Abstract
Single nucleotide polymorphism arrays (SNP-A) have recently been widely applied as a powerful karyotyping tool in numerous translational cancer studies. SNP-A complements traditional metaphase cytogenetics with the unique ability to delineate a previously hidden chromosomal defect, copy neutral loss of heterozygosity (CN-LOH). Emerging data demonstrate that selected hematologic malignancies exhibit abundant CN-LOH, often in the setting of a normal metaphase karyotype and no previously identified clonal marker. In this review, we explore emerging biologic and clinical features of CN-LOH relevant to hematologic malignancies. In myeloid malignancies, CN-LOH has been associated with the duplication of oncogenic mutations with concomitant loss of the normal allele. Examples include JAK2, MPL, c-KIT, and FLT3. More recent investigations have focused on evaluation of candidate genes contained in common CN-LOH and deletion regions and have led to the discovery of tumor suppressor genes, including c-CBL and family members, as well as TET2. Investigations into the underlying mechanisms generating CN-LOH have great promise for elucidating general cancer mechanisms. We anticipate that further detailed characterization of CN-LOH lesions will probably facilitate our discovery of a more complete set of pathogenic molecular lesions, disease and prognosis markers, and better understanding of the initiation and progression of hematologic malignancies.
Introduction
Single nucleotide polymorphism arrays (SNP-A) are currently in use as a powerful genotyping tool for a variety of whole-genome association studies. With resolution as good as one marker per every 100 bp of the genome and quantitative analysis of DNA copy number as well as genotypic information provided by polymorphic SNP markers, an outgrowth of this technology was the realization that SNP-A could potentially provide a powerful karyotyping tool.1 For example, application of SNP-A karyotyping to a large series of pediatric acute lymphocytic leukemia cases identified recurrent subcytogenetic deletions and target genes in the minimal deletion intervals, and new cancer-specific aberrant genetic networks that include PAX5, IKZF1, and CDKN2A.2-4 In addition to the identification of cryptic amplifications and deletions, SNP-A also complements traditional metaphase cytogenetics with the unique ability to delineate a previously underappreciated chromosomal defect, copy neutral loss of heterozygosity (CN-LOH). There are many technical challenges to accurately identifying CN-LOH regions from SNP-A raw data, and CN-LOH results were not reported in the initial acute lymphocytic leukemia (ALL) studies.5 For acute myeloid leukemia (AML) and myelodysplastic syndromes (MDSs), however, even early evaluations using low-density arrays demonstrated CN-LOH as a recurrent finding of potential interest.6 Here, we review the mechanisms that lead to CN-LOH, explore disease-specific CN-LOH patterns and potential biologic implications, and examine the evolving potential clinical significance of this unique genomic finding.
Pathogenesis of CN-LOH
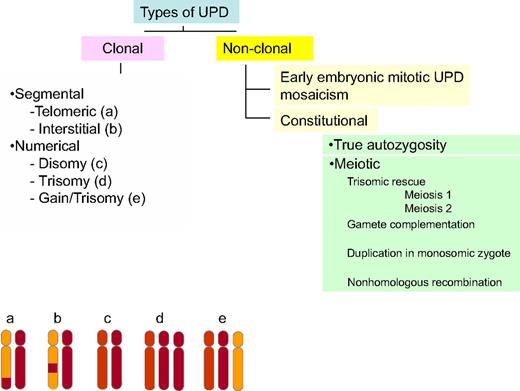
CN-LOH, also often referred as to uniparental disomy (UPD),7 leads to LOH by duplication of a maternal (unimaternal) or paternal (unipaternal) chromosome or chromosomal region and concurrent loss of the other allele. Whereas CN-LOH was initially identified by studying microsatellites, systematic application of SNP-A as a karyotyping tool led to the realization that the previously undetected areas of clonal CN-LOH are frequently encountered in various cancers, including hematologic malignancies.1,8 SNP-A–identified CN-LOH in various malignancies1,9 can be described according to its derivation or its location (Figure 1). CN-LOH, either chromosomal or segmental, can have constitutional or acquired origins. Constitutional UPD is associated with meiotic errors, resulting in developmental diseases, and was first described in humans by Engel10 ; however, it can also be observed in healthy controls, probably because of diverse mechanisms, including early mitotic errors and autozygosity.11-13
Classification of CN-LOH. CN-LOH can be classified by either its origin or its location. CN-LOH can have an acquired, clonal derivation or a constitutional, nonclonal derivation. Nonclonal CN-LOH can be the result of an early embryonic mitotic event, leading to mosaicism, or may be truly constitutional. This constitutional CN-LOH can arise from autozygosity or meiotic events, including trisomic rescue, gamete complementation, duplication of a monosomic chromosome in an aneuploid zygote, or nonhomologous recombination. In addition, CN-LOH can be either segmental or numerical. Segmental CN-LOH arising from one crossing over will be telomeric (a, bottom left), whereas 2 crossing-over events will lead to interstitial CN-LOH (b). CN-LOH can also involve an entire chromosome (numeric; c, d, and e).
Classification of CN-LOH. CN-LOH can be classified by either its origin or its location. CN-LOH can have an acquired, clonal derivation or a constitutional, nonclonal derivation. Nonclonal CN-LOH can be the result of an early embryonic mitotic event, leading to mosaicism, or may be truly constitutional. This constitutional CN-LOH can arise from autozygosity or meiotic events, including trisomic rescue, gamete complementation, duplication of a monosomic chromosome in an aneuploid zygote, or nonhomologous recombination. In addition, CN-LOH can be either segmental or numerical. Segmental CN-LOH arising from one crossing over will be telomeric (a, bottom left), whereas 2 crossing-over events will lead to interstitial CN-LOH (b). CN-LOH can also involve an entire chromosome (numeric; c, d, and e).
Early developmental errors and autozygosity (germline UPD)
Events leading to CN-LOH in developmental disorders help to clarify the pathways that could lead to constitutional UPD observed in healthy persons and distinguish them from acquired UPD (aUPD) in malignancies that, similar to other acquired chromosomal defects, constitute markers of clonality (Figure 1). UPD can occur during meiosis or as a postfertilization mitotic event at a very early embryonic stage, leading to mosaicism.14 Trisomy rescue (loss of a chromosome from a trisomic zygote) of a meiotic nondisjunction error leads to CN-LOH and, depending on the meiotic stage at which the nondisjunction event occurred, results in uniparental isodisomy (meiosis II error) or heterodisomy (meiosis I error).15 In two-thirds of the cases, trisomy rescue would result in a normal diploid, biparental karyotype. A meiosis I nondisjunction error can be followed by a mitotic crossing over between 2 nonuniparental chromatids followed by a loss of chromosomal material originating from a disomic gamete. In a similar setting, UPD can also be associated with an isochromosome arising from meiotic misdivision of the centromeres, with fertilization by a normal gamete and subsequent loss of the homologous chromosome. In addition, fertilization between disomic and nullisomic gametes can produce a similar result. Finally, mitotic misdivision at the centromere in a monosomic zygote can result in CN-LOH (Figure 1).
Theoretically, CN-LOH can lead to either gain of imprinting through duplication of a methylated allele or loss of imprinting through duplication of unmethylated allele.16 For this mechanism to be pathogenic, the CN-LOH region would have to involve genes that are subject to genomic imprinting, such as those in Prader-Willi/Angelman syndromes.17 Detailed descriptions of the discovery of genomic imprinting and pathogenesis of uniparental disomy-related disorders are presented elsewhere.18
Autozygosity is the inheritance of the same ancestral genomic region from both parents. Small stretches (1-5 Mb) of CN-LOH are thought to be the result of autozygosity, in particular in inbred ethnic groups.13 Theoretically, reference samples used as “controls” in some LOH studies may indeed have unexpectedly high genetic similarity from undiscovered consanguinity.19 However, the cause of larger regions of CN-LOH seen in significant proportions of healthy unrelated controls studied in various projects has not been precisely identified, and the size of regions of CN-LOH is expected to get smaller with the increasingly outbred nature of urban human populations.20 Of great interest, forms of constitutional LOH have been implicated in predisposition to malignancies, a fact best illustrated in inbred ethnic populations.21-24 Comparisons of germline SNP-A data of 74 colorectal cancer patients identified that the percentage of those with autozygous segments of 4 Mb or more is at least twice as high as in control groups.25 However, specific locations and disease associations, including their contribution to disease risk, are currently not well defined.
Genesis of aUPD in hematologic malignancy
Extrinsic processes that lead to genomic DNA damage upstream of sites of mitotic recombination have also been implicated in the generation of acquired CN-LOH. Whether treatment with DNA-damaging chemotherapy contributes to the increase in aUPD seen in secondary AML, transformed follicular lymphoma or relapsed versus de novo ALL is unknown. Specific mechanisms responsible for the propensity for chromosomal breaks and subsequent uniallelic strand loss that might occur in MDS or AML before therapy are not well understood, but it is probable that a variety may be involved. These may include acquired or inherited weakness of various components of DNA repair machinery or mitotic spindle machinery or telomere shortening.26 Finally, a supersaturating overwhelming of rate-limiting DNA repair components could theoretically provide another mechanism. The cumulative acquisition of lesions fits well with current models of the association of age with accumulated genetic and epigenetic lesions but remains to be experimentally investigated. For example, whether there is specifically more CN-LOH in disease-associated or even hematologically disease-free aged persons is currently unknown. Chronic lymphocytic leukemia (CLL), myeloproliferative neoplasia (MPN), MDS, and AML are all associated with significant increased incidence with age.
The interpretation of the origin, location, frequency, and clinical implications of CN-LOH ultimately requires an understanding of the potential extrinsic disease-related and intrinsic chromosomal determinants associated with CN-LOH/aUPD development. Unfortunately, there are many more questions than answers related to these issues. Various mechanisms can be responsible for this form of LOH, now found to be very prevalent in various myeloid malignancies. These mechanisms include a cellular attempt to correct a deletion because of mitotic errors, such as anaphase lag through reduplication of the remaining chromosome. In addition, an attempted repair of double-strand breaks resulting in losses whereby the lost region is replaced using the remaining allele as a template can lead to segmental LOH (Figure 2).
Mitotic mechanisms of formation of CN-LOH. (A) CN-LOH can occur as the result of mitotic recombination between homologous chromosomes. Depending on how the chromosomes are sorted during mitosis, daughter cells with CN-LOH can arise. (B) CN-LOH can also arise as the consequence of deletion followed by recombination using the homolog as a template for correction.
Mitotic mechanisms of formation of CN-LOH. (A) CN-LOH can occur as the result of mitotic recombination between homologous chromosomes. Depending on how the chromosomes are sorted during mitosis, daughter cells with CN-LOH can arise. (B) CN-LOH can also arise as the consequence of deletion followed by recombination using the homolog as a template for correction.
Mitotic recombination is considered a major contributor to acquired CN-LOH.27 In the first whole-genome SNP-A study of aUPD in AML, large regions of CN-LOH were identified in 20% of the patients studied; 8 separate nonrandom regions were identified,6 differing in locations from those identified in similar studies of epithelial cancers.28,29 Most regions of homozygosity were partial but extended to the telomere, consistent with UPD occurring as a result of a single mitotic recombination. Subsequent studies of other myeloid malignancies also identified partial aUPD as the most frequent type of CN-LOH in these disorders.1 However, investigations of the molecular basis for somatic NF1 inactivation in childhood leukemias associated with neurofibromatosis type I elegantly illustrated that interstitial UPD can also represent a pathway of aUPD by double-homologous recombination events30 ; 8 of 10 cases had large LOH, half were partial UPD, but 4 had interstitial UPD. Preferred sites of mitotic recombination were also identified, with a clustering of the centromeric and telomeric breakpoints. Similarly, a careful analysis of the sites of aUPD origin in low-risk MDS showed that 43% of UPD regions were localized to within or as part of a previously identified fragile site.31 Fragile locations correspond to known sites of frequent genomic instability. They are associated with the breakpoints of chromosomal aberrations in hematologic malignancies32 and often track with regulatory microRNA amplifications and deletions.33
A high-resolution SNP-A profiling of mantle cell lymphoma (MCL) cell lines and primary tumors34 has provided additional insight into mechanics of aUPD. In MCL, pathognomonic t(11;14)(q13;q32) are associated with secondary chromosomal alterations, including frequent areas of partial UPD, such as UPD17p coinciding with homozygous TP53 inactivation. The breakpoints flanking the genomic alterations, including regions of UPD, were significantly associated with genomic regions enriched in copy number variants and segmental duplications, suggesting that recombination at these regions may play a role in the genetic instability. Thus, copy number variants and segmental duplications may represent DNA breakage-prone regions that may contribute to the generation of chromosomal alterations by facilitating nonallelic homologous recombination, similar to fragile sites.35-37 Additional mechanistic clues have been derived from colon cancer, in which mitotic recombination appears to be important for inactivating tumor suppressor genes. The APC gene located at 5q21 is frequently mutated, often in a homozygous constellation resulting from CN-LOH, through a mitotic recombination mechanism.38 Studies demonstrate that copy number changes in aneuploid/polyploid colorectal tumors generally occur as additional genetic events, whereas LOH by mitotic recombination is an early event that initiates tumorigenesis.39 By mapping mitotic recombination breakpoints between the centromere and APC, they were again found to be nonrandom, with the highest frequency close to locus control regions at 68 to 71 Mb, far from APC. Low copy repeats predispose to chromosomal breakage, perhaps via replication fork stalling, leading to mitotic recombination and ultimately CN-LOH. In contrast, breakpoints involved in APC copy number loss clustered to a different, more centromeric region near a suggested area of the greatest chromosomal fragility. Consequently, it is possible that breakage through the centromere cannot be readily repaired by mitotic recombination because the exposure of pericentromeric repeats produces a chromosome that is prone to nonspecific pairings and recombination.
Specific oncogenes have also been implicated in mechanisms of genomic instability, which make these intriguing candidates to consider as potentially mechanistically involved in the generation of CN-LOH. For example, BRCA1/2 has been linked to UPD seen in ovarian cancer.40 Cyclin D1 is overexpressed in the majority of MCLs, which have abundant aUPD.34 UPD has also been associated with microsatellite instability in AML with normal karyotype.41 The JAK2 V617F mutation is frequently observed in classic MPN, and disease progression is associated with biallelic acquisition of the mutation through mitotic recombination and aUPD. However, in manipulated cell lines and CD34+ cells from patients with JAK2 V617F, an increase in homologous recombination activity in the presence of erythropoietin was observed, without modifications in nonhomologous end joining efficiency.42
Detection of acquired CN-LOH with SNP-A
Traditional karyotyping techniques, including metaphase cytogenetics, cannot detect CN-LOH. In the past, microsatellite analysis was used for detection of CN-LOH. Informative microsatellites present in a germline control sample in a heterozygous constellation were compared with the tumor sample from the same patient to identify LOH. A diploid copy number was shown by karyotyping.43 This approach is cumbersome, and the boundaries of the defect cannot be precisely delineated because of natural limitations of the number of informative microsatellites present in the genome.
SNP-A combines genotyping (classification of a homozygous or heterozygous constellation at a polymorphic locus) and copy number analysis (intensity of hybridization signal).7 A number of general and technical reviews have outlined the conceptual and technical aspects of performing SNP-A karyotyping.1,44 The precision of this technology is increased by the use of bioanalytic algorithms, facilitating a very accurate diagnosis of LOH (Figure 3). Current algorithms are based on the deviation between observed and expected frequencies of homozygous SNPs, which should be approximately 1 of 30 consecutive SNPs tested.45 Consequently, the longer a region of LOH, the higher the number of homozygous SNPs and herewith the statistical improbability of encountering such a situation per chance rises. When a homozygous constellation of genotyping calls is combined with diploid copy number, the diagnosis of CN-LOH can be established (Figure 3). By comparison, comparative genomic hybridization arrays rely solely on analysis of hybridization signals and CN-LOH cannot be detected. The presence of nonclonal diploid cells in the tested DNA sample complicates the SNP-A–based detection algorithm, and in general, the larger the aberrant clone, the better the detection of CN-LOH. In our hands, mixing experiments have shown that even dilution of the clonal, aberrant cells to 30% of the total sample can still allow reliable detection of an acquired abnormality.46
Determination of acquired versus germline nature of CN-LOH. Acquired CN-LOH (red bar, top left) is identified when allelic imbalance (as shown by genotyping calls) with normal copy number (top track) in bone marrow and not CD3+ cells (representing the germline configuration). Top left: An example of acquired CN-LOH of chromosome 7. A region of homozygosity and diploid copy number (as indicated by the red bar) are seen in bone marrow only. Top right: An example of germline CN-LOH of chromosome 20. Runs of homozygosity (red bars) are present in both bone marrow and CD3+ cells. Among a cohort of 1003 healthy controls, CN-LOH was mainly interstitial (bottom left) and ranged in size from 0.3 to 65 Mb (median, 8.7 Mb; bottom center). Bottom right: The distribution of nonclonal, germline CN-LOH in controls on an exemplary chromosome (chromosome 6).
Determination of acquired versus germline nature of CN-LOH. Acquired CN-LOH (red bar, top left) is identified when allelic imbalance (as shown by genotyping calls) with normal copy number (top track) in bone marrow and not CD3+ cells (representing the germline configuration). Top left: An example of acquired CN-LOH of chromosome 7. A region of homozygosity and diploid copy number (as indicated by the red bar) are seen in bone marrow only. Top right: An example of germline CN-LOH of chromosome 20. Runs of homozygosity (red bars) are present in both bone marrow and CD3+ cells. Among a cohort of 1003 healthy controls, CN-LOH was mainly interstitial (bottom left) and ranged in size from 0.3 to 65 Mb (median, 8.7 Mb; bottom center). Bottom right: The distribution of nonclonal, germline CN-LOH in controls on an exemplary chromosome (chromosome 6).
Of utmost importance for the detection of tumor-associated, clonal, acquired CN-LOH is its distinction from germline-encoded forms of CN-LOH, seen in up to 15% of control persons.31 Analysis of 1000 nonclonal control samples (Figure 3; C.O., unpublished results, April 2009) shows that germline regions of CN-LOH usually are smaller (median, 8.7 Mb) and primarily interstitial; whereas in our more than 600 cases of myeloid malignancies, acquired clonal CN-LOH seem virtually uniformly telomeric.46-50 Regions of germline CN-LOH identified in controls show a characteristic size and distribution across the chromosome as show for exemplary chromosome 6 (Figure 3). Interstitial CN-LOH is difficult to explain based on homologous recombination, in particular for LOH regions that are genetically small. Thus, smaller (< 24.6 Mb or 95th percentile of distribution) regions of CN-LOH in particular must be confirmed by analysis of germline DNA to be unambiguously defined as acquired CN-LOH (Figure 3).
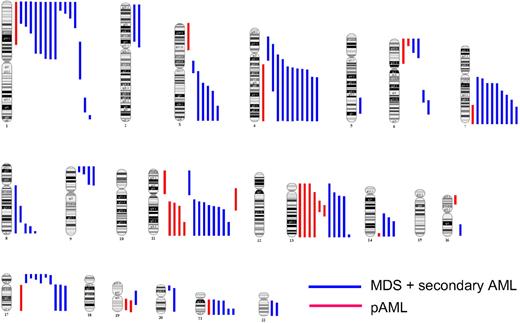
Conceptually, CN-LOH may have several implications. In one way, duplications of chromosomal material constitute UPD as it occurs with the retention of the homologous chromosome, thus implying that UPD without LOH is also possible (Figure 1). In addition, we have observed uniparental trisomy of various chromosomes, including chromosome 21. Although theoretically all chromosomes can be affected, UPD of certain chromosomes is more common. For example, in AML and MDS, chromosomes 4, 7, 11, 13, and 21 show frequently recurrent aUPD (Figure 4).
Genomic distribution of acquired CN-LOH in MDS/secondary AML and primary AML. CN-LOH is nonrandomly distributed across the genome in both MDS/secondary AML (blue lines) and primary AML (red lines), with some chromosomes and chromosomal regions being more frequently affected.
Genomic distribution of acquired CN-LOH in MDS/secondary AML and primary AML. CN-LOH is nonrandomly distributed across the genome in both MDS/secondary AML (blue lines) and primary AML (red lines), with some chromosomes and chromosomal regions being more frequently affected.
Clonal selection of CN-LOH/aUPD
It is increasingly clear that a number of identified and theoretical situations provide a biologic milieu out of which cells may obtain a clonal advantage through the acquisition or expansion of aUPD in hematologic cells. Since the discovery of UPD9p and the corresponding duplication of JAK2 V617F mutations, a number of recurrent areas of UPD associated with homozygous gene mutations have been found,47,51 suggesting that this is a common mechanism leading to homozygosity (Table 1). UPD maps can be constructed for specific malignancies, similar to a map presented in Figure 4 for MDS and AML. This is a new paradigm that brought about the realization that, because many mutations have a pathologic effect only in a homozygous form, areas of CN-LOH point toward the presence of such mutations. It is probable that duplication of a mutated allele is beneficial in the selection process. Thus, the presence of UPD implies at least 2 mutational steps, including acquisition of a mutation followed by homologous recombination as a secondary event resulting in aUPD. Additional steps may be possible as described for patients with concomitant homozygous JAK2 and c-MPL mutations and corresponding areas of CN-LOH.64 However, one could also assume that the initial mutational event is followed by deletion of the normal allele as a secondary step with a subsequent duplication of the retained allele as a tertiary step. Consequently, deletion may reflect an inability to repair genomic losses.
Mutated genes in regions of UPD
| Mutated gene . | Region of UPD . | Described . | References . |
|---|---|---|---|
| MPL | UPD1p | MPN, RARSt | 52 |
| NRAS | UPD1p | JMML, CMML | 49 |
| TET2 | UPD4q | sAML, MPN, MDS/MPD, | 53, 54 |
| JAK2 | UPD9p | PV, ET, IMF, RARSt | 55,–57 |
| CDKN2B | UPD9p | ALL | 4 |
| WT1 | UPD11p | AML | 6 |
| c-CBL | UPD11q | CMML, MDS/MPN, JMML | 49, 50, 58,–60 |
| FLT3ITD | UPD13 | AML | 6, 49 |
| TP53 | UPD17p | MDS, sAML | 61 |
| NF1 | UPD17q | JMML | 62 |
| CEBPa | UPD19q | AML | 6, 63 |
| RUNX1 | UPD21q | AML | 6 |
| Mutated gene . | Region of UPD . | Described . | References . |
|---|---|---|---|
| MPL | UPD1p | MPN, RARSt | 52 |
| NRAS | UPD1p | JMML, CMML | 49 |
| TET2 | UPD4q | sAML, MPN, MDS/MPD, | 53, 54 |
| JAK2 | UPD9p | PV, ET, IMF, RARSt | 55,–57 |
| CDKN2B | UPD9p | ALL | 4 |
| WT1 | UPD11p | AML | 6 |
| c-CBL | UPD11q | CMML, MDS/MPN, JMML | 49, 50, 58,–60 |
| FLT3ITD | UPD13 | AML | 6, 49 |
| TP53 | UPD17p | MDS, sAML | 61 |
| NF1 | UPD17q | JMML | 62 |
| CEBPa | UPD19q | AML | 6, 63 |
| RUNX1 | UPD21q | AML | 6 |
RARSt indicates refractory anemia with ring sideroblasts in transformation; JMML, juvenile myelomonocytic leukemia; sAML, secondary acute myeloid leukemia; PV, polycythemia vera; ET, essential thrombocytopenia; and IMF, idiopathic myelofibrosis.
Principally, we have observed 2 types of recurrent LOH resulting from deletions: those associated with frequent CN-LOH of the corresponding region, for example, UPD7q and del7q, UPD17p and del17, and those that are never or rarely associated with corresponding UPD (Figure 4). In a cohort of more than 600 patients, we have never encountered acquired UPD5q or UPD20q, whereas deletions of the long arm of these chromosomes are frequent. One could stipulate that, should a region deleted from one homolog contain a hemizygous mutation on the other homolog, selection pressure may favor duplication of the mutated copy; consequently, in addition to deletions, corresponding regions of UPD would also be found. However, UPD and deletions affecting the same chromosomal region may also reflect distinct pathogenetic mechanisms. For example, whereas homozygous c-CBL mutations are frequent, hemizygous mutations have not been observed,49 thus implying that deletion and UPD in this region correspond to the presence of different mutations. In contrast, in TET2 mutant cells, homozygous, hemizygous, and heterozygous as well as biallelic mutations were found,53 suggesting that various pathways can be exploited during malignant evolution to inactivate the mutated allele.
As indicated by the identification of homozygous mutations, acquired CN-LOH can convey a selection advantage through total knockout of a tumor suppressor gene by duplication of inactivating mutation (eg, TP53 mutations and UPD17p) or an activating mutation (eg, JAK2 V617F) (Figure 5; Table 1). In addition to mutations, deletions can lead to either a loss or retention of the methylated (silenced) allele, whereas CN-LOH can result in duplication of either the methylated or the unmethylated allele. As a consequence, LOH can lead to either effective knockout or enhanced expression. Clearly, such a mechanism could be operative for genes that can be regulated by methylation-based promoter silencing. The third mechanism by which CN-LOH can become permissive in the context of the selection process is duplication of a minor disease-prone allele present in the germline constellation. Informative loci would have to have a very low homozygous frequency to allow for analysis (Figure 5). A recent remarkable case of aUPD leading to homozygous selection of HLA surface markers that allowed an AML leukemia to escape immune surveillance65 illustrates yet another powerful example of clonal selection and the biologic consequences of CN-LOH as a disease resistance/disease progression mechanism.
Pathogenic actions of LOH, both CN-LOH and deletion. CN-LOH can lead to the duplication of a disease-linked minor germline variant (top line, left) or an acquired mutation (top line, right). It can also lead to increased gene expression by the loss of a negative epigenetic mark (second line, left) or decreased gene expression by the duplication of a repressive epigenetic mark (second line, right). Deletion of chromosomal material can lead to the unveiling of a minor germline variant (third line, left) of acquired mutation (third line, right), resulting in hemizygosity. Furthermore, deletion can affect gene expression: it can lead to increased expression through loss of an imprinted allele, repression by loss of the expressed allele, or haploinsufficiency.
Pathogenic actions of LOH, both CN-LOH and deletion. CN-LOH can lead to the duplication of a disease-linked minor germline variant (top line, left) or an acquired mutation (top line, right). It can also lead to increased gene expression by the loss of a negative epigenetic mark (second line, left) or decreased gene expression by the duplication of a repressive epigenetic mark (second line, right). Deletion of chromosomal material can lead to the unveiling of a minor germline variant (third line, left) of acquired mutation (third line, right), resulting in hemizygosity. Furthermore, deletion can affect gene expression: it can lead to increased expression through loss of an imprinted allele, repression by loss of the expressed allele, or haploinsufficiency.
Clinical significance of CN-LOH in myeloid malignancies
Traditional cytogenetic technologies, including metaphase cytogenetics, fluorescence in situ hybridization, and comparative genomic hybridization, do not detect CN-LOH, and it was not until the description of UPD9p in the setting of MPN55 that the clinical significance of this type of the lesion was begun to be appreciated. With the broader application of SNP-A came the realization as to how widely spread this balanced acquired lesion is in various malignancies, including AML, MDS, MPN, and the overlap syndromes but also ALL, CLL, and multiple myeloma5,31,46-48,55,66-68 (Table 2). Representative examples of SNP-A karyograms of acquired areas of UPD found particularly frequently in myeloid malignancies from our studies are illustrated in Figure 4.
UPD in hematologic disorders
| Disease . | References . | Array . | Frequency, % . | Notes . |
|---|---|---|---|---|
| AML | 47 | 250K | 81 | Secondary, includes MPN |
| 46 | 250K | 23 | Secondary | |
| 69 | 10K | 17 | Primary, UPD13q limited to intermediate-risk group, all UPD lost on remission | |
| 6 | 10K | 12 | Primary, lost on remission | |
| 41 | 10K | 22.7 | All normal karyotype | |
| 70 | 10K | 11; 48 | At diagnosis; at relapse | |
| 49 | 250K | 34 | Secondary | |
| ALL | 5 | 10K | 40; 80 | At diagnosis; at relapse |
| 3 | 500K | 32.5 | ||
| 2 | 100K; 500K | 21 | ||
| 71 | 50K; 250K | 24 | ||
| 72 | 250K | 21; 36 | At diagnosis; at relapse | |
| MM | 68 | 50K | 10 | |
| CLL | 73 | 50K | 7 | Untreated |
| 67 | 10K; 50K | 20 | ||
| MDS | 54 | 250K | 8 | All telomeric, > 21 Mb |
| 66 | 50K | 33 | Low-risk MDS | |
| 46 | 250K | 20 | ||
| 31 | 10K; 50K; 250K | 46 | Low-risk MDS | |
| 74 | 250K | 12 | Normal karyotype by MC | |
| 75 | 500K | 62 | CD34+, all small (< 5 Mb) | |
| 49 | 250K | 18 | ||
| 76 | 50K | 33 | ||
| MDS/MPN | 46 | 250K | 35 | Most prevalent in CMML |
| 47 | 250K | 36; 11 | MDS/MPNu; RARSt | |
| 49 | 250K | 44 | MDS/MPNu, CMML, sAML | |
| 54 | 6.0 | 19 | JMML, UPD11q | |
| 57 | 250K | 8 | JMML, UPD11q | |
| 61 | 100K | 80; 0 | JMML with NF1 mutation; without NF1 mutation | |
| 52 | 250K | 28 | RARSt |
| Disease . | References . | Array . | Frequency, % . | Notes . |
|---|---|---|---|---|
| AML | 47 | 250K | 81 | Secondary, includes MPN |
| 46 | 250K | 23 | Secondary | |
| 69 | 10K | 17 | Primary, UPD13q limited to intermediate-risk group, all UPD lost on remission | |
| 6 | 10K | 12 | Primary, lost on remission | |
| 41 | 10K | 22.7 | All normal karyotype | |
| 70 | 10K | 11; 48 | At diagnosis; at relapse | |
| 49 | 250K | 34 | Secondary | |
| ALL | 5 | 10K | 40; 80 | At diagnosis; at relapse |
| 3 | 500K | 32.5 | ||
| 2 | 100K; 500K | 21 | ||
| 71 | 50K; 250K | 24 | ||
| 72 | 250K | 21; 36 | At diagnosis; at relapse | |
| MM | 68 | 50K | 10 | |
| CLL | 73 | 50K | 7 | Untreated |
| 67 | 10K; 50K | 20 | ||
| MDS | 54 | 250K | 8 | All telomeric, > 21 Mb |
| 66 | 50K | 33 | Low-risk MDS | |
| 46 | 250K | 20 | ||
| 31 | 10K; 50K; 250K | 46 | Low-risk MDS | |
| 74 | 250K | 12 | Normal karyotype by MC | |
| 75 | 500K | 62 | CD34+, all small (< 5 Mb) | |
| 49 | 250K | 18 | ||
| 76 | 50K | 33 | ||
| MDS/MPN | 46 | 250K | 35 | Most prevalent in CMML |
| 47 | 250K | 36; 11 | MDS/MPNu; RARSt | |
| 49 | 250K | 44 | MDS/MPNu, CMML, sAML | |
| 54 | 6.0 | 19 | JMML, UPD11q | |
| 57 | 250K | 8 | JMML, UPD11q | |
| 61 | 100K | 80; 0 | JMML with NF1 mutation; without NF1 mutation | |
| 52 | 250K | 28 | RARSt |
MM indicates multiple myeloma; MDS/MPNu, MDS/MPN unclassifiable; RARSt, refractory anemia with ring sideroblasts in transformation; sAML, secondary AML; and JMML, juvenile myelomonocytic leukemia.
Studies in MDS and AML show that the addition of SNP-A karyotyping to standard metaphase cytogenetics increases the ability to identify a clonal marker from 50% to approximately 80%.46,48,66 This is particularly helpful in cases with normal cytogenetics, which is still associated with a wide range of clinical outcomes in individual patients, such as patients with intermediate-risk AML and a normal karyotype, some mixed MDS/MPN such as chronic myelomonocytic leukemia (CMML),49 and JAK2-negative MPN.58,72 The technical aspects of the SNP-A technology, including not requiring cell culture, provides a significant increase in the number of evaluable samples, particularly in hematologic malignancies associated with bone marrow fibrosis, through the use of peripheral blood cells in selected situations.72
Identification of recurrent CN-LOH raises the question of their clinical impact. Similar to other cytogenetic lesions, areas of aUPD can be mapped as, for example, in myeloid malignancies (Figure 4). Recurrent deletions and gains have an established prognostic and diagnostic value and have been incorporated in many prognostic schemes, including those for multiple myeloma and CLL or the International Prognostic Scoring System in MDS. However, the clinical impact of only a few recurrent areas of CN-LOH has been established. For example, homozygous JAK2 mutations indicate the presence of UPD9p and per inference this “doubled” dose of mutant JAK2 has been shown to be associated with more serious prognosis.56 Similarly, UPD13q associated with homozygous FLT-3 ITD or UPD17p and homozygous TP53 mutation, as well as UPD11q and C-CBL mutations, have been attributed with unfavorable outcomes probably because of the presence of the corresponding mutation.49 However, CN-LOH can by itself convey poor prognosis; we have shown that UPD7q in MDS is associated with equally poor prognosis as is corresponding del7q.48 Assignment of prognostic significance will require systematic application of SNP-A as a routine diagnostic tool complementing metaphase cytogenetics. In particular, in patients with normal cytogenetics or lesions associated with favorable prognosis, detection of UPD may effectively upstage the prognosis and allow for better prognostic resolution explaining heterogeneity of outcomes in patients with otherwise comparable clinical features.
Conclusions
CN-LOH acquired during malignant progression must be distinguished from germline areas of CN-LOH, which may be a result of an early embryonic mitotic homologous recombination event or inherited autozygosity. The latter findings can represent important cancer predisposition events. Acquired CN-LOH, resulting from mitotic homologous recombination, has certain genetic characteristics. Factors that might promote clonal selection with CN-LOH include duplication of acquired epigenetic or mutational events, disease predisposing polymorphisms, or improper imprinting patterns.
First identified in JAK2, c-CBL and TET2 are the most recent examples of mutations found in a homozygous constellation in myeloid malignancies resulting from acquired CN-LOH.
CN-LOH is common in myeloid disorders, even in those patients who are often cytogenetically normal. CN-LOH identified by SNP-A is especially common in mixed MDS/MPN, acute CML, CMML, JAK2-negative MPN, MDS, and AML, all of which have increased incidence with age. For these disorders, CN-LOH provides a novel and often unique molecular marker. As such, this marker has the potential to be diagnostic and prognostic, supported by emerging data from MDS, AML, and CMML, and specific CN-LOH regions, such as 7q, may complement existing methodologies and approaches.
Whether CN-LOH explains the worst prognosis of persons with normal metaphase cytogenetics but abnormal SNP-A karyotype, including aging persons, either as a molecular marker of genomic damage or directly related based on clonal selection of acquired molecular lesions, remains to be formally investigated.
Investigations into the underlying mechanisms generating CN-LOH have great promise for elucidating general cancer mechanisms. They can lead to the discovery of new tumor suppressor genes.
We anticipate that further detailed characterization of CN-LOH lesions will probably facilitate our discovery of a more complete set of pathogenic molecular lesions, discovery of disease and prognosis markers, and better understanding of the initiation and progression of hematologic malignancies.
Acknowledgments
This work was supported by the National Institutes of Health (R01 HL082983 and K24 HL077522, to J.P.M.; DOD MPO48018, to M.A.M.) and the Robert Duggan Cancer Research Foundation.
National Institutes of Health
Authorship
Contribution: C.O., M.A.M., and J.P.M. wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaroslaw P. Maciejewski, Taussig Cancer Institute/R40, 9500 Euclid Ave, Cleveland, OH 44195; e-mail: maciejj@ccf.org.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal