Abstract
Multiple myeloma (MM) is a plasma cell neoplasm that proceeds through a premalignant state of monoclonal gammopathy of unknown significance; however, the molecular events responsible for myelomagenesis remain uncharacterized. To identify cellular pathways deregulated in MM, we addressed that sumoylation is homologous to ubiquitination and results in the attachment of the ubiquitin-like protein Sumo onto target proteins. Sumoylation was markedly enhanced in MM patient lysates compared with normal plasma cells and expression profiling indicated a relative induction of sumoylation pathway genes. The Sumo-conjugating enzyme Ube2I, the Sumo-ligase PIAS1, and the Sumo-inducer ARF were elevated in MM patient samples and cell lines. Survival correlated with expression because 80% of patients with low UBE2I and PIAS1 were living 6 years after transplantation, whereas only 45% of patients with high expression survived 6 years. UBE2I encodes the sole Sumo-conjugating enzyme in mammalian cells and cells transfected with a dominant-negative sumoylation-deficient UBE2I mutant exhibited decreased survival after radiation exposure, impaired adhesion to bone marrow stroma cell and decreased bone marrow stroma cell–induced proliferation. UBE2I confers cells with multiple advantages to promote tumorigenesis and predicts decreased survival when combined with PIAS1. The sumoylation pathway is a novel therapeutic target with implications for existing proteasomal-based treatment strategies.
Introduction
Multiple myeloma (MM) is a neoplasia hallmarked by the clonal expansion of malignant plasma cells (PCs) and the accumulation of a monoclonal immunoglobulin (Ig) or Ig fragment detectable in the serum and/or urine.1-3 MM is the second most commonly diagnosed hematologic malignancy in the Western world, accounts for nearly 20% of all hematologic malignancies, and despite conventional treatment or high-dose therapy with stem cell transplantation is generally considered incurable.4,5 Recent advances in mechanistic understanding and treatment modalities have extended median survival to exceed 6 years, and 10% of MM patients survive beyond 10 years.6-8 Although high-dose therapy and novel agents that include thalidomide, its analog lenalidomide, and proteasome inhibitors have significantly improved prognosis, patient survival remains highly variable and cannot be accurately predicted with current models in part because the cellular pathways that determine patient response to treatment remain unidentified.
In eukaryotes, a highly conserved multienzyme system is used in a sequential process to covalently attach the polypeptide ubiquitin to proteins targeted for degradation.9,10 Ubiquitination is an essential process that maintains cellular homeostasis through dynamic switches in protein functional states. Ubiquitin-protein conjugates are then degraded in by the ATP-dependent 26S proteasome complex.11,12 Deregulation of ubiquitination in tumor models has resulted in malignant transformation and tumor progression.13 In myeloma, proteasomal-dependent catabolism of ubiquitinated substrates has been targeted therapeutically with bortezomib and has demonstrated significant clinical benefit.14-16 Whereas ubiquitination generally leads to proteasome-mediated proteolysis, posttranslational covalent attachment of the small ubiquitin-like modifier (Sumo) to target substrates has been reported to have an additional function and may control subcellular localization, function, or binding of targets to other proteins.17-19 In mammalian cells, 3 Sumo isoforms are expressed as precursors and are cleaved at the C-terminus by sumo-specific proteases. A complex that consists of SAE1 and SAE2 (E1) activates Sumo and transfers it to the conjugating enzyme Ubc9/Ube2I (E2). Interaction of Ube2I with a target protein then induces transfer of the sumo moiety to a lysine residue on the target through isopeptide bond formation. Target selection is facilitated through specific Sumo ligases (E3s), which act as adapters to bind Ube2I and the target protein. E3 ligases include members of the PIAS (protein inhibitor of activated STAT) family, the polycomb group protein Pc2, and RanBP2, which is a component of the nuclear pore complex. In addition, the tumor suppressor ARF triggers sumoylation of many cellular proteins and thus, although not strictly an E3 ligase, appears to function as an inducer of sumoylation. Several identified sumoylation substrates is steadily increasing and has been implicated in critical cellular processes that include cell-cycle regulation, proliferation, apoptosis, drug toxicity, DNA repair, and ubiquitination20-22 and may significantly affect cancer cell development and progression as well as the generation of drug resistance.
Various molecular analyses have suggested that MM, like lymphoma and leukemia, is composed of clinically distinct disease subtypes that possess different molecular signatures. The identification of specific genes that function in myelomagenesis may yield insight into the pathways that are responsive to targeted therapy. Such molecular subtypes should respond differently to treatment and, hence, yield variable prognoses and outcomes. To further the development of prognostically relevant molecular subtypes, gene expression profiling from newly diagnosed patients treated with high-dose melphalan-based tandem transplantations was used to address the role of the sumoylation pathway in MM. We have identified a signature composed of sumoylation pathway components that is enhanced in MM patients and correlates with patient outcome. Induction of the sumoylation pathway confers multiple properties on myeloma cells that promote tumorigenesis.
Methods
Gene expression profiling and data analyses
Gene expression profiling was performed with the Affymetrix U133 Plus 2.0 microarray platform using methods previously described.22 All data used in these analyses were derived with the Affymetrix Microarray Suite GCOS1.1 software. Affymetrix signals were transformed by the log-base 2 for each sample. Supervised cluster analysis of known classes was performed using GeneCluster2 (Broad Institute, Harvard University, Cambridge, MA). Primary MM cells were purified from bone marrow as described previously,23 and all samples applied to microarray contained more than 85% PCs as determined by 2-color flow cytometry (CD138+ and CD45−/dim) performed after selection.
Preparation of total cell lysate from purified normal and patient plasma cells
CD138+ cells from monoclonal gammopathy of unknown significance (MGUS) and MM bone marrow samples were obtained from newly diagnosed, previously untreated patients under protocols with Institutional Review Board approval. Fresh, normal, human adult bone marrow was from AllCells, and CD138+ cells were affinity purified by positive selection using Miltenyi CD138+ microbeads (Miltenyi Biotec) according to the manufacturer's instructions.
RT-PCR of normal, MGUS, and MM patient samples
Total RNA was obtained from MM cell lines as well as purified normal PCs, and MGUS and MM cells from patients. Cells were placed in RNAlater (Ambion) and total RNA was purified according to the manufacturer's instructions using the QIAGEN RNeasy Mini Kit. A total of 150 ng of total RNA was retro-transcribed with Superscript (SS2) EZ Kit of Invitrogen. A total of 1 μL of cDNA was used for each sample. Primers used for polymerase chain reactions (PCRs) were: UBE2I forward, 5′ CATTTGGTTTCGTGGCTGTCC 3′; UBE2I reverse, 5′ TTATGAGGGCGCAAACTTCTT 3′; GADPH forward, 5′ ACCACAGTCCATGCCATCAC 3′; GAPDH reverse, 5′ CCACCACCCTGTTGCTGTA 3′; ACTIN B forward, 5′ CCTCGCCTTTGCCGATCC; and ACTIN B reverse, 5′ GGATCTTCATGAGGTAGTCAGTC 3′. PCRs were performed for 28 cycles for UBE2I and 18 cycles each for GAPDH and ACTIN B. Products were separated on a 1.4% agarose gel supplemented with ethidium bromide in 1× TBE.
Myeloma cell lines
Multiple myeloma cell lines (MMCLs) used in the study were RPMI 8226, U266, ARP1, ARD, OPM1, OPM2, MM1.R, MM1.S, LR5, KMS28-BM, KMS28-PE, NCI-H929, and INA6. MMCLs were cultured in RPMI 1640 that contained 10% fetal bovine serum (Sigma-Aldrich), 2μM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen).
Preparation of cell lysates
Cells were grown in RPMI 1640 supplemented with 10% fetal calf serum, glutamine, and antibiotics. Cells were pelleted at 2500g for 5 minutes at 4°C, washed with ice-cold phosphate-buffered saline (PBS) twice, and resuspended in 50 μL of CelLytic M (Sigma-Aldrich) supplemented with complete mini-ethylenediaminetetraacetic acid-free protease inhibitor cocktail (Roche Diagnostics) and 25mM NEM (Sigma-Aldrich). Cells were pipetted up and down 3 times, rotated for 15 minutes at 4°C, centrifuged at 15 000g for 15 minutes, and supernatant was then removed and used as total cell lysate.
Western blotting
Samples were separated by electrophoresis on Supersep HG TM 5% to 20% gradient gels (Wako). Gels were transferred to nitrocellulose membranes (Invitrogen), blocked with 5% carnation nonfat dry milk (Nestle) in Tris-buffered saline supplemented with 0.5% Tween 20 (Sigma-Aldrich) for 1 hour, and then incubated with antibodies. UBE2I monoclonal antibody was from BD Biosciences and used at a final concentration 1:1000. Sumo-1 rabbit polyclonal antibody (Ab32058) was from Abcam and used at 1:1000 dilution. The anti-myc tag monoclonal antibody, clone 9E10, was from Upstate Biotechnology and used at 1:1000 dilution.
Rabbit polyclonal antibody to human NSE2 (BC100-2506) was obtained from Novus Biologicals and was used at a final concentration of 1:500 and rabbit polyclonal antibody to PIAS1 was from Abcam. Mouse monoclonal antibody to GAPDH (Ab9484) was from Abcam. Secondary antibodies were goat anti–rabbit IgG-HRP (sc-2054) and goat anti–mouse IgG-HRP (sc-2005) lot B1507, both from Santa Cruz Biotechnology and used at a final concentration of 1:2000. Immunoblots were developed using the enhanced chemiluminescence reagent system from GE Healthcare Bio-Sciences and Kodak BioMax MR Film.
Transfection of cell lines
MMCLs were transfected with plasmid DNA using the Amaxa electroporation method according to the manufacturer's instructions. Briefly, the day before transfection, cells were plated at a density of 106 cells/mL in RPMI 1640 media that lacked antibiotics and allowed to grow overnight. For each Amaxa transfection reaction, 106 cells and 2 μg of purified plasmid DNA were used. Cells were then washed with PBS, the pellet resuspended in transfection solution, plasmid DNA added, and then transfected with the Amaxa nucleofector. Immediately after transfection, cells were resuspended in RPMI 1640 media that lacked antibiotics and had been prewarmed to 37°C. Cells were grown for 16 hours, pelleted, placed in complete RPMI 1640 media, and incubated for an additional 24 hours before further use. The viability of cells after Amaxa transfection was 95% as measured by trypan blue staining. Efficiency of transfection was determined by cotransfection of cells with a control plasmid that expressed the fluorescent protein GFP and was estimated at 70%. All media were supplemented with 10% fetal bovine serum, 2mM glutamine, 100 units of penicillin/mL, and 100 μg of streptomycin/mL. Cells were incubated at 37°C in a humidified chamber supplemented with 5% CO2.
Plasmids
Human Ubc9/Ube2I wild-type and a DN mutant that carries the C93S mutation within the Ube2I coding sequence were cloned into the pCMV-Myc vector (Clontech) and were kindly provided by Dr Yin-Yuan Mo (University of Illinois at Chicago, Chicago, IL); (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Quantitation of DNA synthesis by uptake of 3H-thymidine
DNA synthesis was measured by uptake of tritiated thymidine [3H-TdR] (NEN Life Science Products). Briefly, 36 hours after transfection, RPMI 8226 cells that were mock transfected or transfected with plasmids that expressed either wild-type UBE2I or a dominant-negative form, UBE2I-DN, were incubated in 96-well culture plates (Costar) at 2 × 104 cells/well. Cells were incubated alone or in coculture with bone marrow stroma cells (BMSCs) from normal patients, in the absence or presence of different concentrations of bortezomib (Velcade) for 24 hours and 48 hours at 37°C. Cells were pulsed with 3H-TdR (0.5 mCi [0.185 MBq]/well), harvested onto a glass filter with an automatic cell harvester (Cambridge Technology), and counted using an LKB plate reading β-scintillation counter (Wallac). All assays were performed in triplicate.
Detection of apoptosis
Dual staining with fluorescein isothiocyanate (FITC)–labeled annexin V and propidium iodide (PI) was carried out to detect induction of apoptotic cell death; 36 hours after transfection, 106 RPMI 8226 cells transfected mock, UBE2I-wt or UBE2I-DN were treated with 10 Gy γ-radiation and cultured for 0, 24, and 48 hours at 37°C. At each time point, cells were washed with PBS and resuspended in 100 μL of binding buffer containing annexin V–FITC and PI (annexin V–FITC apoptosis detection kit; BD Biosciences PharMingen). After 15 minutes of incubation at room temperature, cells were analyzed using a Coulter flow cytometer for the presence of the annexin V–FITC-positive/PI-negative apoptotic cell population.
Cell adhesion assay
Adhesion of myeloma cells to normal bone marrow was measured using the Vybrant TM Cell Adhesion Assay Kit (V-13181) from Invitrogen according to the manufacturer's instructions. Briefly, 104 BMSCs were placed in a 96-well plate for 12 hours at 37°C. Cells were then washed with PBS, placed in RPMI 1640, and incubated with calcein AM (component A) at a final concentration of 5 μm. After incubation, 105 cells from the calcein suspension were added to the bone marrow adhered to the microtiter plate well. Myeloma cells and bone marrow were incubated for 2 hours; then wells were washed with PBS and read using a fluorescein filter at an excitation of 490nM and an emission of 517nM.
Coimmunoprecipitation
RPMI 8226 cells were grown as described in “Preparation of cell lysates” and treated with 10 Gy γ-radiation, incubated under standard conditions, and at indicated times centrifuged at 2500g at 4°C for 5 minutes. Cells were then washed once with ice-cold PBS, frozen at −80°C, and thawed and lysed in 100 μL of Cel-Lytic M nondenaturing buffer (Sigma-Aldrich) supplemented with 1 tablet/10 mL of protease inhibitor cocktail (Roche) and 50mM NEM (Sigma-Aldrich). Approximately 2 million cells were used for each time point. The cells were rotated at 4°C for 5 minutes, centrifuged at 10 000g for 10 minutes, and the supernatant was removed and constituted the total cell lysate; 50 μL of cell lysate was then removed from each sample, added to a fresh tube, and 3 μL of UBE2I polyclonal antibody (BD Biosciences) and 7 μL of protein A–conjugated Sepharose (Zymed/Invitrogen) were added. The mixture was then rotated at 4°C for 16 hours, washed with 200 μL of ice-cold PBS 3 times, and 20 μL of sodium dodecyl sulfate-sample buffer was then added to the pellet. The sample was then boiled for 3 minutes, centrifuged for 3 minutes at 15 000g, and 10 μL was loaded onto an sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10%-20%) gradient WAKO gels, transferred to a nitrocellulose membrane, and probed with a PIAS1 rabbit polyclonal antibody (Abcam).
Results
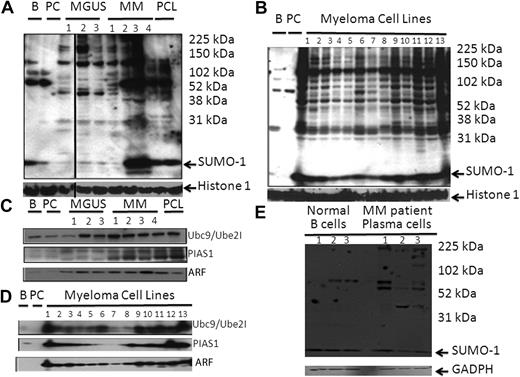
To evaluate the pattern of sumoylation in human nonmalignant and malignant states, whole-cell lysates were initially probed with a Sumo-1 antibody. The pattern of sumoylation was significantly enhanced when MGUS and MM patient samples were compared with that of normal B cells and PCs (Figure 1A). In addition, the pattern of Sumo-1 immunoreactive bands was strikingly similar between individual MGUS and MM patients, although the amount of unconjugated Sumo-1 was variable between patients. Lysates from MMCLs also displayed a set of immunoreactive bands similar to that seen in patient samples (Figure 1B). Western blotting demonstrated a significant elevation of Ube2I in most MGUS and MM patient samples and MMCLs relative to normal B cells and PCs (Figure 1C-D). We also detected a higher level of PIAS1 and ARF in most MGUS and MM patient samples and in the MMCLs. Finally, we compared the pattern of sumoylation in total protein from the lysates obtained from 3 normal B-cell samples and from 3 MM patients (Figure 1E). The relative amount of Sumo-1 was equivalent between the normal and myeloma samples, but the pattern of sumoylation was enhanced in patient samples.
The sumoylation pathway is up-regulated in MM. (A) Sumo-1 immunoblot of protein lysate from myeloma patient PCs. Total lysate was prepared from 105 cells and separated by electrophoresis on 5% to 20% gradient gels. (B) Sumo-1 immunoblot of total protein lysate from MMCLs. Total lysates were analyzed as in panel A. Samples loaded were normal B cell, normal PC, and then samples from the MMCLs, which were: 1, RPMI 8226; 2, U266; 3, ARP1; 4, ARD; 5, OPM1; 6, OPM2; 7, MM1.S; 8, MM1.R; 9, LR5; 10, KMS, 28BM; 11, KMS, 28PE; 12, NCI, H929; and 13, INA6. (C) Sumoylation pathway components on MM patient samples. Total lysate prepared from 105 cells was separated by electrophoresis on 10% to 20% gradient gels. Samples were probed with antisera to Ube2I, PIAS1, and ARF as indicated. (D) Sumoylation pathway components in total lysates were analyzed as in panel A. Samples loaded were normal B cell, normal PC, and then samples from the MMCLs, which were: 1, RPMI 8226; 2, U266; 3, ARP1; 4, ARD; 5, OPM1; 6, OPM2; 7, MM1.S; 8, MM1.R; 9, LR5; 10, KMS-28BM; 11, KMS-28PE; 12, NCI-H929; and 13, INA6. Samples were probed with antisera to Ube2I, PIAS1, Nse2, ARF, and histone 1, respectively. (E) Sumo-1 immunoblot of protein lysate from 3 normal B-cell samples and from 3 myeloma patient PC samples. Total lysate was prepared, separated by electrophoresis on 5% to 20% gradient gels, and probed with a Sumo-1 antibody.
The sumoylation pathway is up-regulated in MM. (A) Sumo-1 immunoblot of protein lysate from myeloma patient PCs. Total lysate was prepared from 105 cells and separated by electrophoresis on 5% to 20% gradient gels. (B) Sumo-1 immunoblot of total protein lysate from MMCLs. Total lysates were analyzed as in panel A. Samples loaded were normal B cell, normal PC, and then samples from the MMCLs, which were: 1, RPMI 8226; 2, U266; 3, ARP1; 4, ARD; 5, OPM1; 6, OPM2; 7, MM1.S; 8, MM1.R; 9, LR5; 10, KMS, 28BM; 11, KMS, 28PE; 12, NCI, H929; and 13, INA6. (C) Sumoylation pathway components on MM patient samples. Total lysate prepared from 105 cells was separated by electrophoresis on 10% to 20% gradient gels. Samples were probed with antisera to Ube2I, PIAS1, and ARF as indicated. (D) Sumoylation pathway components in total lysates were analyzed as in panel A. Samples loaded were normal B cell, normal PC, and then samples from the MMCLs, which were: 1, RPMI 8226; 2, U266; 3, ARP1; 4, ARD; 5, OPM1; 6, OPM2; 7, MM1.S; 8, MM1.R; 9, LR5; 10, KMS-28BM; 11, KMS-28PE; 12, NCI-H929; and 13, INA6. Samples were probed with antisera to Ube2I, PIAS1, Nse2, ARF, and histone 1, respectively. (E) Sumo-1 immunoblot of protein lysate from 3 normal B-cell samples and from 3 myeloma patient PC samples. Total lysate was prepared, separated by electrophoresis on 5% to 20% gradient gels, and probed with a Sumo-1 antibody.
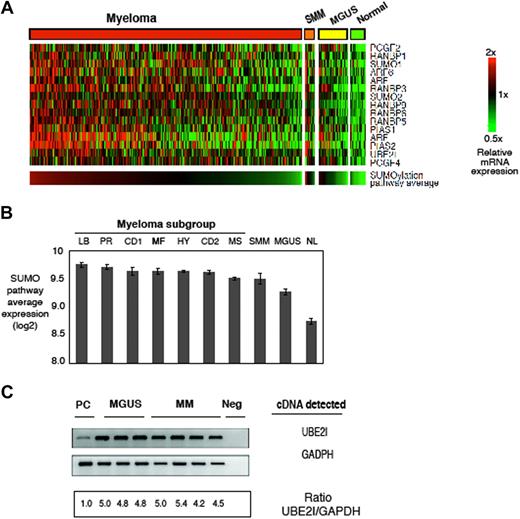
To determine whether the aberrant pattern of sumoylation was driven by transcriptional modulation, we compared expression of genes that regulate sumoylation in the profiles of normal PCs, MGUS, SMM, and MM samples23 (Figure 2A). We detected induction of the Sumo-conjugating enzyme (E2) UBE2I, the sumo ligase (E3) PIAS1, and to a lesser extent the other ligases RanBPs and polycomb gene family, which bind substrates and, coupled with Ube2I, facilitate the attachment of Sumo-1. Unsupervised hierarchic cluster analysis has produced 2 major dendrogram branches with 7 sub-branches, which in turn were strongly influenced by the coordinated overexpression of specific genes, many with anchoring genes, such as c-MAF and MAFB, CCND1, CCND3, ASS, IL6R, MMSET, FGFR3, CCNB2, FRZB, and DKK1.23 We found that expression of the Sumo pathway signature was elevated in all 7 MM sub-branches and also in the SMM group but less so in MGUS samples and not elevated in normal samples (Figure 2B). In addition, the signature was elevated in MMCLs to support the validity of these lines as models of sumoylation pathway dysregulation (data not shown). To validate the expression profiling results, RT-PCR was performed on the MGUS and MM patient samples and indicated a greater than 5-fold induction of UBE2I relative to normal PCs (Figure 2C).
Gene expression profiling of normal, MGUS, smoldering, and myeloma patient plasma cells and generation of a sumoylation pathway signature. (A) Gene expression profiles of sumoylation pathway components from normal, MGUS, SMM (GEO dataset GSE5900), and MM (GEO dataset GSE4204). Samples were median centered and ordered according to average expression of the sumoylation pathway genes. B indicates normal B cells; PC, normal plasma cells; and PCL, plasma cell leukemia. (B) Sumo-pathway signature in the 7 sub-branches of MM, SMM, MGUS, and normal PCs. The sub-branches were classified by expression of MMSET (MS), MAF/MAFB (MF), and proliferation (PR) signatures that constituted high-risk disease, whereas hyperdiploidy (HY), low bone disease (LB), and CCND1/CCND3 translocations (CD1 and CD2) represented low-risk MM. (C) RT-PCR of UBE2I in MGUS and MM patient samples. GAPDH (glyceraldehyde phosphate dehydrogenase) served as a loading control. UBE2I refers to the gene that encodes the SUMO-conjugating enzyme UBE2I; Neg, PCR reaction with no plasma cell cDNA and primers added. Normal plasma cells were purchased from AllCells.
Gene expression profiling of normal, MGUS, smoldering, and myeloma patient plasma cells and generation of a sumoylation pathway signature. (A) Gene expression profiles of sumoylation pathway components from normal, MGUS, SMM (GEO dataset GSE5900), and MM (GEO dataset GSE4204). Samples were median centered and ordered according to average expression of the sumoylation pathway genes. B indicates normal B cells; PC, normal plasma cells; and PCL, plasma cell leukemia. (B) Sumo-pathway signature in the 7 sub-branches of MM, SMM, MGUS, and normal PCs. The sub-branches were classified by expression of MMSET (MS), MAF/MAFB (MF), and proliferation (PR) signatures that constituted high-risk disease, whereas hyperdiploidy (HY), low bone disease (LB), and CCND1/CCND3 translocations (CD1 and CD2) represented low-risk MM. (C) RT-PCR of UBE2I in MGUS and MM patient samples. GAPDH (glyceraldehyde phosphate dehydrogenase) served as a loading control. UBE2I refers to the gene that encodes the SUMO-conjugating enzyme UBE2I; Neg, PCR reaction with no plasma cell cDNA and primers added. Normal plasma cells were purchased from AllCells.
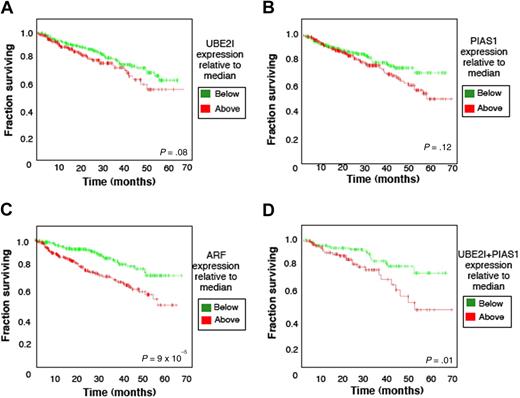
We then postulated that sumoylation-related gene expression would impact the outcome of myeloma patients uniformly treated with melphalan-based high-dose chemotherapy. Interestingly, 67% of patients with UBE2I expression levels below the median survived 6 years, whereas only 55% of those that exhibited UBE2I above the median survived the same period (P = .08, Figure 3A). A similar adverse effect on survival was observed with induction of either PIAS1 (Figure 3B) or ARF (Figure 3C). A 2-gene analysis was then performed and indicated that only 45% of patients who expressed both UBE2I and PIAS1 above the median survived 6 years. Strikingly, the 6-year survival for patients who expressed both genes below the median was nearly 80% (P = .01; Figure 3D). In patients who expressed both UBE2I and ARF above the respective median levels, 6-year survival was significantly superior to those that had levels below the median but was not synergistic as seen with UBE2I and PIAS1 (data not shown).
Overall survival plots for sumoylation pathway components. Kaplan-Meier plots represent 6-year overall survival for MM patients treated with high-dose chemotherapy and stem cell transplantation. Plots indicated the survival of those with indicated gene expression either above or below the median. A total of 551 patients were included in the analyses in (A) with UBE2I, (B) with PIAS1, or (C) with ARF. A total of 350 patients were analyzed (D) with 175 patients having expression of both UBE2I and PIAS1 genes above the median and 175 patients below the median.
Overall survival plots for sumoylation pathway components. Kaplan-Meier plots represent 6-year overall survival for MM patients treated with high-dose chemotherapy and stem cell transplantation. Plots indicated the survival of those with indicated gene expression either above or below the median. A total of 551 patients were included in the analyses in (A) with UBE2I, (B) with PIAS1, or (C) with ARF. A total of 350 patients were analyzed (D) with 175 patients having expression of both UBE2I and PIAS1 genes above the median and 175 patients below the median.
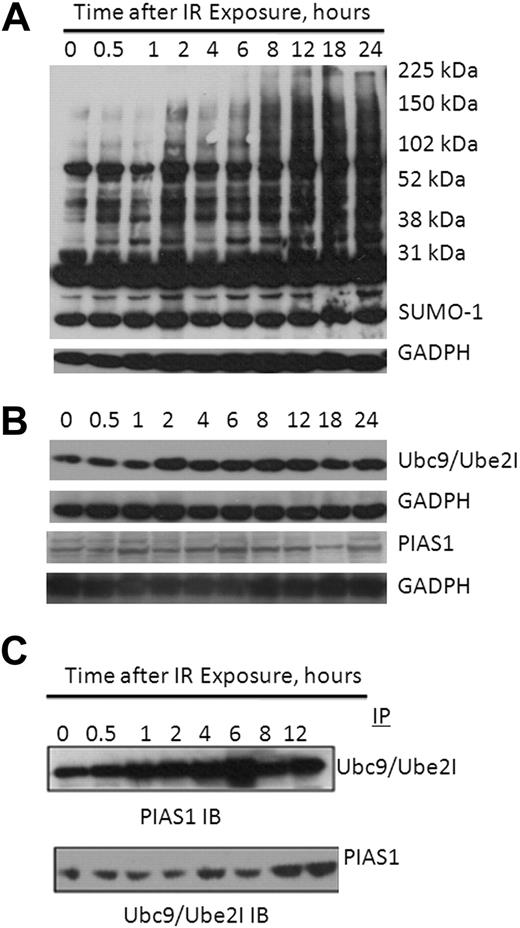
Because we had detected a genomic and clinical correlation between UBE2I and PIAS1 and both genes and their products were elevated in MM patients, we then asked whether a physical association of these proteins was detectable and whether such an association was DNA-damage inducible. RPMI 8226 cells were treated with 10 Gy γ-radiation and lysates probed with a Sumo-1 antibody to demonstrate that sumoylation targets were modified on DNA damage (Figure 4A). Similarly, when lysates were probed with antisera to Ube2I, PIAS1, or ARF, each of the gene products was induced on exposure to radiation as well (Figure 4B). Lysates were then immunoprecipitated with a Ube2I antibody and probed with either a polyclonal antibody specific to either PIAS1 or ARF. A faint amount of PIAS1 was detected in immunoprecipitates without prior γ-radiation exposure consistent with this complex being present in untreated cells, but the association was significantly enhanced as early as 1 hour after radiation exposure (Figure 4C). Similarly, on immunoprecipitation with PIAS1 antisera, we detected an association of UBE2I with PIAS1 without prior treatment and significant enhancement on exposure to γ-radiation (Figure 4C).
DNA damage-induced protein sumoylation and physical association of UBE2I and PIAS1. (A) Sumo-1 immunoblot of RPMI 8226 lysates at indicated times after exposure to 10 Gy γ-radiation. (B) DNA-damage promotes induction of UBE2I and PIAS1. RPMI 8226 lysates at indicated times after exposure to 10 Gy γ-radiation were probed with antisera to either Ube2I (top) or PIAS1 (bottom). (C) Coimmunoprecipitation of PIAS1 with Ube2I. Ube2I immunoprecipitates of RPMI 8226 cells treated with 10 Gy γ-radiation show immunoreactive bands with a PIAS1 pAb (top); conversely, PIAS1 immunoprecipitates probed for MYC detected MYC-Ube2I fusion protein (20 kDa).
DNA damage-induced protein sumoylation and physical association of UBE2I and PIAS1. (A) Sumo-1 immunoblot of RPMI 8226 lysates at indicated times after exposure to 10 Gy γ-radiation. (B) DNA-damage promotes induction of UBE2I and PIAS1. RPMI 8226 lysates at indicated times after exposure to 10 Gy γ-radiation were probed with antisera to either Ube2I (top) or PIAS1 (bottom). (C) Coimmunoprecipitation of PIAS1 with Ube2I. Ube2I immunoprecipitates of RPMI 8226 cells treated with 10 Gy γ-radiation show immunoreactive bands with a PIAS1 pAb (top); conversely, PIAS1 immunoprecipitates probed for MYC detected MYC-Ube2I fusion protein (20 kDa).
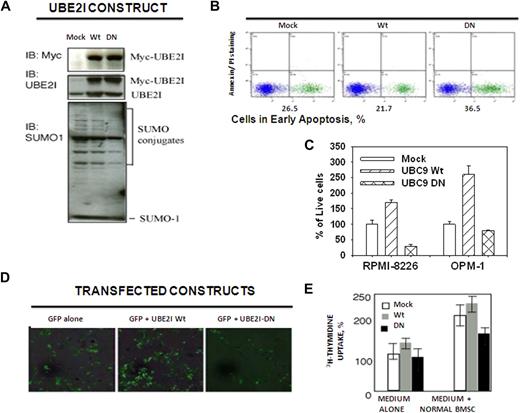
Because UBE2I encodes the sole enzyme responsible for Sumo conjugation, we addressed the biologic properties and downstream events that result from the induction of UBE2I. MMCLs were transfected with plasmids that expressed either a dominant-negative functionally inactive UBE2I mutant (UBE2I-DN) or the wild-type form (UBE2I-wt).24 Both plasmids contained an N-terminal myc peptide tag to confirm transfection and immunoblotting confirmed overexpression of the fusion proteins (Figure 5A top). UBE2I-DN transfectants displayed significantly less total protein sumoylation relative to mock and UBE2I-wt transfected cells (Figure 5A bottom). Cells transfected with UBE2I-DN were treated with 10 Gy γ-radiation to induce DNA damage displayed decreased survival and enhanced apoptosis, whereas those transfected with UBE2I-wt were less sensitive to radiation (Figure 5B). After transfection of UBE2I-DN into both RPMI 8226 and OPM1 cells, transfectants demonstrated impaired MTT activity, whereas those cells transfected with UBE2I-wt exhibited enhanced activity relative to mock-transfected cells (Figure 5C). The capacity of UBE2I-DN transfectants to adhere to BMSCs was also reduced (Figure 5D). Adhesion of myeloma cells to BMSCs not only localizes cells in the bone marrow microenvironment but also triggers interleukin-6 secretion by BMSCs and facilitates cell proliferation.25 The effect of BMSCs to stimulate 3H-thymidine uptake indicated that UBE2I-wt transfected cells displayed a 2-fold induction on interaction with BMSCs (Figure 5E). Conversely, UBE2I-DN transfectants demonstrated diminished proliferative response in the presence of BMSCs.
Effect of UBE2I-dominant-negative mutant on growth, proliferation, and adhesion to BMSCs. (A) Myc-tagged fusion proteins were expressed in RPMI 8226 cells. Mo indicates mock transfected; Wt, UBE2I wild-type; and DN, UBE2I-DN (dominant negative). Lysates were probed with a Myc-tag specific mAb (top), Ube2I-specific mAb (middle), or Sumo-1 polyclonal Ab (bottom). (B) UBE2I-DN increased γ-radiation–induced apoptosis. Early apoptosis was detected by annexin V staining. Plots indicate percentage of RPMI 8226 cells in early apoptosis 24 hours after treatment with 10 Gy γ-radiation. Data are representative of 3 independent experiments. (C) UBE2I-wt and UBE2I-DN resulted in opposite effects on MM cell growth. Values represent the mean of triplicate measurements using the MTT assay. (D) Ube2I is necessary for MM adhesion to bone marrow stroma. Adhesion of GFP-labeled UBE2I transfectants to normal BMSCs is shown by fluorescence microscopy after 96 hours of incubation under standard conditions. Data are representative of 3 independent experiments. Images were viewed with a Zeiss Axiovert 200 inverted epifluorescence microscope using a 20× objective (37°C; cells were in PBS; FITC fluorescent filter). Images were acquired with a Zeiss AxioCam HRc 14-bit color CCD camera and were processed with Axio Vision software (Version 3.1). (E) UBE2I-DN decreased BMSCs-induced uptake of 3H-thymidine. Values represent the mean of triplicate measurements of 3H-thymidine after 96 hours of coculture.
Effect of UBE2I-dominant-negative mutant on growth, proliferation, and adhesion to BMSCs. (A) Myc-tagged fusion proteins were expressed in RPMI 8226 cells. Mo indicates mock transfected; Wt, UBE2I wild-type; and DN, UBE2I-DN (dominant negative). Lysates were probed with a Myc-tag specific mAb (top), Ube2I-specific mAb (middle), or Sumo-1 polyclonal Ab (bottom). (B) UBE2I-DN increased γ-radiation–induced apoptosis. Early apoptosis was detected by annexin V staining. Plots indicate percentage of RPMI 8226 cells in early apoptosis 24 hours after treatment with 10 Gy γ-radiation. Data are representative of 3 independent experiments. (C) UBE2I-wt and UBE2I-DN resulted in opposite effects on MM cell growth. Values represent the mean of triplicate measurements using the MTT assay. (D) Ube2I is necessary for MM adhesion to bone marrow stroma. Adhesion of GFP-labeled UBE2I transfectants to normal BMSCs is shown by fluorescence microscopy after 96 hours of incubation under standard conditions. Data are representative of 3 independent experiments. Images were viewed with a Zeiss Axiovert 200 inverted epifluorescence microscope using a 20× objective (37°C; cells were in PBS; FITC fluorescent filter). Images were acquired with a Zeiss AxioCam HRc 14-bit color CCD camera and were processed with Axio Vision software (Version 3.1). (E) UBE2I-DN decreased BMSCs-induced uptake of 3H-thymidine. Values represent the mean of triplicate measurements of 3H-thymidine after 96 hours of coculture.
Discussion
We have identified a compendium of sumoylation pathway genes that are induced in MM and have shown that UBE2I induction is an early event in myelomagenesis that confers a range of biologic properties that may promote tumorigenesis. These properties include increased proliferation, apoptotic resistance, and adhesion to BMSCs. Because MM may result from an acquired injury or DNA-damaging event to a lymphocyte progenitor destined to become a PC, UBE2I induction may contribute to the survival and clonal selection of such progenitors. The precise targets of Ube2I sumoylation during MM pathogenesis remain speculative and, hence, the multiplicity of Ube2I targets may explain the diverse biologic effects. Numerous targets of the sumoylation pathway have been identified downstream from Ube2I and PIAS1, specifically, STAT family members, p53 and c-Jun.17,18 Importantly, we have recently shown that the tumor suppressor p53 is a major target of sumoylation in MMCLs after DNA damage and is also found sumoylated in MM patient samples but not in normal PCs (J.J.D., manuscript in preparation). Similarly, inhibitors of the 26S proteasome complex that is responsible for ATP+Ub-dependent proteolysis have demonstrated clinical benefit, although the precise substrates stabilized by bortezomib that ultimately lead to cell death remain unidentified. Other potential functions of Ube2I, independent of sumoylation, remain plausible and expand the mechanistic possibilities of function during oncogenesis.26,27
It is reasonable to speculate that the sumoylation of specific substrates is a prerequisite step that precedes ubiquitination and proteasomal degradation. A series of recent publications have reported that poly-Sumo chains may exert a signaling function through a newly discovered class of Ub-pathway E3-ligases. The attachment of poly-Sumo chains to targets promotes ubiquitination and degradation. Two groups demonstrated that in acute promyelocytic leukemia the degradation of PML and PML-RARα is through a sumo-triggered recruitment of the RING finger E3 ligase RNF4. Interaction with RNF4 leads to lysine 48–linked ubiquitination and arsenic-induced proteasomal degradation.28-30 Thus, sumoylation may thereby indirectly influence the degradation, stability, and functional activity of certain targets. The integration of the sumoylation, ubiquitination, and proteasomal pathways is plausible and would further support Ube2I and the sumoylation pathway as therapeutic targets with clinical implications for Ub+proteasomal-based treatment modalities. Because a limited number of MM patients respond to bortezomib, linkage of sumoylation to ubiquitination and proteasomal degradation may provide insights into the mechanism of action and more specific forms of targeted therapy.
In contrast to the Ub-pathway where numerous (> 30) conjugating enzymes exist with distinct substrate specificities, it appears that UBE2I encodes the sole Sumo-conjugating enzyme in mammalian cells. Thus, the fundamental role of UBE2I (and the sumoylation pathway) in critical cellular processes, the correlation with treatment outcome, demonstrable elimination of functionality by a dominant-negative form that prevents homodimerization, and an elevation of the sumo signature in all MM sub-branches collectively render and validate the UBE2I gene product as a highly attractive candidate for therapeutic intervention.
Although MM patients present with similar diagnostic histology, they display widespread genomic variability as well as marked variations in clinical characteristics and survival.31,32 MM subtypes have been identified with concordant expression signatures predominantly driven by recurrent translocations and hyperdiploidy22 and, in concert with the present study, advocate expression profiling as an approach to identify genes and cellular pathways that regulate myelomagenesis. We have used expression profiles to identify genes that may contribute to disease initiation and progression and to identify high-risk groups, namely, those that overexpress UBE2I and PIAS1. After high-dose therapy and stem cell transplantation, the 7 MM subtypes displayed striking differences in event-free and overall survival. Early disease-related death was linked to 70 genes,33 including UBE2I, whereas other genes composed a nuclear factor-κB (NF-κB) signature. The importance of the NF-κB signaling pathway was indicated by the nuclear presence of NF-κB and the sensitivity of MM cell lines to NF-κB inhibition.34,35 Within the BM microenvironment, production of interleukin-6, BAFF, or APRIL may act as potential activators of NF-κB signaling pathways and diverse genetic and epigenetic mechanisms to activate NF-κB signaling have been reported.36,37 Interestingly, NF-κB target gene expression was elevated, although the NF-κB signature was highest among B-cell subsets in normal PCs, possibly the result of BAFF and APRIL-mediated signaling. In contrast, UBE2I was expressed at a relatively low level in normal PCs and was substantially induced in most MMCLs and patient samples from all 7 MM sub-branches. Because the pursuit of novel, therapeutically efficient strategies requires the identification and validation of tumor cell–specific targets that also provide a therapeutic index, the high frequency of UBE2I induction makes it an attractive target for therapeutic intervention. Further understanding of the high degree of genomic heterogeneity and recurrent translocations seen in MM should provide a more integrated view to further decipher myelomagenesis and tumorigenesis in general. The refinement and application of customized, bioinformatic genomic and proteomic tools to newly diagnosed or refractory cases of MM should eventually stratify treatment options. The advent of microarray technology has facilitated the molecular classification of specific cancers, such as MM, into clinically relevant subtypes and should accelerate the transition between empirical and tailored medicine.38
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Y.-Y. Mo (Southern Illinois University) for the gift of UBE2I expression vectors, Wei-Hua Song (Dana-Farber Cancer Institute) for assistance with γ-radiation of cells, Cheng Li and Jie Hu (Harvard Medical School) for assistance with microarray data analysis, and Michael Kuehl (Genetics Branch, National Institutes of Health) for critical reading of the manuscript and insightful comments.
This work was supported in part by the Department of Veterans Affairs Merit Review Awards and the National Institutes of Health (grants RO1-124929, N.C.M.; P50-100007 and PO1-78378, N.C.M., K.C.A.), and a Myeloma Research Fellow SPORE Grant (J.J.D.).
National Institutes of Health
Authorship
Contribution: J.J.D., D.P., R.H.P., and N.C.M. designed the research; J.J.D., D.P., K.L., M.F., and R.H.P. performed the research; P.R.G., B.B., Y.-T.T., K.C.A., and J.D.S. provided essential reagents; and J.J.D., K.L., C.M.A., and N.C.M. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for J.J.D. is National Cancer Institute, Medical Oncology Branch, Bethesda, MD.
Correspondence: James J. Driscoll, MD, PhD, National Cancer Institute, Medical Oncology Branch, Bethesda, MD 20892; e-mail: driscollj@mail.nih.gov; or Nikhil C. Munshi, MD, Dana-Farber Cancer Institute, Jerome Lipper Multiple Myeloma Center, Boston, MA 02115; e-mail: nikhil_munshi@dfci.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal