Abstract
To further unravel the molecular pathogenesis of T-cell acute lymphoblastic leukemia (T-ALL), we performed high-resolution array comparative genomic hybridization on diagnostic specimens from 47 children with T-ALL and identified monoallelic or biallelic LEF1 microdeletions in 11% (5 of 47) of these primary samples. An additional 7% (3 of 44) of the cases harbored nonsynonymous sequence alterations of LEF1, 2 of which produced premature stop codons. Gene expression microarrays showed increased expression of MYC and MYC targets in cases with LEF1 inactivation, as well as differentiation arrest at an early cortical stage of thymocyte development characterized by expression of CD1B, CD1E, and CD8, with absent CD34 expression. LEF1 inactivation was associated with a younger age at the time of T-ALL diagnosis, as well as activating NOTCH1 mutations, biallelic INK4a/ARF deletions, and PTEN loss-of-function mutations or activating mutations of PI3K or AKT genes. These cases generally lacked overexpression of the TAL1, HOX11, HOX11L2, or the HOXA cluster genes, which have been used to define separate molecular pathways leading to T-ALL. Our findings suggest that LEF1 inactivation is an important step in the molecular pathogenesis of T-ALL in a subset of young children.
Introduction
Wider use of intensive chemotherapy has improved outcomes in patients with T-cell acute lymphoblastic leukemia (T-ALL), but such treatment is toxic and fails in approximately 25% of children and 50% to 70% of adults.1,2 Moreover, understanding of the molecular mechanisms that drive the aberrant proliferation and survival of malignant T lymphoblasts remains deficient, impeding efforts to uncover novel targets for molecularly directed therapies. We have shown that T-ALL can be subclassified according to the dominant pattern of oncogene expression, with each subtype characterized by developmental arrest at a specific stage of T-cell differentiation.3 Although overexpression of TAL1, HOX11, HOX11L2, LYL1, or the HOXA cluster is sufficient to identify most T-ALL subtypes,3-6 17% of the cases in our original study were not classifiable by the expression of known oncogenes,3 indicating that critical molecular abnormalities remain to be identified in leukemic T lymphoblasts.
LEF1 is a member of the lymphoid enhancer factor/T-cell factor (LEF/TCF) family of DNA-binding transcription factors, which interact with nuclear β-catenin in the WNT signaling pathway.7 LEF1 has also been shown to mediate key aspects of transforming growth factor β (TGFβ) signaling during craniofacial morphogenesis in the mouse8 and directly interacts with Smad4, a key mediator of TGFβ signaling, during the establishment of the Spemann organizer in early amphibian embryogenesis.9 The intracellular domain of NOTCH1 has also been shown to function as a coactivator of LEF1, leading to the up-regulation of target genes distinct from those activated by β-catenin binding.10 Interestingly, LEF1 has been shown to function in vivo as either an oncogene or a tumor suppressor in different cellular contexts. Transplantation of Lef1-transduced bone marrow leads to acute myeloid leukemia and B-precursor ALL in the mouse.11 Conversely, N-terminal LEF1 mutations that impair β-catenin binding are commonly found in human sebaceous skin tumors,12 and expression of an N-terminal–deleted Lef1 mutant that lacks the β-catenin binding domain leads to sebaceous skin tumors in the mouse.13
Here, we identify a new molecular subtype of human T-ALL defined by inactivation of LEF1 through a combination of monoallelic or biallelic microdeletions and gene-specific mutations that are predicted to lead to the premature termination of translation.
Methods
Patient samples
Diagnostic specimens were collected (with informed consent in accordance with the Declaration of Helsinki and institutional review board approval) from children with T-ALL who were treated on Children's Oncology Group study P9404 or Dana-Farber Cancer Institute (DFCI) study 00-01 clinical trials, which tested similar therapeutic regimens based on an identical backbone, including postinduction consolidation with asparaginase and anthracycline.14,15 Mononuclear cells were purified by Ficoll-Hypaque centrifugation before cryopreservation. All specimens consisted of more than 90% lymphoblasts. Genomic DNA was extracted with the PureGene kit or the DNeasy blood and tissue kit according to the manufacturer's instructions (QIAGEN). Samples initially extracted with the PureGene kit were repurified with the DNeasy kit before array comparative genomic hybridization (CGH) analysis.
Microarray-based comparative genomic hybridization
Microarray-based CGH (array CGH) was performed on genomic DNA with Human Genome CGH 244A microarrays (Agilent Technologies), as previously described.16-18 Feature extraction data were obtained with Agilent G2567AA feature extraction software, normalized with a LOcally-WEighted regression Scatterplot Smoother available in an R package developed by the Lynda Chin laboratory (http://genomic.dfci.harvard.edu/Tools/Agilent_1.0.2.tar.gz), and segmented with the BioConductor DNAcopy package (http://www.bioconductor.org/packages/2.2/bioc/html/DNAcopy.html). Samples 36 and 37 were excluded from analysis because the CGH quality controls failed. The CGH log2 copy number ratio for heterozygous deletion was defined as −0.5 to −1.5 (corresponding to 35%-70% of normal copy number), whereas log2 copy number ratios less than −1.5 (corresponding to < 35% of normal copy number) were defined as homozygous deletions. Array CGH data are available on the Gene Expression Omnibus (GEO) website under accession numbers GSE14959 and GSE7615 (http://www.ncbi.nlm.nih.gov/geo/; see supplemental Table 1 sheet 2, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Mutation detection
Sequencing was performed on the entire coding region of LEF1 together with key exons of several oncogenes and tumor suppressors known to be mutated in human T-ALL. These included exons 26, 27, and 34 of NOTCH1, exons 9 and 10 of FBXW7, the entire coding region of PTEN, exons 9 and 20 of PIK3CA (encoding the catalytic subunit of class IA phosphatidylinositol 3-kinase [PI3K]), exons 12 and 13 of PIK3R1 (encoding the regulatory subunit of class IA PI3K), coding exon 2 of the AKT1-3 genes, and exons 1 to 2 of NRAS and KRAS.18-21 All sequencing of primary patient samples was performed at Agencourt Bioscience Corporation.
Gene expression analyses
Gene expression microarrays were previously performed on 40 of the primary T-ALL patient samples analyzed in our study, comprising the cases treated on the Children's Oncology Group P9404 study. Gene expression profiling was performed with Affymetrix U133 Plus 2.0 arrays as previously described.14 The expression profiling data are available in the National Center for Biotechnology Information Gene Expression Omnibus under accession no. GSE14618 (see supplemental Table 1 sheet 2). Expression data were extracted from CEL files with the use of dChip 2007 software (http://biosun1.harvard.edu/complab/dchip/) and normalized to case T-ALL 15. Case T-ALL 01 was excluded from the gene expression analyses because the effect of the Asp85Asn missense LEF1 mutation in this sample on the pathogenic function of LEF1 in T-ALL is unknown.
Gene Set Enrichment Analysis (GSEA) was performed as previously described,22,23 using the gene sets from the Molecular Signatures Database.24 The “FERRANDO HOX11/EARLY CORTICAL” gene set was created based on a previously published gene expression signature of T-ALL cases that shows HOX11 overexpression and developmental arrest at an early cortical stage of thymocyte differentiation.3 GSEA was performed on the unfiltered gene expression dataset, as recommended.
Differential expression analysis was performed with GenePattern software (http://www.broad.mit.edu/cancer/software/genepattern/index.html).25 The gene expression data were first filtered with the GenePattern “PreprocessDataset” module with the use of the following parameters: minimum fold change, 3; minimum delta, 100; threshold, 20; and ceiling, 20 000. Differential expression analysis was then performed with the “ComparativeMarkerSelection” module, using a number of permutations of 0 to calculate asymptotic P values, as recommended when there are less than 10 samples in any class.
Cell lines
T-ALL cell lines were obtained from American Type Culture Collection or Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH and were cultured in RPMI-1640 containing 1% l-glutamine and 10% fetal bovine serum (Sigma-Aldrich). The results of array CGH of our T-ALL cell lines with the use of Agilent 44K arrays have been previously published16,17 and are available on the GEO website under accession no. GSE7615 (http://www.ncbi.nlm.nih.gov/geo/).
Short hairpin RNA knockdown
Short hairpin RNA (shRNA) knockdown analyses were performed as previously described.26-28 Briefly, double-stranded DNAs including the 21mer shRNA sequence were cloned into the lentivirus vector pLKO1-puro. After cotransfection into the 293 packaging cell line with packaging plasmid delta 8.9 and envelope plasmid VSV-G, supernatants containing the lentivirus were collected and used to infect the T-ALL cell lines Jurkat, RPMI-8402, PEER and SUP-T7. Knockdown efficacy was tested by Western blotting. shRNA target sequences were as follows: LEF1 shRNA1, GCAGCTATCAACCAGATTCTT; LEF1 shRNA2, CGGGTACATAATGATGCCAAA; LEF1 shRNA3, CCACACTGACAGTGACCTAAT; LEF1 shRNA4, GCACGGAAAGAAAGACAGCTA; LEF1 shRNA5, CCATCAGATGTCAACTCCAAA.
Methyl thiazolyl tetrazolium assay
The methyl thiazolyl tetrazolium assay was performed as previously described.28
Western blotting
For analysis of LEF1 knockdown in T-ALL cell lines, protein lysates were prepared with RIPA buffer (Upstate) supplemented with complete mini protease inhibitor (0.5 tablet/100 mL; Roche Applied Science). Protein lysate (30 μg) was mixed with Laemmli 4× sodium dodecyl sulfate sample buffer (Boston Bioproducts) and β-mercaptoethanol in appropriate proportions and incubated at 100°C for 3 minutes before being run on a 10% polyacrylamide gel at 20 mA for 1.5 hours. Blots were then transferred to a nitrocellulose membrane (Millipore) at 350 mA for 1 hour, blocked with 5% milk in Tris-buffered saline with 0.1% Tween, and probed with mouse anti-LEF1 antibody (1:1000; NA64; Calbiochem), and rabbit anti–α-tubulin antibody (1:1000; 2144; Cell Signaling Technology). Signal was detected with SuperSignal West Dura substrate (Pierce Protein Research Products, Thermo Fisher Scientific).
RNA extraction and quantitative reverse transcription–polymerase chain reaction
Total RNA was extracted from T-ALL cell lines after infection with GFP shRNA, LEF1 shRNA4, or LEF1 shRNA5 with the use of Trizol reagent according to the manufacturer's instructions (Invitrogen). cDNA was synthesized from 2 μg of total RNA with the use of the SuperScript First-Strand Synthesis System for real-time polymerase chain reaction (RT-PCR) according to the manufacturer's instructions (Invitrogen). Three percent of the cDNA reaction volume was then taken for each quantitative RT-PCR reaction, performed with the SYBR Green PCR Core Reagents kit (Applied Biosystems) and an Applied Biosystems 7300 Real Time PCR System instrument (Applied Biosystems) according to the manufacturer's instructions. Quantitative RT-PCR primer sequences were as follows: MYC forward, CTCTCCGTCCTCGGATTCT; MYC reverse, CAACATCGATTTCTTCCTCATC; β-actin forward, CTGGCACCCAGCACAATG; β-actin reverse, GCCGATCCACACGGAGTACT. Note that all RT-PCR primer pairs used span intron/exon boundaries.
Results
Recurrent LEF1 genetic alterations in T-ALL
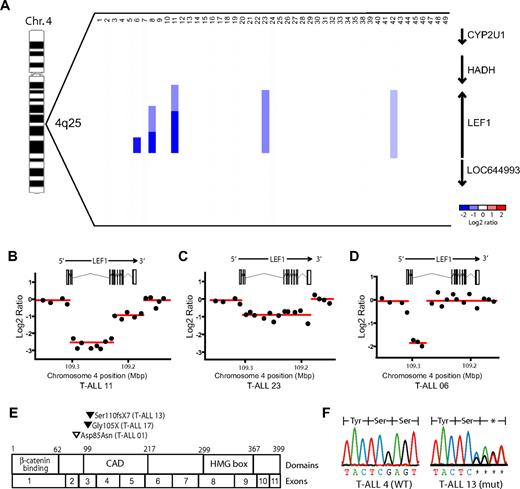
To gain further insight into the molecular pathogenesis of T-ALL, we performed array CGH on genomic DNA extracted from the diagnostic specimens of 47 pediatric patients with T-ALL, as previously described.16,18 Analysis of the CGH data showed recurrent LEF1 microdeletions in 11% (5 of 47; 90% exact binomial confidence interval, 4%-21%) of the primary clinical samples (Figure 1A). These deletions were highly focal and involved no other known genes. They were homozygous in 3 cases, 2 of which (T-ALL 08 and 11) showed distinct regions of homozygous versus heterozygous loss at the LEF1 locus (Figure 1A-B), indicating that the deletion of each allele was a distinct genetic event. Analysis of the raw CGH data showed that the deletions unequivocally disrupted the LEF1 coding sequence, as shown by the deletion of CGH probes spanning multiple LEF1 exons (Figure 1A-D). No focal copy number alterations involving other LEF/TCF family members were identified.
LEF1 microdeletions and truncating mutations are recurrent genetic alterations in T-ALL. Array CGH was performed on genomic DNA from diagnostic specimens collected from 47 children with T-ALL. (A) dChip plot of the segmented CGH log2 copy number ratios at the LEF1 genomic locus. Recurrent microdeletions involving LEF1 and no other known genes were identified in 5 (11%) of the 47 primary T-ALL samples. Note that cases 36 and 37 were excluded because CGH quality controls failed. (B-D) Raw CGH log2 copy number ratio data (black dots) shown for 3 representative cases, together with the genomic location of LEF1 exons. The segmented data plotted in panel A are shown as red lines. The y-axis is log2 of the copy number ratio (0 = no copy number change). (E) Sequencing of LEF1 genomic coding sequence identified heterozygous nonsynonymous sequence alterations in 3 (7%) of the 44 T-ALL cases analyzed. Black arrowheads denote the location of predicted truncating mutations, whereas the white arrowhead denotes the missense mutation identified. (F) Sequence chromatograms for representative mutant and wild-type samples, showing the presence of a heterozygous frameshift mutation in sample T-ALL 13.
LEF1 microdeletions and truncating mutations are recurrent genetic alterations in T-ALL. Array CGH was performed on genomic DNA from diagnostic specimens collected from 47 children with T-ALL. (A) dChip plot of the segmented CGH log2 copy number ratios at the LEF1 genomic locus. Recurrent microdeletions involving LEF1 and no other known genes were identified in 5 (11%) of the 47 primary T-ALL samples. Note that cases 36 and 37 were excluded because CGH quality controls failed. (B-D) Raw CGH log2 copy number ratio data (black dots) shown for 3 representative cases, together with the genomic location of LEF1 exons. The segmented data plotted in panel A are shown as red lines. The y-axis is log2 of the copy number ratio (0 = no copy number change). (E) Sequencing of LEF1 genomic coding sequence identified heterozygous nonsynonymous sequence alterations in 3 (7%) of the 44 T-ALL cases analyzed. Black arrowheads denote the location of predicted truncating mutations, whereas the white arrowhead denotes the missense mutation identified. (F) Sequence chromatograms for representative mutant and wild-type samples, showing the presence of a heterozygous frameshift mutation in sample T-ALL 13.
To detect other genetic lesions contributing to LEF1 inactivation, we sequenced the LEF1 genomic coding sequence in 44 of the 47 primary T-ALL samples, identifying nonsynonymous sequence alterations in 7% (3 of 44) of these cases (Figure 1E-F; supplemental Table 1). Two of the mutations were in exon 3; thus, they were predicted to truncate all known LEF1 isoforms because of premature termination of translation before the context-dependent activation domain and the high-mobility group DNA-binding domain.29,30 The third sequence alteration, found in exon 2, was predicted to result in a missense Asp85Asn mutation of undetermined significance in an exon that is absent from some LEF1 variants.30 None of these alterations represent known single nucleotide polymorphisms (SNPs) based on the National Center for Biotechnology Information (build 129)31 or the HapMap (rel27) SNP32 databases.
Sequencing of NOTCH1 exons 26, 27, and 34 showed nonsynonymous sequence alterations involving the heterodimerization and/or PEST domains in 7 of the 8 cases harboring LEF1 alterations (supplemental Table 1), indicating that LEF1 inactivation collaborates with NOTCH1 activating mutations in the pathogenesis of human T-ALL. All cases harboring LEF1 alterations also had homozygous deletions of the CDKN2A gene, encoding p14ARF and p16INK4a, whereas 6 of these 8 cases also harbored PTEN, PI3K, or AKT mutations (supplemental Table 1).
In addition to the reported pathogenic role of LEF1 inactivation in human and murine sebaceous skin tumors,12,13 this transcription factor can also induce acute myeloid leukemia and B-precursor ALL when overexpressed in murine bone marrow cells.11 Furthermore, β-catenin, a transcriptional cofactor for LEF1,7 can induce T-ALL when aberrantly stabilized in murine thymocytes.33 In an effort to determine whether a subset of T-ALL cases is characterized by overexpression of LEF1 or activation of the WNT/β-catenin pathway, we examined the gene expression microarray data that were previously published on 40 of the 47 primary cases analyzed in our study,14 but we found no evidence for a distinguishable subset of primary T-ALL showing LEF1 overexpression or of a recently described gene expression signature characteristic of WNT/β-catenin pathway activation.34 Examination of the top quintile of cases with highest LEF1 expression (n = 9) showed no distinguishing features in terms of clinical features, cell surface marker expression, or associated genetic abnormalities (data not shown).
MYC overexpression and cortical T-cell developmental arrest in LEF1-inactivated T-ALL
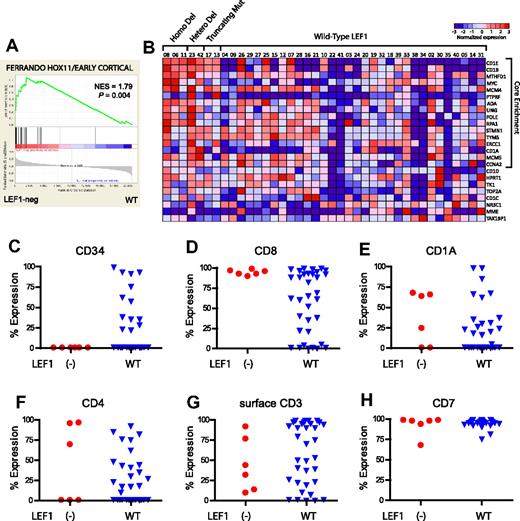
Using previously generated expression microarray data on 40 of these 47 cases,14 we analyzed the gene expression changes associated with LEF1 inactivation (deletions or truncating mutations). LEF1 inactivation was generally mutually exclusive of the overexpression of the known T-ALL oncogenes TAL1, HOX11, and HOX11L2 or of genes in the HOXA cluster (Figure 2A), which were used to define the currently recognized molecular subtypes of T-ALL.3,4 These cases also did not reiterate the gene expression profile associated with the recently reported early T-cell precursor form of high-risk ALL,35 in which LEF1 expression was found to be low. We noted that LEF1-inactivated cases had increased levels of MYC expression (Figure 2A-B), even compared with cases with intact LEF1 that also harbored NOTCH1 mutations (Figure 2C), suggesting that LEF1 inactivation may promote the transcriptional up-regulation of MYC by NOTCH1. Furthermore, GSEA identified a strong association between LEF1 inactivation and the overexpression of MYC targets, because 3 of the 4 gene sets most highly enriched in LEF1-inactivated T-ALL were those representing MYC target genes (Figure 2D-F; supplemental Table 2).
LEF1-inactivated T-ALL is characterized by the overexpression of MYC and of MYC targets. Gene expression profiling was previously performed on 40 of the 47 T-ALL cases analyzed in our study, using Affymetrix U133 Plus 2.0 microarrays. (A) Heatmap showing the expression pattern of known T-ALL oncogenes, based on the expression microarrays applied. Probe sets that showed no significant expression (defined as expression values < 100) in any T-ALL sample were excluded. Low expression values were truncated to 30. Note that data from the [1561651_s_at] TAL1 probe set were excluded because we have found that expression measured by this probe set does not correlate with TAL1 RNA levels (data not shown). (B-C) Expression of MYC by LEF1 and NOTCH1 status. P values were calculated by the Wilcoxon rank-sum test. (D-F) Gene set enrichment analysis showed that 3 of the 4 gene sets most highly up-regulated in LEF1-inactivated T-ALL represent MYC target gene sets.
LEF1-inactivated T-ALL is characterized by the overexpression of MYC and of MYC targets. Gene expression profiling was previously performed on 40 of the 47 T-ALL cases analyzed in our study, using Affymetrix U133 Plus 2.0 microarrays. (A) Heatmap showing the expression pattern of known T-ALL oncogenes, based on the expression microarrays applied. Probe sets that showed no significant expression (defined as expression values < 100) in any T-ALL sample were excluded. Low expression values were truncated to 30. Note that data from the [1561651_s_at] TAL1 probe set were excluded because we have found that expression measured by this probe set does not correlate with TAL1 RNA levels (data not shown). (B-C) Expression of MYC by LEF1 and NOTCH1 status. P values were calculated by the Wilcoxon rank-sum test. (D-F) Gene set enrichment analysis showed that 3 of the 4 gene sets most highly up-regulated in LEF1-inactivated T-ALL represent MYC target gene sets.
Differential gene expression analysis identified CD1E and CD1B as the 2 most consistently up-regulated genes in LEF1-inactivated T-ALL cases (supplemental Figure 1). Expression of CD1 in T-ALL is linked to developmental arrest at a cortical stage of T-cell differentiation,36 an observation also made in HOX11-overexpressing cases,3 even though LEF1-inactivated T-ALL cases lacked detectable HOX11 expression (Figure 2A). We thus performed GSEA with a gene set based on the previously reported HOX11-positive early cortical T-ALL gene expression signature3 and found a high degree of similarity between this signature and that of LEF1-inactivated T-ALL (Figure 3A-B; P = .004). Furthermore, analysis of the T-ALL cell surface immunophenotype obtained at the time of diagnosis in all cases in which these data were available showed that all LEF1-inactivated cases lacked CD34, expressed CD8, and had variable expression of CD1A, CD4, and surface CD3 (Figure 3C-H). Taken together, these data indicate that LEF1-inactivated T-ALL is characterized by developmental arrest at an early cortical stage of T-cell differentiation characterized by CD1B, CD1E, and CD8 positivity, but without CD34 expression.
LEF1-inactivated T-ALL is characterized by developmental arrest at an aberrant cortical stage of T-cell differentiation. (A) Gene set enrichment analysis shows that the published gene expression signature of HOX11-positive T-ALL cases, showing T-cell developmental arrest at an early cortical stage of thymocyte differentiation, closely resembles that of LEF1-inactivated T-ALL. (B) Heatmap depicting the results of the gene set enrichment analysis for the HOX11-positive early cortical signature depicted in Figure 1A. (C-H) Results of flow cytometry to detect T-cell surface markers are shown for all cases in which such data were available. Percent Expression denotes the percentage of blasts that were positive for expression of each immunophenotypic marker. Note that the CD1A antibody does not appear to cross-react with CD1B and CD1E, which were highly expressed in all LEF1-inactivated cases analyzed by expression microarray. Taken together, these data indicate that LEF1-inactivated cases of T-ALL show developmental arrest at an aberrant CD1E/CD1B+, CD8+, CD34− cortical stage of T-cell development.
LEF1-inactivated T-ALL is characterized by developmental arrest at an aberrant cortical stage of T-cell differentiation. (A) Gene set enrichment analysis shows that the published gene expression signature of HOX11-positive T-ALL cases, showing T-cell developmental arrest at an early cortical stage of thymocyte differentiation, closely resembles that of LEF1-inactivated T-ALL. (B) Heatmap depicting the results of the gene set enrichment analysis for the HOX11-positive early cortical signature depicted in Figure 1A. (C-H) Results of flow cytometry to detect T-cell surface markers are shown for all cases in which such data were available. Percent Expression denotes the percentage of blasts that were positive for expression of each immunophenotypic marker. Note that the CD1A antibody does not appear to cross-react with CD1B and CD1E, which were highly expressed in all LEF1-inactivated cases analyzed by expression microarray. Taken together, these data indicate that LEF1-inactivated cases of T-ALL show developmental arrest at an aberrant CD1E/CD1B+, CD8+, CD34− cortical stage of T-cell development.
Clinical features associated with LEF1 inactivation
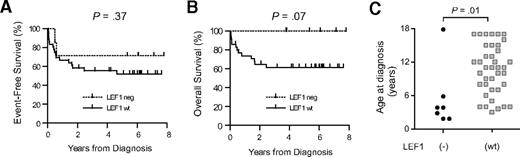
Analysis of the available clinical data showed that LEF1 inactivation was not a significant predictor of event-free survival in children with T-ALL treated in the Children's Oncology Group P9404 or Dana-Farber Cancer Institute 00-01 studies (Figure 4A), indicating that LEF1 status does not predict response to contemporary first-line therapy for T-ALL. However, LEF1 inactivation was associated with a trend toward improved overall survival (Figure 4B; P = .07), suggesting the need for further studies to determine whether these cases may indeed have a more favorable response to salvage therapy for T-ALL. In addition, LEF1 inactivation was associated with a younger age at the time of diagnosis (Figure 4C; P = .01; supplemental Table 1), but not with sex, white blood cell count, central nervous system involvement, or the presence of an anterior mediastinal mass at the time of diagnosis.
Clinical features associated with LEF1 inactivation. (A-B) Kaplan-Meier analysis of event-free survival rates shows that LEF1 status is not a significant predictor of response to initial therapy; however, there was a trend toward improved overall survival in patients with LEF1-inactivated T-ALL, suggesting that this molecular subtype of the disease may be more responsive to salvage therapy for relapsed T-ALL. (C) LEF1 inactivation is associated with a younger age at the time of T-ALL diagnosis. P value was calculated by the Wilcoxon rank-sum test.
Clinical features associated with LEF1 inactivation. (A-B) Kaplan-Meier analysis of event-free survival rates shows that LEF1 status is not a significant predictor of response to initial therapy; however, there was a trend toward improved overall survival in patients with LEF1-inactivated T-ALL, suggesting that this molecular subtype of the disease may be more responsive to salvage therapy for relapsed T-ALL. (C) LEF1 inactivation is associated with a younger age at the time of T-ALL diagnosis. P value was calculated by the Wilcoxon rank-sum test.
LEF1 knockdown analyses
LEF/TCF family members are known to bind MYC gene regulatory elements and up-regulate MYC transcription in the setting of active β-catenin signaling, but LEF/TCFs can also act as transcriptional repressors in the absence of active β-catenin.37 In an effort to test the hypothesis that LEF1 is a tumor suppressor in a subset of human T-ALL cases because it acts as a transcriptional repressor of MYC, we examined the status of LEF1 in cultured human T-ALL cell lines, but we found no evidence of LEF1 deletions, mutations in exons 1 to 3, or the absence of LEF1 protein by Western blot analysis in any of the 16 T-ALL cell lines analyzed (data not shown). We thus tested the effect of 5 LEF1 shRNA constructs and identified 2 that resulted in effective knockdown of LEF1 by Western blotting, which were then used to knock down LEF1 in 4 T-ALL cell lines (supplemental Figure 2A). LEF1 knockdown failed to increase MYC expression by quantitative RT-PCR, cell survival by MTT assay, cell cycle progression, or apoptosis in the cell lines examined (supplemental Figure 2B-F; data not shown). Gene expression microarrays were then performed on 3 of these cells lines before and after LEF1 knockdown, and, although LEF1 expression was significantly decreased after LEF1 knockdown (supplemental Figure 3), there were no other genes whose expression was altered with a false discovery rate less than 43% (data not shown). Further studies in the appropriate developmental context are needed to identify the mechanism by which LEF1 inactivation contributes to the molecular pathogenesis of T-ALL.
Discussion
In this article we identify recurrent monoallelic or biallelic LEF1 microdeletions and truncating mutations in T-ALL and thus provide compelling evidence that LEF1 inactivation is selected for during T-cell leukemogenesis. LEF1-inactivated T-ALL was characterized by distinctive clinical features, including a younger age at diagnosis, and a trend toward improved overall survival (but not event-free survival) in children treated with contemporary T-ALL combination therapy. This suggests the need for additional studies to address whether LEF1-inactivated T-ALL may indeed respond well to salvage therapy for T-ALL, which typically includes allogeneic bone marrow transplantation. Furthermore, this subtype of T-ALL was associated with specific genetic alterations, including frequent mutations in NOTCH1 and the PTEN-PI3K-AKT pathway, and the overexpression of MYC and its downstream target genes. However, these cases generally lacked overexpression of TAL1, HOX11, HOX11L2, or of the HOXA/MEIS1 cluster, indicating a distinct combination of molecular abnormalities leading to human T-ALL.
LEF1-inactivated T-ALLs have a gene expression signature that suggests developmental arrest at a cortical stage of differentiation, which is also seen in HOX11-positive T-ALL. Cortical arrest in T-ALL has been defined on the basis of CD1A immunophenotypic positivity,36 and CD1A-E genes are normally expressed from the late double-negative to the double-positive stages of thymocyte differentiation.38,39 LEF1-inactivated T-ALL cases were characterized by the overexpression of CD1B and CD1E, CD8 positivity, variable CD4, and the lack of CD34 expression, reflecting developmental arrest at the transition from a CD8+ immature single positive (ISP) stage to the double-positive stage. Although murine ISP cells are characteristically CD8+, human ISPs are generally CD4+,38 suggesting developmental arrest at an aberrant ISP stage in LEF1-inactivated T-ALL.
LEF1 is expressed throughout normal human T-cell development, particularly at the CD1+ cortical stages.39 Studies in knockout mice have shown that Lef1 loss exacerbates the T-cell developmental defects induced by Tcf1 deficiency, resulting in T-cell developmental arrest at the ISP stage.40,41 However, Lef1 deficiency alone does not lead to detectable T-cell abnormalities in the mouse,40 and there were no focal TCF1 DNA copy number alterations by CGH nor decreased TCF1 expression by microarray in any of the primary T-ALL samples analyzed.
The mechanism by which the loss of LEF1 expression contributes to T-ALL pathogenesis is unclear. One possibility is that LEF1 inactivation may act in concert with NOTCH1 activation to promote the up-regulation of MYC expression. Indeed, most cases of T-ALL with loss of LEF1 have NOTCH1 mutations, and we have identified overexpression of MYC and of MYC targets as prominent abnormalities in these cases. LEF/TCF family members have been shown to directly bind MYC gene regulatory elements and to up-regulate MYC transcription in the setting of active WNT/β-catenin signaling,42,43 whereas LEF/TCF family members can also act as transcriptional repressors when bound to transducin-like enhancer of split (TLE) family members in the absence of active β-catenin.37 The loss of LEF1 activity might prevent the recruitment of TLE corepressors to the MYC promoter, which otherwise would occur if the cases lacked activation of β-catenin. Moreover, LEF1 binds to SMAD4 downstream of TGFβ signaling,8,9 a known tumor suppressor pathway that is frequently inactivated in human T-ALL,44 providing another route by which LEF1 inactivation could promote the transcriptional activation of MYC. Thus, the available evidence favors relief of transcriptional repression of MYC as the most plausible mechanism driving the selection for LEF1 inactivation in T-ALL, hence allowing MYC to be maximally overexpressed downstream of mutations that activate NOTCH1 signaling. Further experiments in the appropriate cellular context will be needed to test this hypothesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the children with T-ALL and their families, as well as members of the Children's Oncology Group and Dana-Farber Cancer Institute Acute Lymphoblastic Leukemia Consortium member institutions, for the samples analyzed in these studies. We thank Jason Berndt, Andy Chien, and Randall Moon for assistance with gene expression analyses for WNT pathway activation in primary T-ALL. We thank John Gilbert for editorial assistance.
This work was supported by the National Institutes of Health (grant 5P01CA68484; S.E.S. and A.T.L.; and grant 1K08CA133103; A.G.); by an award from the Wine Advocate Fund for Philanthropy of The Community Foundation of the National Capital Region; the William Lawrence Foundation (A.G.); and the American Society of Hematology-Amos Medical Faculty Development program (A.G.). Children's Oncology Group (COG) cell banking and sample distribution were supported by the COG 9900 cell biology study and grants CA98543, CA114766, and CA98413. S.P.H. is the Ergen Family Chair in Pediatric Cancer at The Children's Hospital, Aurora, CO. The array CGH analyses were performed and supported in part by the Belfer Institute for Applied Cancer Science.
National Institutes of Health
Authorship
Contribution: A.G. designed, performed, and analyzed the research and wrote the paper; T.S. performed and analyzed the research; R.G. and W.M. designed and performed the research; S.D. and D.N. analyzed data; J.Z., A.P., and L.C. developed vital CGH analytical tools and analyzed data; S.S.W., R.S.L., M.J.B., L.B.S., S.P.H., and S.E.S. provided vital reagents and analyzed data; C.J. supervised research and analyzed data; and A.T.L. supervised research, analyzed data, and cowrote the manuscript.
Conflict-of-interest disclosure: C.J. is a consultant for Wintherix Inc. The remaining authors declare no competing financial interests.
Correspondence: A. Thomas Look, Dana-Farber Cancer Institute, Mayer 630, 44 Binney St, Boston, MA 02115; e-mail: thomas_look@dfci.harvard.edu

![Figure 2. LEF1-inactivated T-ALL is characterized by the overexpression of MYC and of MYC targets. Gene expression profiling was previously performed on 40 of the 47 T-ALL cases analyzed in our study, using Affymetrix U133 Plus 2.0 microarrays. (A) Heatmap showing the expression pattern of known T-ALL oncogenes, based on the expression microarrays applied. Probe sets that showed no significant expression (defined as expression values < 100) in any T-ALL sample were excluded. Low expression values were truncated to 30. Note that data from the [1561651_s_at] TAL1 probe set were excluded because we have found that expression measured by this probe set does not correlate with TAL1 RNA levels (data not shown). (B-C) Expression of MYC by LEF1 and NOTCH1 status. P values were calculated by the Wilcoxon rank-sum test. (D-F) Gene set enrichment analysis showed that 3 of the 4 gene sets most highly up-regulated in LEF1-inactivated T-ALL represent MYC target gene sets.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/14/10.1182_blood-2009-07-234377/4/m_zh89991050590002.jpeg?Expires=1769081922&Signature=L9Rc6f7R1wflD5w4wb0wCdQY~lLXkFB5U1uoaL2aua~-3BAFx4jy3nvLXaoUhWEMLPTzl32UgEpJylLm9jCc-QZmuBpgpc9fYWXznYJI7e8yxDQlSJqiXHbFFme3dwElcOgJMafynrXoetXpCFYPCIZRCPh6bFX2vYBL374fEcIU7EwXKGMvtOlZotxwIyg4Vtr1btcWyPJoqKIDCQ3WkRNcL7JrqTNxTJal-naufa5eg2Z3SNxsV7PGagxgd8C5V~OhRHtCwloTykbMdS7R1d65Lz86Rey-T-tVjo50yOlDLFkX3d67d5u59-9jiO94UmItbeaC-tU2PrT9Bnfzug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)