Abstract
Mutations of nicotinamide adenine dinucleotide phosphate-dependent isocitrate dehydrogenase gene (IDH1) have been identified in patients with gliomas. Recent genome-wide screening also revealed IDH1 mutation as a recurrent event in acute myeloid leukemia (AML), but its clinical implications in AML are largely unknown. We analyzed 493 adult Chinese AML patients in Taiwan and found 27 patients (5.5%) harboring this mutation. IDH1 mutation was strongly associated with normal karyotype (8.4%, P = .002), isolated monosomy 8 (P = .043), NPM1 mutation (P < .001), and French-American-British M1 subtype (P < .001), but inversely associated with French-American-British M4 subtype (P = .030) and expression of HLA-DR, CD13, and CD14 (P = .002, .003, and .038, respectively). There was no impact of this mutation on patient survival. Sequential analysis of IDH1 mutation was performed in 130 patients during follow-ups. None of the 112 patients without IDH1 mutation at diagnosis acquired this mutation at relapse. In all 18 IDH1-mutated patients studied, the mutation disappeared in complete remission; the same mutation reappeared in all 11 samples obtained at relapse. We conclude that IDH1 is associated with distinct clinical and biologic characteristics and seems to be very stable during disease evolution.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease in terms of clinical presentation, biologic characteristics, and response to treatment. The classification for this disease has been continuously refined as more and more molecular markers are identified. Recently, genetic mutations involving NPM1 and CEBPA have been incorporated as references in the World Health Organization classification of AML.1
As an effort to identify novel molecular alterations in this disease, whole genome sequencing on AML patients was performed, comparing the genomes of leukemia cells and the matching normal counterparts.2,3 Although few recurrent genetic alterations were found in these studies, mutation at arginine 132 (R132) in nicotinamide adenine dinucleotide phosphate-dependent isocitrate dehydrogenase 1 (IDH1) was identified as a new recurrent genetic alteration, with an incidence of 8.3% and 16% of total AML patients and those with a normal karyotype, respectively, in a Western cohort.3 This mutation was first identified in glioma via a comprehensive sequencing on the brain tumors.4 Subsequent studies have shown a very high incidence of mutation in this gene (up to 70%) in lower-grade gliomas.5-7
To characterize the clinical and biologic features of AML with IDH1 mutations, which have not been fully clarified yet, we analyzed 493 adult patients with de novo AML. We also analyzed sequential marrow samples from 130 patients to evaluate the stability of IDH1 mutations during disease progression. We found that this mutation occurred with a much lower incidence in our cohort than in the Western cohort first reported.3 Our preliminary results implied that IDH1 mutation may be a relatively stable disease marker, but more sensitive methods are necessary to confirm this observation. Moreover, our comprehensive analysis revealed several unreported and distinct clinical and biologic features in IDH1-mutated AML.
Methods
Patients
A total of 674 adults (≥ 18 years of age) with de novo AML were newly diagnosed in the National Taiwan University Hospital from 1995 to 2007. We focused on a total of 493 patients (73%) who had available cryopreserved bone marrow samples and complete clinical and laboratory data for analyses. These 493 patients seem to be representative of the whole group of patients because their clinical features and treatment outcomes were similar to those of the total 674 patients (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The bone marrow cells were serially collected at diagnosis, after chemotherapy, and at relapse. The mononuclear cells were isolated via Ficoll-Hypaque gradient centrifugation and then cryopreserved until use. This study was approved by the Institutional Review Board of the National Taiwan University Hospital.
Mutation analysis
Determination of mutations in various genes, including FLT3/ITD, FLT3/D835, MLL/PTD, CEBPA, NPM1, PTPN11, NRAS, KRAS, JAK2, KIT, AML1(RUNX1), and WT1 mutations, was performed as described previously.8-13 IDH1 R132 mutation was analyzed by direct sequencing of the polymerase chain reaction (PCR) product as described previously.4
Gene cloning
When IDH1 mutations at diagnosis were no longer seen in relapsed samples by direct sequencing, we cloned the PCR products spanning the mutation hotspot by TA cloning (Yeastern Biotech), followed by sequencing of individual clones to search for any mutation.
Immunophenotyping
A panel of monoclonal antibodies, including myeloid-associated antigens (CD13, CD33, CD11b, CD15, CD14, and CD41a), lymphoid-associated antigens (CD2, CD5, CD7, CD19, CD10, and CD20), and the lineage-nonspecific antigens (HLA-DR, CD34, and CD56), was used to determine the immunophenotypes of leukemia cells as described previously.12
Cytogenetic analysis
Bone marrow cells were harvested directly or after 1 to 3 days of unstimulated culture, and the metaphase chromosomes were banded by the G-banding method as described earlier.14
Statistics
The χ2 test was used to calculate the significance of association between IDH1 mutations and other discrete parameters, such as immunophenotypes, cytogenetics, and mutations of a specific gene. Fisher exact test was used to compare the IDH1 mutation incidences between different cohorts. Continuous variables were tested with Mann-Whitney method. Kaplan-Meier survival curve was calculated with SPSS software (Version 14).
Results
IDH1 R132 mutations
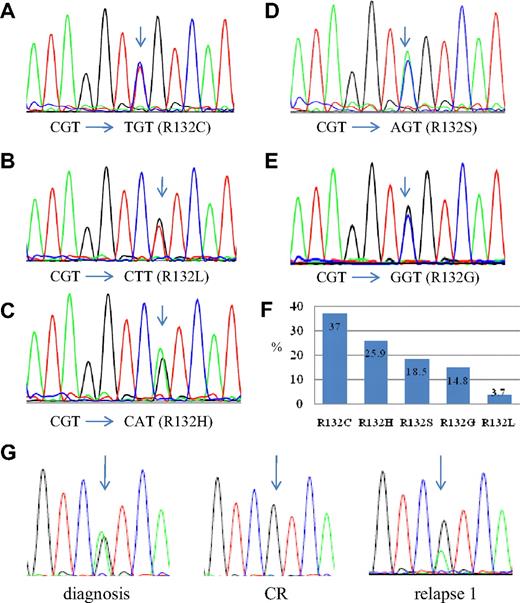
These 493 patients include 281 males and 212 females, with a median age of 53 years (range, 18-90 years). The IDH1 R132 mutation occurs in 27 (5.5%) of these 493 patients. The basic clinical and biologic data of these patients are listed in Table 1. There were 5 types of mutations, including R132C (CGT to TGT, 10 of 27, 37.0%), R132H (CGT to CAT, 7 of 27, 25.9%), R132S (CGT to AGT, 5 of 27, 18.5%), R132G (CGT to GGT, 4 of 27, 14.8%), and R132L (CGT to CTT, 1 of 27, 3.7%; Figure 1). All patients with the mutation were heterozygous and retained a wild-type allele.
The mutation patterns and interaction with other gene alterations in 27 patients with IDH1 mutations
| UPN . | Age/sex . | FAB . | Karyotype . | IDH1 mutation . | Other genetic mutations* . | |
|---|---|---|---|---|---|---|
| n.t. change . | a.a. change . | |||||
| 1 | 75/F | 2 | N | CAT | R132H | NPM1 |
| 2 | 75/F | 1 | +8 | GGT | R132G | NPM1, FLT3/ITD, FLT3/TKD |
| 3 | 72/M | 1 | +8 | TGT | R132C | — |
| 4 | 72/M | 1 | N | TGT | R132C | RUNX1 |
| 5 | 71/F | 2 | no mitosis | AGT | R132S | NPM1, FLT3/ITD |
| 6 | 68/M | 2 | N | CAT | R132H | NPM1, FLT3/ITD, FTL3/TKD |
| 7 | 65/F | 1 | N | TGT | R132C | NPM1, FLT3/ITD |
| 8 | 65/M | 1 | Add(1)(p13), +8 | TGT | R132C | — |
| 9 | 62/F | 2 | N | CAT | R132H | NPM1 |
| 10 | 60/M | 2 | N | AGT | R132S | MLL/PTD |
| 11 | 60/F | 1 | t(9;22) | TGT | R132C | — |
| 12 | 57/M | 1 | N | TGT | R132C | — |
| 13 | 55/M | 1 | +21 | AGT | R132S | NPM1, FLT3/ITD |
| 14 | 50/M | 2 | N | GGT | R132G | NPM1, FLT3/ITD, NRAS |
| 15 | 48/M | 5 | N | CAT | R132H | NPM1 |
| 16 | 47/M | 1 | N | TGT | R132C | NPM1, PTPN11 |
| 17 | 47/M | 2 | N | CAT | R132H | CEBPA, FLT3/TKD, MLL/PTD |
| 18 | 39/M | 1 | N | TGT | R132C | PTPN11 |
| 19 | 38/F | 5 | N | CAT | R132H | NPM1, NRAS |
| 20 | 38/F | 2 | N | GGT | R132G | NPM1, FLT3/ITD |
| 21 | 35/M | 1 | N | CTT | R132L | — |
| 22 | 34/F | 1 | N | AGT | R132S | NPM1, FLT3/ITD |
| 23 | 28/F | 1 | +8 | AGT | R132S | NPM1, FLT3/ITD |
| 24 | 25/F | 2 | N | CAT | R132H | FLT3/ITD, MLL/PTD, RUNX1 |
| 25 | 26/F | 4 | N | GGT | R132G | NPM1, NRAS |
| 26 | 70/M | 4 | N | TGT | R132C | — |
| 27 | 35/F | 1 | N | TGT | R132C | NRAS |
| UPN . | Age/sex . | FAB . | Karyotype . | IDH1 mutation . | Other genetic mutations* . | |
|---|---|---|---|---|---|---|
| n.t. change . | a.a. change . | |||||
| 1 | 75/F | 2 | N | CAT | R132H | NPM1 |
| 2 | 75/F | 1 | +8 | GGT | R132G | NPM1, FLT3/ITD, FLT3/TKD |
| 3 | 72/M | 1 | +8 | TGT | R132C | — |
| 4 | 72/M | 1 | N | TGT | R132C | RUNX1 |
| 5 | 71/F | 2 | no mitosis | AGT | R132S | NPM1, FLT3/ITD |
| 6 | 68/M | 2 | N | CAT | R132H | NPM1, FLT3/ITD, FTL3/TKD |
| 7 | 65/F | 1 | N | TGT | R132C | NPM1, FLT3/ITD |
| 8 | 65/M | 1 | Add(1)(p13), +8 | TGT | R132C | — |
| 9 | 62/F | 2 | N | CAT | R132H | NPM1 |
| 10 | 60/M | 2 | N | AGT | R132S | MLL/PTD |
| 11 | 60/F | 1 | t(9;22) | TGT | R132C | — |
| 12 | 57/M | 1 | N | TGT | R132C | — |
| 13 | 55/M | 1 | +21 | AGT | R132S | NPM1, FLT3/ITD |
| 14 | 50/M | 2 | N | GGT | R132G | NPM1, FLT3/ITD, NRAS |
| 15 | 48/M | 5 | N | CAT | R132H | NPM1 |
| 16 | 47/M | 1 | N | TGT | R132C | NPM1, PTPN11 |
| 17 | 47/M | 2 | N | CAT | R132H | CEBPA, FLT3/TKD, MLL/PTD |
| 18 | 39/M | 1 | N | TGT | R132C | PTPN11 |
| 19 | 38/F | 5 | N | CAT | R132H | NPM1, NRAS |
| 20 | 38/F | 2 | N | GGT | R132G | NPM1, FLT3/ITD |
| 21 | 35/M | 1 | N | CTT | R132L | — |
| 22 | 34/F | 1 | N | AGT | R132S | NPM1, FLT3/ITD |
| 23 | 28/F | 1 | +8 | AGT | R132S | NPM1, FLT3/ITD |
| 24 | 25/F | 2 | N | CAT | R132H | FLT3/ITD, MLL/PTD, RUNX1 |
| 25 | 26/F | 4 | N | GGT | R132G | NPM1, NRAS |
| 26 | 70/M | 4 | N | TGT | R132C | — |
| 27 | 35/F | 1 | N | TGT | R132C | NRAS |
UPN indicates unique patient number; N, normal karyotype; NM, no mitosis; n.t., nucleotide; and a.a., amino acid.
The gene alterations studied included FLT3/ITD, FLT3/TKD and MLL/PTD and mutations of NRAS, KRAS, KIT, PTPN11, WT1, NPM1, JAK2, RUNX1, and CEBPA.
Sequences of IDH1 mutations. (A-E) The 5 types of IDH1 mutation. The arrow indicates the mutated nucleotide. (F) The percentages of each type of mutation. R132C comprises 37% of all types of IDH1 mutation. Only 1 of 27 IDH1-mutated patients (3.7%) has R132L. (G) The sequential study of IDH1 mutation in patient 9. The arrow points to R132H mutation in the patient.
Sequences of IDH1 mutations. (A-E) The 5 types of IDH1 mutation. The arrow indicates the mutated nucleotide. (F) The percentages of each type of mutation. R132C comprises 37% of all types of IDH1 mutation. Only 1 of 27 IDH1-mutated patients (3.7%) has R132L. (G) The sequential study of IDH1 mutation in patient 9. The arrow points to R132H mutation in the patient.
Sequential studies of IDH1 mutations
The IDH1 R132 mutation was serially studied in 130 patients, including 18 patients with and 112 patients without IDH1 mutation at diagnosis, with DNA sequencing. In all 112 patients without IDH1 mutation, none acquired this mutation at disease relapse. In addition, the IDH1 mutations disappeared at complete remission in all 18 patients who harbored the mutations at diagnosis and had serial samples available for analysis (Table 2); the same mutations as those at diagnosis reappeared in 9 of the 11 samples obtained at relapse but were lost in the remaining 2 (the second relapse of patient 13 and the first relapse of patient 17; Table 2). These 2 marrow samples contained 11.2% and 16.8% blasts, respectively, indicating an early relapse. These numbers of blasts may be below the sensitivity of detection by direct DNA sequencing. We thus performed TA cloning of the PCR products spanning the IDH1 mutation hotspot and analyzed individual clones of each of these 2 samples by sequencing. We found that the mutation did exist in these 2 relapsed samples (in 1 of 17 clones and 1 of 12 clones, respectively). Thus, IDH1 mutation seems to be quite stable during disease evolution.
Results of sequential studies of IDH1 mutations and chromosomal changes in the patients with the mutation at diagnosis
| Case no./interval, mo* . | Status . | Karyotype . | IDH1 mutation† . |
|---|---|---|---|
| 1 | Diagnosis | N | + |
| 0.5 | CR1 | N | No mutation |
| 4 | Diagnosis | N | + |
| 1.4 | CR1 | N | No mutation |
| 15.8 | Relapse 1 | del(13q) | + |
| 0.9 | CR2 | N | No mutation |
| 7 | Diagnosis | N | + |
| 1.3 | CR1 | N | No mutation |
| 11.5 | Relapse 1 | N | + |
| 4.1 | CR2 | ND | No mutation |
| 11.8 | Relapse 2 | ND | + |
| 8 | Diagnosis | Add (1)(p13), +8 | + |
| 1.1 | CR1 | ND | No mutation |
| 9 | Diagnosis | N | + |
| 3.7 | CR1 | ND | No mutation |
| 6.2 | Relapse 1 | ND | + |
| 10 | Diagnosis | N | + |
| 1.1 | CR1 | ND | No mutation |
| 8.1 | Relapse 1 | ND | + |
| 4.3 | CR2 | ND | No mutation |
| 2.2 | Relapse 2 | ND | + |
| 11 | Diagnosis | t (9;22)(q34;q11) | + |
| 6.3 | CR1 | ND | No mutation |
| 10.1 | Relapse 1 | ND | + |
| 12 | Diagnosis | N | + |
| 2.3 | CR1 | ND | No mutation |
| 3.0 | Relapse 1 | ND | + |
| 1.0 | CR2 | ND | No mutation |
| 13 | Diagnosis | +21 | + |
| 2.4 | CR1 | N | No mutation |
| 18.3 | Relapse 1 | N | + |
| 3.9 | CR2 | N | No mutation |
| 36.9 | Relapse 2 | N | +‡ |
| 14 | Diagnosis | N | + |
| 2.5 | CR1 | N | No mutation |
| 16 | Diagnosis | N | + |
| 0.9 | CR1 | N | No mutation |
| 17 | Diagnosis | N | + |
| 1.8 | CR1 | ND | No mutation |
| 10.1 | Relapse 1 | ND | +‡ |
| 19 | Diagnosis | N | + |
| 1.5 | CR1 | ND | No mutation |
| 20 | Diagnosis | N | + |
| 1.6 | CR1 | ND | No mutation |
| 21 | Diagnosis | N | + |
| 1.1 | CR1 | N | No mutation |
| 22 | Diagnosis | N | + |
| 7.0 | CR1 | ND | No mutation |
| 25 | Diagnosis | N | + |
| 6.1 | CR1 | ND | No mutation |
| 27 | Diagnosis | N | + |
| 0.8 | CR1 | N | No mutation |
| Case no./interval, mo* . | Status . | Karyotype . | IDH1 mutation† . |
|---|---|---|---|
| 1 | Diagnosis | N | + |
| 0.5 | CR1 | N | No mutation |
| 4 | Diagnosis | N | + |
| 1.4 | CR1 | N | No mutation |
| 15.8 | Relapse 1 | del(13q) | + |
| 0.9 | CR2 | N | No mutation |
| 7 | Diagnosis | N | + |
| 1.3 | CR1 | N | No mutation |
| 11.5 | Relapse 1 | N | + |
| 4.1 | CR2 | ND | No mutation |
| 11.8 | Relapse 2 | ND | + |
| 8 | Diagnosis | Add (1)(p13), +8 | + |
| 1.1 | CR1 | ND | No mutation |
| 9 | Diagnosis | N | + |
| 3.7 | CR1 | ND | No mutation |
| 6.2 | Relapse 1 | ND | + |
| 10 | Diagnosis | N | + |
| 1.1 | CR1 | ND | No mutation |
| 8.1 | Relapse 1 | ND | + |
| 4.3 | CR2 | ND | No mutation |
| 2.2 | Relapse 2 | ND | + |
| 11 | Diagnosis | t (9;22)(q34;q11) | + |
| 6.3 | CR1 | ND | No mutation |
| 10.1 | Relapse 1 | ND | + |
| 12 | Diagnosis | N | + |
| 2.3 | CR1 | ND | No mutation |
| 3.0 | Relapse 1 | ND | + |
| 1.0 | CR2 | ND | No mutation |
| 13 | Diagnosis | +21 | + |
| 2.4 | CR1 | N | No mutation |
| 18.3 | Relapse 1 | N | + |
| 3.9 | CR2 | N | No mutation |
| 36.9 | Relapse 2 | N | +‡ |
| 14 | Diagnosis | N | + |
| 2.5 | CR1 | N | No mutation |
| 16 | Diagnosis | N | + |
| 0.9 | CR1 | N | No mutation |
| 17 | Diagnosis | N | + |
| 1.8 | CR1 | ND | No mutation |
| 10.1 | Relapse 1 | ND | +‡ |
| 19 | Diagnosis | N | + |
| 1.5 | CR1 | ND | No mutation |
| 20 | Diagnosis | N | + |
| 1.6 | CR1 | ND | No mutation |
| 21 | Diagnosis | N | + |
| 1.1 | CR1 | N | No mutation |
| 22 | Diagnosis | N | + |
| 7.0 | CR1 | ND | No mutation |
| 25 | Diagnosis | N | + |
| 6.1 | CR1 | ND | No mutation |
| 27 | Diagnosis | N | + |
| 0.8 | CR1 | N | No mutation |
The data of sequential studies on 112 patients without IDH1 mutation at diagnosis were not shown in this table; none acquired IDH1 mutation at relapse in these patients.
CR indicates complete remission; N, normal; and ND, no data.
Between 2 consecutive studies.
All patients with IDH1 mutations are heterozygous.
The IDH1 mutations in these 2 relapsed samples could not be detected by sequencing of PCR products but were confirmed by sequencing of the individual clones.
Correlation of IDH1 mutations with clinical features and biologic characteristics
A total of 476 patients, including 26 with IDH1 mutations, had cytogenetic data. The IDH1 mutation was mainly seen in patients with a normal karyotype (Table 3). Among the patients with a normal karyotype, 8.8% (20 of 227) showed the IDH1 mutation, compared with 2.4% in those with chromosomal abnormalities (6 of 249). Indeed, 23 of the 27 patients with IDH1 mutation had either normal karyotype (n = 20) or isolated trisomy 8 (+8, n = 3); one did not have metaphase chromosome for analysis, and the remaining 3 patients harbored combined add(1)(p13) and +8, t(9;22), and +21, respectively. Therefore, the IDH1 mutation was significantly associated with a normal karyotype (P = .002) and isolated trisomy 8 (P = .043). No IDH1 mutation was seen in patients with t(8;21), t(15;17), inv(16), and other recurrent chromosomal abnormalities, such as t(6;9), t(7;11), or deletions involving chromosome 5 or 7 (Table 3). The IDH1 mutation occurred equally in males and females and distributed evenly among different age groups (Table 3). More than half the patients with the IDH1 mutation (14 of 27, 51.9%) had AML M1 subtype (P < .001). In contrast, only 2 of these patients have M4 subtype, compared with 119 of the 457 patients with wild-type IDH1 (2 of 27 vs 119 of 457, P = .030, excluding 9 patients with undetermined French-American-British [FAB] subtype; Table 3). There was no significant association between IDH1 mutation and the initial hemoglobin levels, white blood cell counts, blast percentages, and platelet counts in the peripheral blood and lactate dehydrogenase levels in serum (data not shown).
Clinical data, FAB types, and cytogenetic changes in AML patients with IDH1 mutations
| Variant . | Total no. (n = 493) . | Mutation (n = 27, 5.50%) . | Wild-type (n = 466, 94.50%) . | P . | ||
|---|---|---|---|---|---|---|
| No. . | Percentage . | No. . | Percentage . | |||
| Age, y | .517 | |||||
| 18-29 | 60 | 3 | 5 | 57 | 95 | — |
| 30-39 | 77 | 6 | 7.79 | 71 | 92.21 | — |
| 40-49 | 89 | 3 | 3.41 | 86 | 96.59 | — |
| 50-59 | 76 | 3 | 3.95 | 73 | 96.05 | — |
| 60-69 | 80 | 6 | 7.5 | 74 | 92.5 | — |
| 70-79 | 78 | 6 | 7.69 | 72 | 92.31 | — |
| 80 or older | 33 | 0 | 0 | 33 | 100 | — |
| Sex | .578 | |||||
| Male | 281 | 14 | 4.98 | 267 | 94.02 | — |
| Female | 212 | 13 | 6.13 | 199 | 93.87 | — |
| FAB | ||||||
| M0 | 10 | 0 | 0.0 | 10 | 100 | .437 |
| M1 | 112 | 14 | 12.5 | 98 | 87.5 | < .001 |
| M2 | 169 | 9 | 5.32 | 160 | 94.68 | .858 |
| M3 | 37 | 0 | 0 | 37 | 100 | .124 |
| M4 | 121 | 2 | 1.65 | 119 | 98.35 | .030 |
| M5 | 23 | 2 | 8.70 | 21 | 91.30 | .505 |
| M6 | 12 | 0 | 0 | 12 | 100 | .394 |
| M7 | 0 | 0 | 0 | 0 | 100 | — |
| Undetermined | 9 | 0 | — | 9 | — | — |
| Cytogenetics | ||||||
| Abnormal | ||||||
| t(15;17) | 37* | 0 | 0 | 37 | 100 | .134 |
| t(8;21) | 37 | 0 | 0.0 | 37 | 100 | .128 |
| Inv(16) | 17 | 0 | 0.0 | 17 | 100 | .313 |
| t(7;11) | 10 | 0 | 0.0 | 10 | 100 | .442 |
| del(7) | 30 | 0 | 0.0 | 30 | 100 | .174 |
| del(5) | 26 | 0 | 0.0 | 26 | 100 | .207 |
| Isolated trisomy 8 | 19 | 3 | 17.65 | 16 | 82.35 | .043 |
| Others | 79 | 3 | 2.82 | 76 | 97.18 | — |
| Subtotal | 249† | 6 | 6.1 | 243 | 93.9 | — |
| Normal | 227 | 20 | 8.4 | 207 | 91.6 | .002 |
| Variant . | Total no. (n = 493) . | Mutation (n = 27, 5.50%) . | Wild-type (n = 466, 94.50%) . | P . | ||
|---|---|---|---|---|---|---|
| No. . | Percentage . | No. . | Percentage . | |||
| Age, y | .517 | |||||
| 18-29 | 60 | 3 | 5 | 57 | 95 | — |
| 30-39 | 77 | 6 | 7.79 | 71 | 92.21 | — |
| 40-49 | 89 | 3 | 3.41 | 86 | 96.59 | — |
| 50-59 | 76 | 3 | 3.95 | 73 | 96.05 | — |
| 60-69 | 80 | 6 | 7.5 | 74 | 92.5 | — |
| 70-79 | 78 | 6 | 7.69 | 72 | 92.31 | — |
| 80 or older | 33 | 0 | 0 | 33 | 100 | — |
| Sex | .578 | |||||
| Male | 281 | 14 | 4.98 | 267 | 94.02 | — |
| Female | 212 | 13 | 6.13 | 199 | 93.87 | — |
| FAB | ||||||
| M0 | 10 | 0 | 0.0 | 10 | 100 | .437 |
| M1 | 112 | 14 | 12.5 | 98 | 87.5 | < .001 |
| M2 | 169 | 9 | 5.32 | 160 | 94.68 | .858 |
| M3 | 37 | 0 | 0 | 37 | 100 | .124 |
| M4 | 121 | 2 | 1.65 | 119 | 98.35 | .030 |
| M5 | 23 | 2 | 8.70 | 21 | 91.30 | .505 |
| M6 | 12 | 0 | 0 | 12 | 100 | .394 |
| M7 | 0 | 0 | 0 | 0 | 100 | — |
| Undetermined | 9 | 0 | — | 9 | — | — |
| Cytogenetics | ||||||
| Abnormal | ||||||
| t(15;17) | 37* | 0 | 0 | 37 | 100 | .134 |
| t(8;21) | 37 | 0 | 0.0 | 37 | 100 | .128 |
| Inv(16) | 17 | 0 | 0.0 | 17 | 100 | .313 |
| t(7;11) | 10 | 0 | 0.0 | 10 | 100 | .442 |
| del(7) | 30 | 0 | 0.0 | 30 | 100 | .174 |
| del(5) | 26 | 0 | 0.0 | 26 | 100 | .207 |
| Isolated trisomy 8 | 19 | 3 | 17.65 | 16 | 82.35 | .043 |
| Others | 79 | 3 | 2.82 | 76 | 97.18 | — |
| Subtotal | 249† | 6 | 6.1 | 243 | 93.9 | — |
| Normal | 227 | 20 | 8.4 | 207 | 91.6 | .002 |
— indicates not applicable.
Including one patient with t(8;15;17).
Excluding patients with coexistence of the listed abnormalities, without mitosis, or without cytogenetic analysis.
We also analyzed mutations in CEBPA, NPM1, AML1(RUNX1), WT1, PTPN11, NRAS, and KRAS, and FLT3/ITD, FLT3/D835, and MLL/PTD. The comparison of these genetic alterations between the patients with and without the IDH1 mutation is summarized in Table 4. The IDH1 mutation was strongly associated with mutation of NPM1 (P < .001) and had a trend to be associated with FLT3/ITD (P = .083) but not other mutations (Table 4).
Comparison of other genetic alterations between AML patients with and without the IDH1 mutation
| Variant . | Percentage of patients with the gene mutation . | P . | ||
|---|---|---|---|---|
| Total patients (n = 493) . | IDH1-mutated patients (n = 27) . | IDH1 wild-type patients (n = 466) . | ||
| NPM1 | 21.4 (105/493) | 55.6 (15/27) | 19.4 (90/466) | < .001 |
| CEBPA | 13.2 (65/493) | 3.7 (1/27)) | 13.8 (64/466) | .134 |
| FLT3/ITD | 23.4 (115/493) | 37.0 (10/27) | 22.6 (105/466) | .083 |
| RUNX1 | 12.2 (60/493) | 7.4 (2/27) | 12.4 (58/466) | .436 |
| WT1 | 6.3 (31/493) | 0 (0/27) | 6.7 (31/466) | .166 |
| MLL/PTD | 5.7 (28/493) | 11.1 (3/27) | 5.4 (25/466) | .210 |
| FLT3/D835 | 7.5 (37/493) | 11.1 (3/27) | 7.3 (34/466) | .465 |
| KIT | 2.4 (12/493) | 0 (0/27) | 2.6 (12/466) | .398 |
| JAK2 | 0.6 (3/493) | 0 (0/27) | 0.6 (3/466) | .676 |
| PTPN11 | 4.1 (20/493) | 7.4 (2/27) | 3.9 (18/466) | .364 |
| NRAS | 11.5 (57/493) | 14.8 (4/27) | 11.4 (53/466) | .586 |
| KRAS | 3.3 (16/493) | 0 (0/27) | 3.4 (16/466) | .328 |
| Variant . | Percentage of patients with the gene mutation . | P . | ||
|---|---|---|---|---|
| Total patients (n = 493) . | IDH1-mutated patients (n = 27) . | IDH1 wild-type patients (n = 466) . | ||
| NPM1 | 21.4 (105/493) | 55.6 (15/27) | 19.4 (90/466) | < .001 |
| CEBPA | 13.2 (65/493) | 3.7 (1/27)) | 13.8 (64/466) | .134 |
| FLT3/ITD | 23.4 (115/493) | 37.0 (10/27) | 22.6 (105/466) | .083 |
| RUNX1 | 12.2 (60/493) | 7.4 (2/27) | 12.4 (58/466) | .436 |
| WT1 | 6.3 (31/493) | 0 (0/27) | 6.7 (31/466) | .166 |
| MLL/PTD | 5.7 (28/493) | 11.1 (3/27) | 5.4 (25/466) | .210 |
| FLT3/D835 | 7.5 (37/493) | 11.1 (3/27) | 7.3 (34/466) | .465 |
| KIT | 2.4 (12/493) | 0 (0/27) | 2.6 (12/466) | .398 |
| JAK2 | 0.6 (3/493) | 0 (0/27) | 0.6 (3/466) | .676 |
| PTPN11 | 4.1 (20/493) | 7.4 (2/27) | 3.9 (18/466) | .364 |
| NRAS | 11.5 (57/493) | 14.8 (4/27) | 11.4 (53/466) | .586 |
| KRAS | 3.3 (16/493) | 0 (0/27) | 3.4 (16/466) | .328 |
Immunophenotyping study showed that the IDH1 mutation was inversely associated with the expression of HLA-DR, CD13, and CD14 on leukemia cells (P = .002, .003, and .038, respectively; Table 5). The expression of other markers, such as CD34, CD11b, CD33, CD15, CD19, CD7, CD56, and CD2, was not significantly different between the 2 groups of patients with and without the IDH1 mutation. Combining the results of both morphology and immunophenotyping, which can always be available immediately at the time of diagnosis, approximately 23.5% (8 of 34) of the patients with M1 subtype and absence of HLA-DR expression are expected to harbor IDH1 mutation, compared with 4.3% (19 of 445) in other patients (P < .001; excluding 14 patients who had no FAB subtype or HLA-DR expression data for analysis). We did not find significant impact of IDH1 mutation on the disease-free or overall survival by either univariate or multivariate analyses (hazard ratio = 1.043; 95% confidence interval, 0.582-1.870, P = .887; and hazard ratio = 0.820, 95% confidence interval, 0.378-1.777, P = .614, respectively; supplemental Table 2), even subgrouping the patients based on karyotype or mutation status of NPM1 or FLT3, probably because of the small number of patients harboring this mutation in this study.
Comparison of immunophenotyping between AML patients with and without the IDH1 mutation
| Variant . | Percentage of patients with the antigen expression . | P . | ||
|---|---|---|---|---|
| Total patients . | IDH1-mutated patients . | IDH1 wild-type patients . | ||
| HLA-DR | 67.4 (328/487) | 40.7 (11/27) | 68.9 (317/460) | .002 |
| CD34 | 64.3 (302/470) | 55.6 (15/27) | 65.2 (287/440) | .308 |
| CD13 | 94.3 (445/472) | 81.5 (22/27) | 95.1 (423/445) | .003 |
| CD33 | 91.3 (430/471) | 92.6 (25/27) | 91.2 (405/444) | .805 |
| CD11b | 31.1 (46/148) | 28.6 (2/7) | 31.2 (44/141) | .882 |
| CD14 | 13.1 (60/458) | 0 (0/27) | 13.9 (60/431) | .038 |
| CD15 | 43.8 (204/466) | 25.9 (7/27) | 44.9 (197/439) | .054 |
| CD19 | 6.7 (31/465) | 7.4 (2/27) | 6.6 (29/438) | .874 |
| CD7 | 19.9 (92/463) | 8.3 (2/24) | 20.5 (90/439) | .146 |
| CD2 | 4.2 (19/454) | 3.8 (1/26) | 4.2 (18/428) | .929 |
| CD56 | 22.8 (96/421) | 8.7 (2/23) | 23.6 (94/398) | .097 |
| Variant . | Percentage of patients with the antigen expression . | P . | ||
|---|---|---|---|---|
| Total patients . | IDH1-mutated patients . | IDH1 wild-type patients . | ||
| HLA-DR | 67.4 (328/487) | 40.7 (11/27) | 68.9 (317/460) | .002 |
| CD34 | 64.3 (302/470) | 55.6 (15/27) | 65.2 (287/440) | .308 |
| CD13 | 94.3 (445/472) | 81.5 (22/27) | 95.1 (423/445) | .003 |
| CD33 | 91.3 (430/471) | 92.6 (25/27) | 91.2 (405/444) | .805 |
| CD11b | 31.1 (46/148) | 28.6 (2/7) | 31.2 (44/141) | .882 |
| CD14 | 13.1 (60/458) | 0 (0/27) | 13.9 (60/431) | .038 |
| CD15 | 43.8 (204/466) | 25.9 (7/27) | 44.9 (197/439) | .054 |
| CD19 | 6.7 (31/465) | 7.4 (2/27) | 6.6 (29/438) | .874 |
| CD7 | 19.9 (92/463) | 8.3 (2/24) | 20.5 (90/439) | .146 |
| CD2 | 4.2 (19/454) | 3.8 (1/26) | 4.2 (18/428) | .929 |
| CD56 | 22.8 (96/421) | 8.7 (2/23) | 23.6 (94/398) | .097 |
Discussion
Although the IDH1 mutation at R132 has been extensively studied in glioma,4-7 little information about this mutation in AML is available.3 In this study, we performed comprehensive analyses of the clinical and biologic characteristics of AML harboring this mutation.
Compared with the first report of the IDH1 mutation in AML patients by Mardis et al, who focused on a Western cohort, the incidence of this gene mutation seems lower (approximately half) in our Chinese population3 ; however, the difference did not reach statistical significance (8.0% vs 5.5%; P = .216 by Fisher exact test). Even the analysis is restricted to the subgroup of patients with a normal karyotype; the difference is not significant either (16.3% vs 8.4%; P = .091 by Fisher exact test). Indeed, in the study of the IDH1 mutation in gliomas, Yan et al (in the United States) screened the IDH1 mutation in a variety of other malignant diseases but did not find any mutation in 45 samples from AML patients.5 The difference shown in our study may be just related to many confounding factors, such as small numbers of mutated patients in each group, different age distribution, or distinct cytogenetic categories. Thus, more studies are needed to determine the real incidence of IDH1 mutation in AML and determine whether there is racial difference in its occurrence.
Exogenous expression of the IDH1 mutants in cultured oligodendroglioma cells did not change the baseline enzymatic activity, suggesting that the mutant IDH1 protein did not disturb the corresponding endogenous IDH1.5 However, a recent mechanistic study demonstrated a dominant negative function of the mutant.15 The affinity of the mutant IDH1 protein to its substrate, isocitrate, was low. In addition, the heterodimerization of the mutant and wild-type IDH1 proteins greatly reduces the affinity of isocitrate to the wild-type IDH1 part of this heterodimerized protein, thus leading to reduction of the reaction product, α-ketoglutarate, which is important for degradation of hypoxia-inducing factor (HIF). It is thus hypothesized that the IDH1 mutation causes HIF up-regulation, which then facilitates growth of gliomas.
Although the hypothesis seems plausible in brain tumors, the underlying pathophysiologic effects of IDH1 mutations in AML remain to be investigated. It may be impossible to extend the observation in brain tumors to other malignancies for several reasons. First, the IDH1 mutation seems highly specific to subgroups of gliomas; the mutation is mainly found in grade 2 and 3 gliomas, but not in World Health Organization grade 1 pilocytic astrocytoma or other solid tumors.5,16 The narrow spectrum of malignancies in which the IDH1 mutation is detected strongly argues for the presence of a highly specific and a yet-to-be-defined pathophysiologic mechanism. Second, besides the obvious difference of incidence of this mutation in glioma and AML, the mutation patterns are also dissimilar. In both this study and the previous report,3 R132C, rather than R132H, was the most common mutation in AML, in contrast to the situation in gliomas, where R132H constituted up to 88.2% of all mutations, whereas R132C composed only 4%.3,5 Third, accumulation of HIF seems to induce leukemia cell differentiation and growth arrest, in contrast to the solid tumors, in which HIF seems to enhance tumor growth.17-19 In addition, the expression of HIF1α-targeted genes was not significantly altered in AML patients with the IDH1 mutation, whereas these genes were upgraded in glioblastoma cell line overexpressing R132H mutant.3 Thus, the underlying mechanisms linking this mutation to AML are different from those in glioma and need to be investigated.
In this study, the IDH1 mutation was demonstrated to be frequently accompanied with concurrent alterations of other genes, especially NPM1 and FLT3/ITD, in the same person (Tables 1, 4), a finding compatible with a 2-hit theory in the leukemogenesis.20,21 Further mechanistic studies are necessary to delineate the pathologic effects of the concert actions of these mutations in AML cells.
Although the IDH1 mutation confers better prognosis to patients with gliomas,5,22 it seems to have a possible adverse effect on overall survival among AML patients with normal karyotype and wild-type NPM1.3 However, we did not find any prognostic significance of IDH1 mutation on our patients, even when we compared the subgroups of patients based on their karyotypes, presence of NPM1 mutation or FLT3/ITD (data not shown). Studies on larger patient cohorts are necessary to clarify the prognostic significance of the IDH1 mutation in AML.
In contrast to the instability of FLT3/ITD during disease evolution, we found that the IDH1 mutation seemed rather stable; none of the patients with wild-type IDH1 at diagnosis acquired novel mutation during clinical follow-up, and the same mutation remained in all relapsed samples. Although the IDH1 mutations in 2 relapsed marrow samples could not be detected by direct DNA sequencing, in which only 11.2% and 16.8% blasts were present, respectively, they were detectable by a more sensitive gene cloning technique. We expect that this mutation would be another potential marker for motoring of minimal residual disease.
In conclusion, we have demonstrated that IDH1 mutations occur in approximately 5.5% and 8.4% of total adult AML patients and the subgroup with normal karyotype, respectively. Whether the incidences of this mutation in AML are correlated with ethnicities remains to be determined by future studies. The IDH1 mutation distributes evenly among different ages and does not show a gender preference. The IDH1 mutation is strongly associated with the NPM1 mutation, normal karyotype, isolated trisomy 8, and FAB M1 subtype, but inversely associated with the M4 subtype and expression of HLA-DR, CD13, and CD14 antigens. This mutation seems very stable during disease progression and is a potential marker for minimal residual disease monitoring.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Science Council (Taiwan; grants NSC 96-2628-B002-013-MY2 and 98-2314-B-002-033-MY3; and NSC 97-2314-B002-015-MY3, H.-F.T.), National Health Research Institute (Taiwan; NHRI-EX97-9731BI), and YongLin Healthcare Foundation (W.-C.C.).
Authorship
Contribution: W.-C.C. and H.-F.T. designed the experiment and wrote the paper; W.-C.C., H.-A.H., C.-Y.C., Y.-N.H., Y.-C. Chang, F.-Y.L., M.-C.L., C.-W.L., M.-H.T., and C.-F.H. performed the experiment; and J.-L.T., M.Y., W.T., B.-S.K., S.-J.W., S.-Y.H., S.-C.H., and Y.-C. Chen provided important materials.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hwei-Fang Tien, Division of Hematology, Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan; e-mail: hftien@ntu.edu.tw; or Wen-Chien Chou, Department of Laboratory Medicine and Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan; e-mail: wchou@ntu.edu.tw.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal