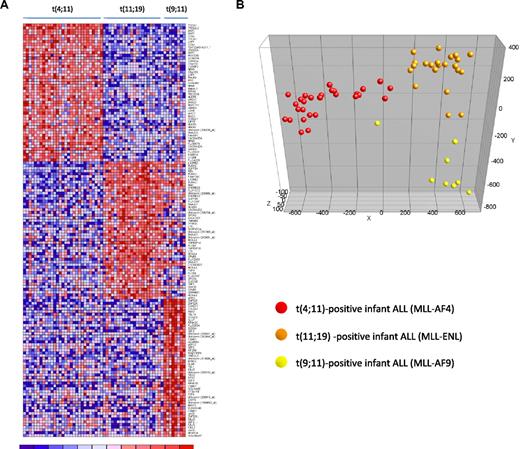
Accumulating evidence suggests that MLL translocations cause deregulated gene expression as a result of translocation-specific histone modifications, which may in part be influenced by the translocation partner gene.26,27 Therefore, we asked whether distinct gene expression profiles could be identified associated with the type of MLL translocation. For this we separated our MLL-rearranged infant ALL samples according to the type of translocation, ie, t(4;11) (n = 29), t(11;19) (n = 22), or t(9;11) (n = 8), and determined the differentially expressed probe sets for each subgroup (compared with the other 2 subgroups combined). In total, 1229 probe sets were significantly differentially expressed between the 3 MLL-rearranged subgroups (FDR < 0.01). Figure 5A shows a heatmap visualizing the 50 most significantly up-regulated probe sets for each of the MLL-rearranged subgroups. Probe set identifications and descriptions, gene names, log-fold changes, and P values are listed in supplemental Table 7. PCA showed that based on these 150 probe sets, t(4;11), t(11;19) and t(9;11)-positive infant ALL cases cluster completely separate form one another (Figure 5B).
Gene expression–based separation of MLL-rearranged infant ALL subtypes. (A) Heatmap demonstrating differential gene expression between t(4;11) (n = 29), t(11;19) (n = 22), and t(9;11)-positive (n = 8) MLL-rearranged infant ALL samples, based on the 50 most significantly up-regulated probe sets for each patient group (compared with the other patient groups combined). Columns represent patient samples, and rows represent the gene names corresponding to the probe sets. Normalized gene expression is depicted in red (high expression) or blue (low expression). (B) PCA plot clustering the t(4;11) (red dots), t(11;19) (orange dots), and t(9;11) (yellow dots) according to these 150 selected probe sets.
Gene expression–based separation of MLL-rearranged infant ALL subtypes. (A) Heatmap demonstrating differential gene expression between t(4;11) (n = 29), t(11;19) (n = 22), and t(9;11)-positive (n = 8) MLL-rearranged infant ALL samples, based on the 50 most significantly up-regulated probe sets for each patient group (compared with the other patient groups combined). Columns represent patient samples, and rows represent the gene names corresponding to the probe sets. Normalized gene expression is depicted in red (high expression) or blue (low expression). (B) PCA plot clustering the t(4;11) (red dots), t(11;19) (orange dots), and t(9;11) (yellow dots) according to these 150 selected probe sets.
Subdivision of t(4;11)-positive infant ALL based on the presence or absence of HOXA expression
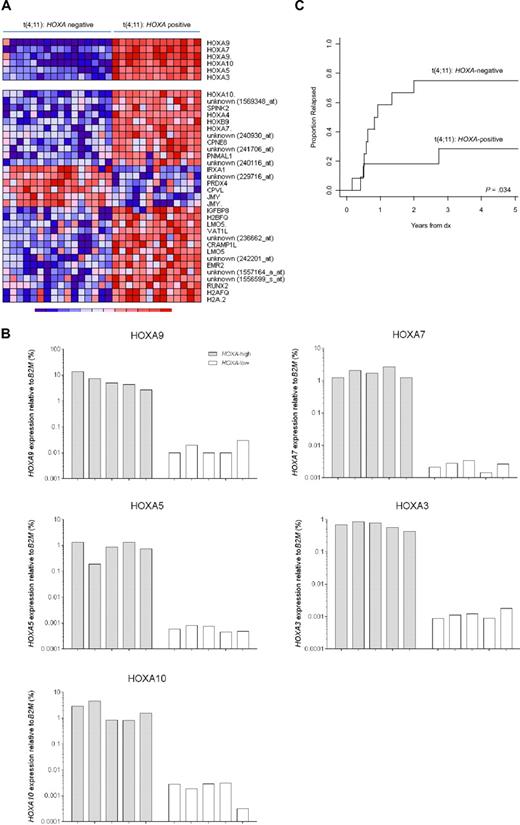
Finally, we asked whether gene expression profiles existed that subdivided MLL-rearranged infant ALL samples even among patients characterized by the same type of MLL translocation. Translocation t(4;11), giving rise to the MLL-AF4 fusion protein, is by far the most common MLL translocation among infant ALL patients (found in ∼ 50% of all cases).7 As such, t(4;11)-positive infant ALL represents the largest subgroup of MLL-rearranged infant ALL cases in this study. Therefore, we particularly chose our t(4;11)-positive gene expression profiles to explore differential gene expression among t(4;11)-positive infant ALL cases. For this, the SD of the expression of each probe set was calculated among all t(4;11)-positive cases (n = 29), to identify probe sets with the largest variation, possibly indicating differential expression among these patients. Surprisingly, 6 probe sets corresponding to HOXA9, HOXA7, HOXA10, HOXA5, and HOXA3, appeared to display pronounced standard deviations, and consistently separated 2 subgroups of t(4;11)-positive infant ALL samples uniformly characterized either by the presence (n = 13) or absence (n = 16) of HOXA expression (Figure 6A top panel). To validate these findings, quantitative reverse-transcribed PCR was applied to quantify HOXA9, HOXA7, HOXA10, HOXA5, and HOXA3 expression relative to the housekeeping gene B2M in primary t(4;11)-positive infant ALL samples characterized by either high (n = 5) or low (n = 5) HOXA expression (Figure 6B). Adopting this separation, we compared the gene expression profiles and identified an additional 31 probe sets to be differently expressed between these subgroups (Figure 6A bottom panel). Several of these probe sets represented other homeobox genes, such as HOXA4, HOXB9, and IRXA1 (or IRX1) or denoted additional probe sets for HOXA10 and HOXA7. Probe set identifications and descriptions, gene names, log-fold changes, and P values are listed in supplemental Table 8.
HOXA-based subclustering of t(4;11)-positive infant ALL samples. (A) Heatmap visualizing 2 clusters among t(4;11)-positive infant ALL samples (n = 29) based on the present or absent of HOXA9, HOXA10, HOXA7, HOXA5, and HOXA3 expression (upper panel). Apart from the 6 probe sets initially separating both patient groups, and additional 31 probe sets (lower panel) appeared to be significantly (FDR < 0.01) differentially expressed between HOXA-negative (n = 16) and HOXA-positive (n = 13) t(4;11)-positive infant ALL. (B) HOXA9, HOXA10, HOXA7, HOXA5, and HOXA3 expression as determined by quantitative reverse-transcribed PCR analyses in t(4;11)-positive infant ALL samples characterized by high (n = 5) or low (n = 5) HOXA expression according to the microarray data. (C) Relapse-free survival curves for HOXA-negative (n = 12) and HOXA-positive (n = 11) t(4;11)-positive infant ALL patients, demonstrating a significantly higher relapse incidence in t(4;11)-positive infant ALL patients lacking HOXA expression (P = .034). Because of a lack of data availability or exclusion of patients who died before entering the INTERFANT-99 treatment protocol, relapse-free survival could only be plotted for 23 of the 29 t(4;11)-positive infant ALL cases.
HOXA-based subclustering of t(4;11)-positive infant ALL samples. (A) Heatmap visualizing 2 clusters among t(4;11)-positive infant ALL samples (n = 29) based on the present or absent of HOXA9, HOXA10, HOXA7, HOXA5, and HOXA3 expression (upper panel). Apart from the 6 probe sets initially separating both patient groups, and additional 31 probe sets (lower panel) appeared to be significantly (FDR < 0.01) differentially expressed between HOXA-negative (n = 16) and HOXA-positive (n = 13) t(4;11)-positive infant ALL. (B) HOXA9, HOXA10, HOXA7, HOXA5, and HOXA3 expression as determined by quantitative reverse-transcribed PCR analyses in t(4;11)-positive infant ALL samples characterized by high (n = 5) or low (n = 5) HOXA expression according to the microarray data. (C) Relapse-free survival curves for HOXA-negative (n = 12) and HOXA-positive (n = 11) t(4;11)-positive infant ALL patients, demonstrating a significantly higher relapse incidence in t(4;11)-positive infant ALL patients lacking HOXA expression (P = .034). Because of a lack of data availability or exclusion of patients who died before entering the INTERFANT-99 treatment protocol, relapse-free survival could only be plotted for 23 of the 29 t(4;11)-positive infant ALL cases.
Interestingly, the relapse-free survival varied significantly between both subgroups (P = .034), with t(4;11)-positive infant ALL patients negative for HOXA expression being at extreme high risk of disease relapse (Figure 6C). The 1-year cumulative relapse incidence for HOXA-positive patients was 18.2% (± 12.3%) and for HOXA-negative patients 58.3% (± 15.4%). In a Cox model on the hazard of relapse, HOXA-negative t(4;11)-positive infant ALL patients had a significantly (P = .036) 4.17-fold increased hazard ratio (95% confidence interval, 1.10-15.81) compared with HOXA-positive patients. However, as indicated by the relatively large 95% confidence interval, these findings should be interpreted with caution because of the small sample size. Nonetheless, a possible explanation for the pronounced difference in relapse-free survival between both t(4;11)-positive patient groups may lie in the genes that discriminate between them. For example, high-level PRDX4 (Peroxiredoxin 4) expression, such as that found in HOXA-negative t(4;11)-negative infant ALL samples (Figure 6A), has been associated with metastasizing colon cancer.28 In case PRDX4 also contributes to tumor progression and metastasis in MLL-rearranged ALL, up-regulated PRDX4 expression may contribute to the worse outcome of HOXA-negative t(4;11)-negative infant ALL patients compared with patients who do show HOXA expression.
Discussion
MLL-rearranged ALL samples display unique and ample deregulated expression profiles that are clearly distinguishable from profiles found in other specific ALL subtypes.1-3,12,13 However, the number of MLL-rearranged infant ALL cases in these studies were small, inevitably leading to the analyses of these samples as a single patient group regardless of the type of MLL translocation. The most common MLL translocations among infant ALL patients are translocation t(4;11), t(11;19), and t(9;11), and the possible existence of specific gene expression profiles underlying these different MLL translocations remains unacknowledged. In addition, the aforementioned profiling studies made tremendous progress in classifying unique types of genetically distinct ALL subgroups, but infant ALL cases carrying germline MLL genes were never studied in these analyses. Therefore, the present study was designed to explore the possible existence of MLL translocation specific gene expression profiles, and evaluates how MLL germline infant ALL genetically relates to MLL-rearranged infant ALL and ALL in children older than 1 year.
Establishing the integrity of our data, we took 2 previously published gene expression profiles associated with MLL-rearranged ALL and applied these signatures to our MLL-rearranged infant ALL samples compared with pediatric precursor B-ALL samples. For both published signatures, approximately 80% of the probe sets in both of the signatures appeared significantly differentially expressed in our MLL-rearranged infant ALL samples, demonstrating that our dataset is consistent with other datasets reported earlier. The approximately 20% of the probe sets in both signatures that did not show differential expression in our samples may be explained by slight differences or biases in the composition of the patient cohorts in which these signatures were originally identified. For example, the signature reported by Armstrong et al,12 was based predominantly on t(4;11) and t(11;19)-positive cases, whereas no t(9;11)-positive cases were included. Moreover, this patient cohort also included MLL-rearranged ALL samples from children older than one year of age, as well as a few adult patients. Likewise, in the study of Yeoh et al,2 the inclusion criteria of MLL-rearranged ALL samples were solely based on the presence of an MLL translocation regardless of age. Our MLL-rearranged ALL cohort consists entirely of infants younger than one year in which all 3 common MLL translocations found among infant ALL patients are represented.
Given the superior number of probe sets on the HU133plus2.0 GeneChips (used in the present study) over the first generation HU95A chips used in earlier studies,2,12 we also compared MLL-rearranged infant ALL with MLL translocation-negative noninfant pediatric precursor B-ALL samples, based on our data. This comparison demonstrated that high-resolution HU133plus2.0 data are capable of separating these patient groups even more convincingly than already shown earlier and revealed differential expression of genes that have not been associated with MLL-rearranged ALL before, which may therefore provide further insights into this aggressive type of leukemia, on top of recent progress in understanding mechanism by which MLL fusions alter gene expression. The most important breakthrough in our comprehension of MLL translocation induced transformation has been the notion that, because of the loss of MLL-specific histone methyltransferase activity necessary for H3K4 methylation, MLL fusions recruit alternative histone methyltransferases (eg, DOT1L) that subsequently establish H3K79 methylation. In turn, H3K79 methylation results in accessible chromatin at inappropriate loci, allowing the abnormal en presumably pathogenic activation (expression) of associated genes.27,29 From this respect, MLL fusion proteins are often regarded as activating oncogenic molecules. In line with this assumption, we here show that approximately 7000 probe sets are significantly up-regulated in MLL-rearranged infant ALL compared with noninfant pediatric precursor B-ALL samples. On the other hand, we found an equal amount of probe sets to be significantly down-regulated in MLL-rearranged infant ALL, indicating that the considerably deregulated gene expression patterns in this disease are not necessarily characterized by an overrepresentation of activated genes but show that down-regulated gene expression is at least as common. In concordance with this, we recently found MLL-rearranged infant ALL samples to display vast amounts of genome-wide gene promoter methylation that appeared to be associated with the transcriptional silencing of the affected genes.30 Thus, whereas the mechanisms by which MLL fusions activate gene expression are currently being elucidated, the mechanisms by which MLL fusions deactivate gene expression remain to be studied.
As infant ALL samples carrying germline MLL genes have not yet been properly analyzed as a single patient group, we compared gene expression profiles of these patients against MLL-rearranged infant ALL and noninfant pediatric precursor B-ALL profiles. A completely unsupervised clustering analysis revealed that MLL germline infant ALL resembles neither MLL-rearranged infant ALL nor pediatric precursor B-ALL lacking known genetic abnormalities. Based on this unsupervised analysis using all probe sets present on the HU133plus2.0 GeneChip, the MLL germline infant ALL samples seem more closely related to MLL-rearranged infant ALL samples (of the same age) than to the precursor ALL samples, also carrying germline MLL genes, derived from children older than one year. This finding possibly reflects the influences of very young age, at which ALL (in the absence of MLL rearrangements) apparently develops after alternative mechanisms, giving rise to a characteristic gene expression profile. In other words, MLL germline infant ALL may represent a unique biologic entity. Alternatively, these patients could also display a gene expression profile that is more similar than one of the established ALL subtypes not included in the present study.
Since the observation that MLL-rearranged ALL displays a highly characteristic gene expression profile,12 scientists have been searching for the mechanisms driving deregulated transcription induced by the MLL fusion. As the MLL gene itself has specific histone methyltransferase activity,31,32 which is lost during fusion of MLL to one of its translocation partner genes, MLL translocations probably result in altered chromatin structures resulting from aberrant histone modifications. This may, to a large extent, explain the characteristic gene expression patterns uniformly associated with MLL-rearranged leukemia. However, influence of the translocation partner gene should not be ignored. A growing body of evidence implies that many of the MLL fusion partners are part of transcriptional regulation networks that also function through chromatin remodeling,33 and not necessarily lead to similar changes. For instance, although the recruitment of the histone methyltransferase DOT1L has been well established for MLL-AF4 fusions, it is certainly not unthinkable that other MLL fusion partners recruit histone methyltransferases other than DOT1L, leading to alternative chromatin modifications. In any case, apart from basal deregulation of gene expression driven by the interruption of the MLL gene that is shared by all MLL-rearranged leukemias, the fusion partner seems to determine additional changes in gene expression characteristic for the type of MLL translocation. As shown in the present study, MLL-rearranged infant ALL samples carrying translocations t(4;11), t(11;19), or t(9;11) indeed display translocation specific gene expression signatures that clearly separate these samples into 3 distinct patient groups. In line with these findings, we recently found that these different MLL translocations also specify distinct genome-wide promoter methylation patterns.30 Hypothetically, these data may collectively imply that MLL-rearranged leukemias transform by dramatically changing epigenetic landscapes induced and guided by the type of MLL fusion protein, which initially triggers abnormal chromatin remodeling and subsequently alters genome-wide DNA methylation patterns and transcription, all in favor of the development of leukemia.
Finally, we asked whether distinct gene expression profiles could also be hidden among infant ALL patients carrying the same type of MLL translocation. Interestingly, we found the presence of 2 separate clusters among our t(4;11)-positive ALL samples, distinguishable by either the presence or absence of HOXA9, HOXA7, HOXA10, HOXA5, and HOXA3 expression. Moreover, the separation of both t(4;11)-positive infant ALL subgroups was not based on moderate variations in HOXA expression but rather divided patients either firmly expressing or completely lacking HOXA gene expression. These findings confirm a similar observation recently reported by Trentin et al,34 who showed that, based on the localization of the MLL breakpoints and the absence or presence of AF4-MLL (the reciprocal fusion transcript of MLL-AF4), and the presence or absence of HOXA expression, t(4;11)-positive ALL samples can be subdivided into 2 separate genetic subgroups. However, in contrast to the data from Trentin et al,34 who identified hundreds of genes to be associated with either high or low HOXA expression, we only found 27 probe sets to significantly discriminate between t(4;11)-positive infant ALL patients expressing either high or low HOXA levels. Nevertheless, these findings are particularly remarkable, as HOXA overexpression is thought to be a hallmark of MLL-rearranged leukemias,12,35 and HOXA9 expression has recently been postulated to be required for leukemia survival in MLL-rearranged leukemia cell lines and primary MLL-rearranged AML samples.36 Surprisingly, our data revealed that the absence of HOXA expression appears to be of significant clinical importance, as these patients are at extreme high risk of disease relapse, even within a patient group already characterized by a poor prognosis. Collectively, these observations challenge the dogma that HOXA9 is consistently highly expressed in all MLL-rearranged leukemias, and demonstrate that HOXA9 is not per se required for the maintenance of MLL-rearranged infant ALL, as t(4;11)-positive infant ALL patients lacking HOXA9 expression seem to be burdened by a more aggressive leukemia with a high risk of early relapse. Thus, in contrast to recent suggestions that suppression of HOXA9 may represent an attractive therapeutic approach in AML, targeting HOXA9 in t(4;11)-positive infant ALL appears not to be an option. Finally, these data clearly indicate that variations in gene expression patterns among MLL-rearranged infant ALL cases are not limited to the type of MLL translocation alone but continue to extend beyond translocation-specific subgroups, at least in case of translocation t(4;11).
Taken together, the present study demonstrates that the distinct gene expression profiles associated with MLL-rearranged infant ALL are more heterogeneous and complicated than ostensibly shown earlier and, to a certain extent, are dependent on the MLL translocation partner genes. In addition, based on our gene expression profiles, infant ALL patients lacking MLL translocations differ both from MLL-rearranged infant ALL and noninfant pediatric precursor B-ALL patients. The expression signatures reported here potentially constitute new and additional insights in the genetic makeup of both MLL-rearranged and MLL germline infant ALL. The work at hand now is to unravel the biologic meaning of these signatures and implement these novel pieces of the puzzle into currently ongoing studies on the complex biology of this malignancy. Eventually, these profiles should reveal novel therapeutic targets, uncover yet unidentified regulators of leukemogenesis and leukemia maintenance, and perhaps may become useful in future gene expression–based classification of pediatric ALL.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the members and participating hospitals of the INTERFANT-99 study for supporting our research by generously providing leukemic samples.
Members of the INTERFANT-99 study are as follows: M. Campbell (Programa Infantil Nacional de Drogas Atineoplasicas [PINDA]), M. Felice (Argentina), A. Ferster (Children's Leukemia Group [CLCG]), I. Hann and A. Vora (UK Children's Cancer Study Group [UKCCSG]), L. Hovi (Nordic Society of Paediatric Haematology and Oncology [NOPHO]), G. Janka-Schaub (Cooperative Study Group for Treatment of ALL [COALL]), C.K. Li (Hong Kong), G. Mann (Berlin-Frankfurt-Munster Group-Austria [BFM-A]), T. LeBlanc (French ALL Group [FRALLE]), R. Pieters (Dutch Childhood Oncology Group [DCOG]), G. de Rossi and A. Biondi (Associazione Italiana Ematologia Oncologia Pediatrica [AIEOP]), J. Rubnitz (St Jude Children's Research Hospital [SJCRH]), M. Schrappe (Berlin-Frankfurt-Munster Group-Germany [BFM-G]), L. Silverman (Dana-Farber Cancer Institute [DFCI]), J. Stary (Czech Paediatric Haematology [CPH]), R. Suppiah (Australian and New Zealand Children's Haematology/Oncology Group [ANZCHOG]), T. Szczepanski (Polish Paediatric Leukemia and Lymphoma Study Group [PPLLSG]), and M. Valsecchi and P. de Lorenzo (Trial Operating Center [CORS]).
This work was supported by Kinderen Kankervrij (KIKA) and supported in part by the Dutch Cancer Society (grant EMCR 2005-2662; R.W.S.).
Authorship
Contribution: R.W.S. performed and designed research and wrote the manuscript; P.S., J.A.P.H., M.H.v.d.L., and D.J.P.M.S. performed research; R.X.d.M. performed and guided statistical analyses; P.d.L. collected and processed patient information and reviewed the manuscript; M.G.V. collected and processed patient information and reviewed the manuscript; and R.P. designed research and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ronald W. Stam, Erasmus MC–Sophia Children's Hospital, Pediatric Oncology/Hematology, Room Sp2456, Dr. Molewaterplein 60, P.O. Box 2060, 3000 CB Rotterdam, The Netherlands; e-mail: r.stam@erasmusmc.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal