Abstract
Graft-versus-host disease and graft rejection are major complications of allogeneic HLA-mismatched stem cell transplantation or organ transplantation that are caused by alloreactive T cells. Because a range of acute viral infections have been linked to initiating these complications, we hypothesized that the cross-reactive potential of virus-specific memory T cells to allogeneic (allo) HLA molecules may be able to mediate these complications. To analyze the allo-HLA reactivity, T cells specific for Epstein-Barr virus, cytomegalovirus, varicella zoster virus, and influenza virus were tested against a panel of HLA-typed target cells, and target cells transduced with single HLA molecules. Eighty percent of T-cell lines and 45% of virus-specific T-cell clones were shown to cross-react against allo-HLA molecules. The cross-reactivity of the CD8 and CD4 T-cell clones was directed primarily against HLA class I and II, respectively. However, a restricted number of CD8 T cells exhibited cross-reactivity to HLA class II. T-cell receptor (TCR) gene transfer confirmed that allo-HLA reactivity and virus specificity were mediated via the same TCR. These results demonstrate that a substantial proportion of virus-specific T cells exert allo-HLA reactivity, which may have important clinical implications in transplantation settings as well as adoptive transfer of third-party virus-specific T cells.
Introduction
HLA disparity between donor and recipient increases the risk and the severity of graft-versus-host disease (GVHD) after stem cell transplantation (SCT). The risk of graft rejection is also significantly increased in HLA-mismatched compared with HLA-matched SCTs and solid organ transplantations. The negative effect of HLA disparity on the clinical outcome of transplantations is the result of high frequencies of alloreactive T cells. In HLA-mismatched mixed lymphocyte reactions, the frequency of reactive T cells was demonstrated to be a 1000-fold higher than the frequency of T cells reactive in HLA-identical mixed lymphocyte reactions.1,2 By testing alloreactive T cells against panels of third-party target cells expressing different HLA molecules1,3-5 and against target cells blocked with different HLA antibodies,6-8 it was determined that the recognition exhibited by alloreactive T cells is directed against non–self-HLA (allo-HLA) molecules, and that the frequency of allo-HLA–reactive T cells ranged from 1% to 10%.
During thymic development, T cells undergo an instruction process of positive and negative selection that results in the composition of a mature T-cell repertoire that is selected on the basis of tolerance for self-HLA molecules presenting self-peptides.9,10 However, during thymic development, T cells never encounter allo-HLA molecules, and therefore no selection based on tolerance for allo-HLA molecules occurs. We therefore hypothesize that every antigen-specific self-HLA–restricted T-cell could potentially cross-react with non–self-HLA molecules and exert allo-HLA reactivity.
Although it was shown that alloreactivity is equally presented in the naive and memory T-cell populations,11 the ability of T cells to exhibit allo-HLA reactivity could have especially serious consequences when exerted by memory T cells. Memory T cells lack the requirement for costimulation,12,13 and therefore allo-HLA reactivity of memory T cells can be efficiently triggered by nonprofessional antigen-presenting cells after HLA-mismatched SCT or solid organ transplantation. Based on the restricted T-cell receptor (TCR) repertoire of virus-specific memory T cells,14-17 the number of different virus-specific T cells will be limited, but the total number of virus-specific T cells with an identical TCR will be much higher in the memory pool compared with the naive compartment. T cells directed against latent viruses, such as Epstein-Barr virus (EBV) and cytomegalovirus (CMV), are present at high frequencies in blood of healthy persons and patients.18-21 Therefore, if certain virus-specific T cells with cross-reactive potential against the mismatched allo-HLA molecule are triggered by viral activation and expanded in the memory pool, these T cells will react against the mismatched HLA molecule, and may induce severe GVHD or graft rejection.
Studies by Burrows et al first illustrated that virus-specific T cells exert allo-HLA reactivity by demonstrating that EBV-EBNA3A–specific HLA-B8–restricted T cells cross-react with HLA-B44.22,23 T cells specific for HSV-VP13/14 presented in HLA-A2 were also found to cross-react with HLA-B44,24 and CD4 T cells specific for tetanus toxoid presented in HLA-DR3 were found to be cross-reactive against HLA-DR4.25 In addition, the association between reactivation of viral infections during organ transplantation and increased graft rejection26 supports the hypothesis that virus-specific T cells exhibit allo-HLA–reactive potential.
In this study, we investigate the ability of a large panel of virus-specific T cells to exert allo-HLA reactivity. We determined the cross-reactive potential of virus-specific T cells to allo-HLA molecules by screening single viral antigen-specific T-cell lines and clones against a panel of EBV-transformed B cells (EBV-LCLs), together covering almost all prevalent HLA class I and II molecules, as well as single HLA-transduced target cells. The tested CD8 and CD4 virus-specific memory T cells were specific for Epstein-Barr virus (EBV), cytomegalovirus (CMV), varicella zoster virus (VZV), and influenza virus (Flu). Most virus-specific T-cell lines and 45% of the virus-specific T-cell clones were demonstrated to be cross-reactive against allo-HLA molecules. TCR gene transfer demonstrated that the virus specificity and the cross-reactivity to allo-HLA molecules were mediated by the same TCR. These results demonstrate that T cells specific for different viruses exert cross-reactivity to allo-HLA molecules, and illustrate the high frequency of T cells able to exert allo-HLA reactivity.
Methods
Cell collection and preparation
After informed consent was given in accordance with the Declaration of Helsinki, peripheral blood (PB) was obtained from different persons. All experiments were approved by the Leiden University Medical Center ethics committee. Mononuclear cells (MNCs) were isolated by Ficoll-Isopaque separation and cryopreserved. Stable EBV-LCLs were generated using standard procedures. EBV-LCLs and K562 cells were cultured in Iscove modified Dulbecco medium (IMDM; Lonza) and 10% fetal bovine serum (FBS; Lonza). Phytohemagglutinin (PHA) blasts were generated by stimulation of PB-MNCs with PHA (0.8 μg/mL; Murex Biotec Limited) in IMDM, 5% FBS, 5% human serum, and interleukin-2 (IL-2; 120 IU/mL). K562 and EBV-LCLs expressing single allo-HLA molecules were generated by transduction with retroviral vectors encoding for the allo-HLA molecules or by transfection of allo-HLA molecules.27,28 For the isolation of T cells, B cells, and monocytes, peripheral blood mononuclear cells of healthy donors were stained with either anti-CD3, anti-CD19, or anti-CD14 magnetic-activated cell sorting beads (Miltenyi Biotec), respectively, and isolated according to the manufacturer's instructions. CD40 ligand (CD40L)–activated B cells were generated by culturing the CD19+ fraction for 3 days on CD40L-transduced murine fibroblasts29 in medium containing CpG (10 μg/mL) and IL-4 (500 IU/mL; Schering-Plough). Monocyte-derived DCs were generated by culturing the CD14+ fraction in medium containing activating cytokines as described previously.30 Fibroblasts were cultured from skin biopsies in Dulbecco modified Eagle medium (Lonza) with 1 g/L glucose (BioWhittaker) and 10% FBS.
Generation of virus-specific T-cell lines and clones
PB-MNCs from healthy persons were stained with tetramer and anti-CD8 monoclonal antibody (mAb) for 1 hour at 4°C and washed once. The tetramers used were constructed as described previously31 and are shown in Table 1. Tetramer-positive, CD8+ T cells were sorted at 50 cells per well for the generation of lines or a single cell per well for the generation of clones into U-bottom microtiter plates containing 100 μL of feeder mixture. Sorting was performed at 4°C using the FACSVantage (BD). The feeder mixture consisted of IMDM, 5% FBS, 5% human serum, IL-2 (120 IU/mL), PHA, and 30 Gy irradiated allogeneic third-party PB-MNCs (0.5 × 106/mL). Proliferating T-cell clones were selected and further expanded by nonspecific stimulation every 14 days using the previously mentioned feeder mixture. The viral specificity of the expanded lines and clones was confirmed by tetramer staining, cytotoxicity, and cytokine production assays. Polyclonality or monoclonality of the T-cell lines and clones was analyzed by TCR Vβ analysis using the TCR Vβ kit (Beckman Coulter).
Tetramers used for the generation of virus specific T-cell lines and clones
| Virus . | Viral antigen . | HLA . | Epitope . |
|---|---|---|---|
| CMV | pp50 | HLA-A*0101 | VTEHDTLLY |
| CMV | pp65 | HLA-A*0101 | YSEHPTFTSQY |
| CMV | pp65 | HLA-A*0201 | NLVPMVATV |
| CMV | pp65 | HLA-B*0702 | RPHERNGFTVL |
| CMV | pp65 | HLA-B*03501 | IPSINVHHY |
| CMV | pp65 | HLA-DRB1*0101 | KYQEFFWDANDIYRI |
| CMV | IE1 | HLA-B*0801 | ELRRKMMYM |
| EBV | EBNA3A | HLA-A*0301 | RLRAEAQVK |
| EBV | EBNA3A | HLA-B*0702 | RPPIFIRRL |
| EBV | EBNA3A | HLA-B*0801 | FLRGRAYGL |
| EBV | BMLF1 | HLA-A*0201 | GLCTLVAML |
| EBV | BRLF1 | HLA-A*0301 | RVRAYTYSK |
| EBV | BZLF1 | HLA-B*0801 | RAKFKQLL |
| EBV | LMP2 | HLA-A*0201 | CLGGLLTMV |
| FLU | IMP | HLA-A*0201 | GILGFVFTL |
| FLU | HA | HLA-DRB1*0401 | PKYVKQNTLKLAT |
| VZV | IE62 | HLA-A*0201 | ALWALPHAA |
| Virus . | Viral antigen . | HLA . | Epitope . |
|---|---|---|---|
| CMV | pp50 | HLA-A*0101 | VTEHDTLLY |
| CMV | pp65 | HLA-A*0101 | YSEHPTFTSQY |
| CMV | pp65 | HLA-A*0201 | NLVPMVATV |
| CMV | pp65 | HLA-B*0702 | RPHERNGFTVL |
| CMV | pp65 | HLA-B*03501 | IPSINVHHY |
| CMV | pp65 | HLA-DRB1*0101 | KYQEFFWDANDIYRI |
| CMV | IE1 | HLA-B*0801 | ELRRKMMYM |
| EBV | EBNA3A | HLA-A*0301 | RLRAEAQVK |
| EBV | EBNA3A | HLA-B*0702 | RPPIFIRRL |
| EBV | EBNA3A | HLA-B*0801 | FLRGRAYGL |
| EBV | BMLF1 | HLA-A*0201 | GLCTLVAML |
| EBV | BRLF1 | HLA-A*0301 | RVRAYTYSK |
| EBV | BZLF1 | HLA-B*0801 | RAKFKQLL |
| EBV | LMP2 | HLA-A*0201 | CLGGLLTMV |
| FLU | IMP | HLA-A*0201 | GILGFVFTL |
| FLU | HA | HLA-DRB1*0401 | PKYVKQNTLKLAT |
| VZV | IE62 | HLA-A*0201 | ALWALPHAA |
CMV indicates cytomegalovirus; EBV, Epstein-Barr virus; FLU, influenza virus; and VZV, varicella zoster virus.
Allo-HLA reactivity of the virus-specific T-cell lines and clones
In the interferon-γ (IFNγ) production assays, 5000 T cells were cocultured with 20 000 stimulator cells in a final volume of 150 μL IMDM culture medium supplemented with 100 IU/mL IL-2. After 18 hours of incubation, supernatants were harvested and IFNγ production was measured by standard enzyme-linked immunosorbent assay (ELISA; CLB). In the cytotoxicity assays, the virus-specific T-cell clones were tested in a standard 6-hour 51Cr-release assay32 against EBV-LCLs at an effector to target ratio of 10:1.
TCR gene transfer
The TCRAV and TCRBV gene usage of the BRLF1/HLA-A3 clone 19 and the VZV-IE62/HLA-A2–specific T-cell clone 7 was determined using reverse transcriptase–polymerase chain reaction and sequencing.27 Retroviral vectors were constructed that encoded the TCRα chain in combination with green fluorescent protein and the TCRβ chain in combination with the marker gene ΔNGF-R.27 CMV-IE1/HLA-A1–specific T cells derived from an HLA-A*0301–negative healthy person were transduced with the TCRα and TCRβ of the BRLF1/HLA-A3 clone 19. CMV-pp50/HLA-A1–specific T cells derived from peripheral blood of a healthy person negative for HLA-A*0201 and HLA-A*0205 were transduced with the retroviral vectors encoding for the TCRα and TCRβ chain of the VZV-specific T-cell clone.27 The TCR-transduced T cells were sorted on basis of double positivity for green fluorescent protein and ΔNGF-R, and after expansion the T cells were tested for viral specificity and allo-HLA reactivity in stimulation assays.
Results
Alloreactivity of virus-specific T-cell lines
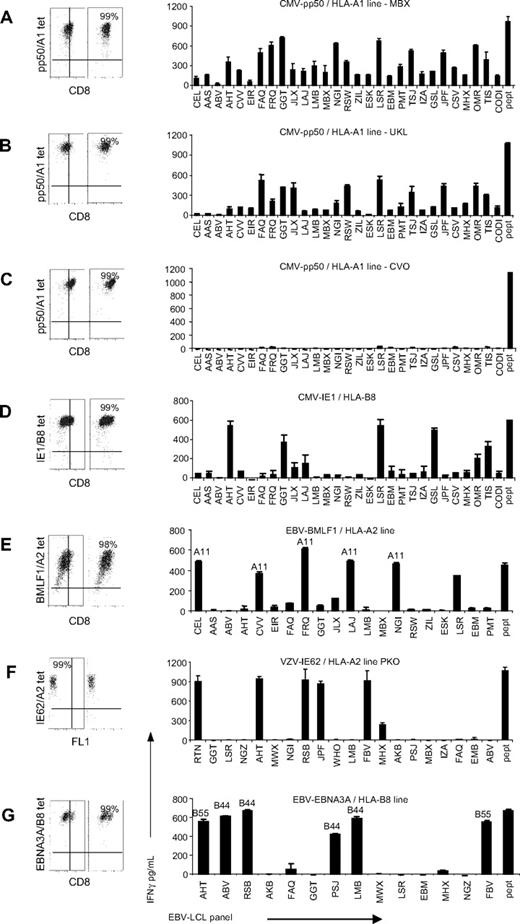
To investigate the ability of virus-specific T cells to exert alloreactivity, virus-specific T-cell lines were tested against a panel of EBV-LCLs, together covering almost all frequently occurring HLA class I and II molecules. The HLA-typing of the EBV-LCLs used in the panel is listed in Table 2. The virus-specific T-cell lines were generated by isolation of tetramer-positive CD8+ T cells by fluorescence-activated cell sorting and subsequent expansion. Tetramer and CD8 staining confirmed the purity of the virus-specific lines and showed that all T-cell lines were more than 98% tetramer positive (Figure 1A-G). In total, 11 virus-specific lines were tested, derived from 9 different donors, specific for 7 different antigens of CMV, EBV, and VZV and restricted to 3 different HLA molecules. In the figures in which EBV-specific T-cell lines were tested, the EBV-LCLs expressing the HLA molecules to which the T-cell lines were restricted were not shown, to present only the alloreactivity of the T cells and not the virus specificity. All LCLs were tested for IFNγ production in the absence of T cells and did not show production of IFNγ (data not shown). In Figure 1A through G, 7 representative lines are shown. Of the 11 tested virus-specific T-cell lines, 9 were shown to be alloreactive, because these lines produced IFNγ upon stimulation with at least 1 of the EBV-LCLs of the panel. Of the 11 tested lines, 2 exerted no alloreactivity against the EBV-LCLs tested in our panel (including Figure 1C). The recognition pattern of the virus-specific lines that demonstrated alloreactivity ranged from recognition of almost all EBV-LCLs, as shown in Figure 1A and B by the CMV-pp50/HLA-A1–specific lines of patients MBX and UKL, to recognition of a limited number of EBV-LCLs, as shown by the IE1/HLA-B8–, BMLF1/HLA-A2–, VZV-IE62/HLA-A2–, and EBNA3A/HLA-B8–specific lines (Figure 1D, E, F, and G, respectively). The pattern of allorecognition of some of the T-cell lines suggested that the alloreactivity was directed against 1 or several of the allo-HLA molecules presented by the EBV-LCL panel. The BMLF1/HLA-A2–specific line showed high reactivity against 6 EBV-LCLs, of which 5 EBV-LCLs expressed HLA-A*1101. All HLA-A*1101–expressing EBV-LCLs within this panel were highly recognized by this T-cell line. The EBV-LCLs recognized by the EBNA3A/HLA-B8–specific line expressed either HLA-B*4402 or HLA-B*5501. TCR Vβ analysis of the lines demonstrated that the complexity of the TCR composition was correlated with the broadness of the alloreactivity. T-cell lines that did not show alloreactivity expressed maximally 2 different TCR Vβ chains. T-cell lines with limited alloreactivity expressed 1 to 4 different TCR Vβ chains, and lines that recognized almost all EBV-LCLs expressed at least 8 different TCR Vβ chains. This relation between the clonal composition and the recognition of the lines suggests that the alloreactivity of the lines is the sum of the alloreactivity of the various clonal populations present within the lines.
HLA expression of the EBV-LCL panel
| ID . | HLA class I . | HLA class II . | ||||
|---|---|---|---|---|---|---|
| A . | B . | C . | DR . | DQ . | DP . | |
| CEL | 1101 | 1502, 4001 | 0401, 0801 | 0901 | 0202, 0303 | 0501 |
| AAS | 3101, 6601 | 2705, 4102 | 0202, 1703 | 1101, 1303 | 0301 | 0401, 0402 |
| ABV | 0301, 2902 | 0702, 4403 | 0702, 1601 | 0701, 1454 | 0502, 0202 | 0401, 1101 |
| AHT | 2402, 2501 | 5501, 1501 | 0303 | 0806, 1501 | 0501, 0602 | 0401 |
| CVV | 11, 31* | 57*, 1501 | 3*, 0602 | 0401, 7* | 0301, 0303 | 0201, 0401 |
| EIR | 0101, 2402 | 3502, 3701 | 0401, 0602 | 0404, 1104 | 0301, 0402 | 0301, 0401 |
| FAQ | 2301, 6802 | 1402, 3801 | 0802, 1203 | 1301, 1303 | 0301, 0603 | 0201 |
| FRQ | 0101, 1101 | 0801, 4402 | 0501, 0701 | 0301, 0401 | 0201, 0301 | 0101, 0401 |
| GGT | 2601, 3101 | 1401, 4901 | 0701, 0802 | 0101, 0701 | 0202, 0504 | 0402, 1101 |
| JLX | 0101, 6802 | 0801, 5301 | 0401, 0701 | 0301, 1302 | 0201, 0604 | 0101, 0401 |
| LAJ | 1101, 2402 | 1302, 5501 | 0303, 0602 | 0701, 1601 | 0202, 0502 | 0401, 1701 |
| LMB | 2902 | 4402, 5101 | 1402, 1601 | 0701, 0801 | 0202, 0402 | 0401, 1101 |
| MBX | 0101 | 0801, 1517 | 0701 | 1202, 0301 | 201, 0301 | 0101, 0401 |
| NGI | 1101, 2402 | 0801, 3906 | 0701 | 0301, 0801 | 0201, 0402 | 0101, 1401 |
| RSW | 3001, 6802 | 4201 | 1701 | 0302 | 0402 | 0101, 0402 |
| ZIL | 2402, 2601 | 5601, 5801 | 0101, 0701 | 0101, 0804 | 0402, 0501 | 0201, 0301 |
| ESK | 23, 66* | 51, 72* | 2, 18* | 12, 13* | 1, 3* | ND |
| LSR | 3201, 6801 | 3503, 5201 | 1202, 1203 | 1502, 1602 | 0502, 0601 | 0401, 1401 |
| EBM | 2301 | 1401 | 0802 | 0401 | 0302 | 0201 |
| PMT | 0301, 3301 | 1402 | 0802 | 0102 | 0501 | 0301, '0401 |
| TSJ | 2, 24 | 1501, 75 | 4 | 12, 15 | 6, 0301 | ND |
| IZA | 0201, 2402 | 0801, 4001 | 0304, 0701 | 0301, 1301 | 0603 | 0401, 1401 |
| GSL | 2, 3* | 47, 1501* | 3, 6* | 11, 13* | 1*, 0301 | ND |
| JPF | 0201, 0205 | 4002, 1501 | 0202, 0304 | 0701, 1104 | 0301, 0303 | 0402, 1401 |
| CSV | 0201, 3101 | 5101, 5501 | 0303, 1402 | 0404, 1001 | 0501, 0301 | 0401, 0402 |
| MHX | 0101, 0205 | 1801, 5001 | 0602, 0701 | 0701, 0901 | 0201, 0303 | 0201, 0301 |
| OMR | 201 | 4501 | 1601 | 1301 | 0603 | 0101 |
| TIS | 0206, 0207 | 4601 | 0102, 0801 | 0901 | 0303 | 1301 |
| CODI | 2*, 8001 | 58, 70* | 2, 6* | 17, 11* | 2, 7* | 1, 4* |
| AKB | 0101, 0201 | 3701, 3901 | 0602, 0702 | 0101, 1001 | 501 | 0201, 0401 |
| PSJ | 0201, 3001 | 1302, 4402 | 0501, 0602 | 0401, 0701 | 0202, 0301 | 0402, 1401 |
| MWX | 0101, 3401 | 1521, 3503 | 0403, 1203 | 0101, 1502 | 0501, 0601 | 0601, 1301 |
| NGZ | 1101, 2401 | 5201, 4002 | 0202, 1202 | 0101, 1101 | 0501, 0301 | 0201, 0301 |
| RSB | 0201, 0301 | 5701, 4402 | 0602, 0704 | 0701, 1101 | 0301, 0303 | 0201, 0401 |
| WOH | 1, 28 | 8, 27 | 2, 7 | ND | ND | ND |
| FBV | 0201, 1101 | 0702, 5501 | 0303, 0702 | 1454, 1501 | 0503, 0602 | 0401 |
| RTN | 0101, 1101 | 0801, 5701 | 0602, 0701 | 0301, 0701 | 0201, 0301 | 0401, 1401 |
| ID . | HLA class I . | HLA class II . | ||||
|---|---|---|---|---|---|---|
| A . | B . | C . | DR . | DQ . | DP . | |
| CEL | 1101 | 1502, 4001 | 0401, 0801 | 0901 | 0202, 0303 | 0501 |
| AAS | 3101, 6601 | 2705, 4102 | 0202, 1703 | 1101, 1303 | 0301 | 0401, 0402 |
| ABV | 0301, 2902 | 0702, 4403 | 0702, 1601 | 0701, 1454 | 0502, 0202 | 0401, 1101 |
| AHT | 2402, 2501 | 5501, 1501 | 0303 | 0806, 1501 | 0501, 0602 | 0401 |
| CVV | 11, 31* | 57*, 1501 | 3*, 0602 | 0401, 7* | 0301, 0303 | 0201, 0401 |
| EIR | 0101, 2402 | 3502, 3701 | 0401, 0602 | 0404, 1104 | 0301, 0402 | 0301, 0401 |
| FAQ | 2301, 6802 | 1402, 3801 | 0802, 1203 | 1301, 1303 | 0301, 0603 | 0201 |
| FRQ | 0101, 1101 | 0801, 4402 | 0501, 0701 | 0301, 0401 | 0201, 0301 | 0101, 0401 |
| GGT | 2601, 3101 | 1401, 4901 | 0701, 0802 | 0101, 0701 | 0202, 0504 | 0402, 1101 |
| JLX | 0101, 6802 | 0801, 5301 | 0401, 0701 | 0301, 1302 | 0201, 0604 | 0101, 0401 |
| LAJ | 1101, 2402 | 1302, 5501 | 0303, 0602 | 0701, 1601 | 0202, 0502 | 0401, 1701 |
| LMB | 2902 | 4402, 5101 | 1402, 1601 | 0701, 0801 | 0202, 0402 | 0401, 1101 |
| MBX | 0101 | 0801, 1517 | 0701 | 1202, 0301 | 201, 0301 | 0101, 0401 |
| NGI | 1101, 2402 | 0801, 3906 | 0701 | 0301, 0801 | 0201, 0402 | 0101, 1401 |
| RSW | 3001, 6802 | 4201 | 1701 | 0302 | 0402 | 0101, 0402 |
| ZIL | 2402, 2601 | 5601, 5801 | 0101, 0701 | 0101, 0804 | 0402, 0501 | 0201, 0301 |
| ESK | 23, 66* | 51, 72* | 2, 18* | 12, 13* | 1, 3* | ND |
| LSR | 3201, 6801 | 3503, 5201 | 1202, 1203 | 1502, 1602 | 0502, 0601 | 0401, 1401 |
| EBM | 2301 | 1401 | 0802 | 0401 | 0302 | 0201 |
| PMT | 0301, 3301 | 1402 | 0802 | 0102 | 0501 | 0301, '0401 |
| TSJ | 2, 24 | 1501, 75 | 4 | 12, 15 | 6, 0301 | ND |
| IZA | 0201, 2402 | 0801, 4001 | 0304, 0701 | 0301, 1301 | 0603 | 0401, 1401 |
| GSL | 2, 3* | 47, 1501* | 3, 6* | 11, 13* | 1*, 0301 | ND |
| JPF | 0201, 0205 | 4002, 1501 | 0202, 0304 | 0701, 1104 | 0301, 0303 | 0402, 1401 |
| CSV | 0201, 3101 | 5101, 5501 | 0303, 1402 | 0404, 1001 | 0501, 0301 | 0401, 0402 |
| MHX | 0101, 0205 | 1801, 5001 | 0602, 0701 | 0701, 0901 | 0201, 0303 | 0201, 0301 |
| OMR | 201 | 4501 | 1601 | 1301 | 0603 | 0101 |
| TIS | 0206, 0207 | 4601 | 0102, 0801 | 0901 | 0303 | 1301 |
| CODI | 2*, 8001 | 58, 70* | 2, 6* | 17, 11* | 2, 7* | 1, 4* |
| AKB | 0101, 0201 | 3701, 3901 | 0602, 0702 | 0101, 1001 | 501 | 0201, 0401 |
| PSJ | 0201, 3001 | 1302, 4402 | 0501, 0602 | 0401, 0701 | 0202, 0301 | 0402, 1401 |
| MWX | 0101, 3401 | 1521, 3503 | 0403, 1203 | 0101, 1502 | 0501, 0601 | 0601, 1301 |
| NGZ | 1101, 2401 | 5201, 4002 | 0202, 1202 | 0101, 1101 | 0501, 0301 | 0201, 0301 |
| RSB | 0201, 0301 | 5701, 4402 | 0602, 0704 | 0701, 1101 | 0301, 0303 | 0201, 0401 |
| WOH | 1, 28 | 8, 27 | 2, 7 | ND | ND | ND |
| FBV | 0201, 1101 | 0702, 5501 | 0303, 0702 | 1454, 1501 | 0503, 0602 | 0401 |
| RTN | 0101, 1101 | 0801, 5701 | 0602, 0701 | 0301, 0701 | 0201, 0301 | 0401, 1401 |
The panel of EBV-transformed B cells (EBV-LCLs) was composed of HLA-typed EBV-LCLs, which together covered almost all frequently occurring HLA molecules. The HLA typing was determined mainly molecularly; however, some EBV-LCLs were only serologically typed.
ND indicates that the HLA expression was not determined.
HLA expression was determined by serologic typing.
Alloreactivity of virus-specific T-cell lines. Eleven virus-specific T-cell lines, of which 7 are shown in this figure, were stimulated with a panel of EBV-LCLs for 18 hours and IFNγ production was measured by ELISA. In experiments in which EBV-specific T-cell lines were tested, we excluded the EBV-LCLs expressing the HLA molecules to which the T-cell lines were restricted. The purity of the virus-specific lines was analyzed by tetramers and CD8 staining, and all T-cell lines proved to be more than 98% pure. As a positive control, the lines were tested against EBV-LCLs expressing the HLA-restricting molecule of the viral epitope, loaded with the viral peptide recognized by the T-cell lines (pept). (A) The CMV-pp50/HLA-A1–specific lines of patient MBX recognized almost all EBV-LCLs. (B) The CMV-pp50/HLA-A1–specific line of patient UKL showed broad alloreactivity. (C) Two of the 10 tested T-cell lines exerted no alloreactivity against the tested EBV-LCLs of which 1, the pp50/HLA-A1–specific line, is shown. (D) The CMV-IE1/HLA-B8 recognized a limited number of EBV-LCLs. (E) The BMLF1/HLA-A2–specific line showed high reactivity against all HLA-A11–positive EBV-LCLs and 1 HLA-A11–negative EBV-LCL. (F) The VZV-IE62/HLA-A2–specific line of patient PKO recognized a limited number of EBV-LCLs. (G) EBNA3A/HLA-B8–specific line recognized EBV-LCLs expressing either HLA-B44 or HLA-B55. Experiments were performed in duplicate, mean values are shown ± SD.
Alloreactivity of virus-specific T-cell lines. Eleven virus-specific T-cell lines, of which 7 are shown in this figure, were stimulated with a panel of EBV-LCLs for 18 hours and IFNγ production was measured by ELISA. In experiments in which EBV-specific T-cell lines were tested, we excluded the EBV-LCLs expressing the HLA molecules to which the T-cell lines were restricted. The purity of the virus-specific lines was analyzed by tetramers and CD8 staining, and all T-cell lines proved to be more than 98% pure. As a positive control, the lines were tested against EBV-LCLs expressing the HLA-restricting molecule of the viral epitope, loaded with the viral peptide recognized by the T-cell lines (pept). (A) The CMV-pp50/HLA-A1–specific lines of patient MBX recognized almost all EBV-LCLs. (B) The CMV-pp50/HLA-A1–specific line of patient UKL showed broad alloreactivity. (C) Two of the 10 tested T-cell lines exerted no alloreactivity against the tested EBV-LCLs of which 1, the pp50/HLA-A1–specific line, is shown. (D) The CMV-IE1/HLA-B8 recognized a limited number of EBV-LCLs. (E) The BMLF1/HLA-A2–specific line showed high reactivity against all HLA-A11–positive EBV-LCLs and 1 HLA-A11–negative EBV-LCL. (F) The VZV-IE62/HLA-A2–specific line of patient PKO recognized a limited number of EBV-LCLs. (G) EBNA3A/HLA-B8–specific line recognized EBV-LCLs expressing either HLA-B44 or HLA-B55. Experiments were performed in duplicate, mean values are shown ± SD.
The results demonstrate that 80% of the tested virus-specific T-cell lines were able to exert alloreactivity. Some virus-specific lines showed a pattern of alloreactivity suggestive of allo-HLA reactivity. However, for most of the virus-specific cell lines, allo-HLA reactivity could not be determined because the exerted alloreactivity was very broad.
Allo-HLA reactivity of virus-specific T-cell clones
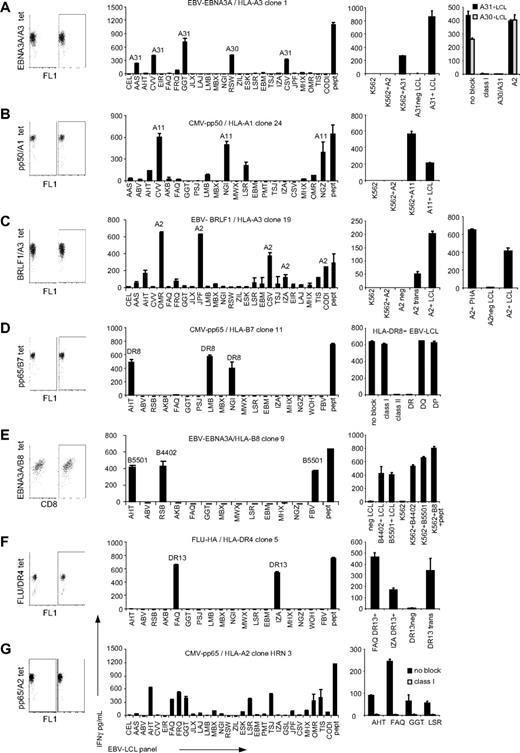
Because we were unable to determine allo-HLA reactivity with the oligoclonal virus-specific T-cell lines, further characterization of the allo-HLA reactivity of virus-specific T cells was performed with T-cell clones. For this purpose, tetramer-positive T cells were sorted at a single cell per well and expanded. The specificity of the T-cell clones was confirmed by tetramer staining, and the TCR Vβ usage of the clones was analyzed using the TCR Vβ kit. The T-cell clones as shown in Table 3 were different based on either their different origin, TCR Vβ usage, or recognition pattern. In total, 41 virus-specific CD8 and CD4 T-cell clones were tested against the EBV-LCL panel. These virus-specific T-cell clones were derived from 16 persons, specific for 13 different CMV, EBV, VZV, and Flu antigens and restricted to 8 different HLA molecules. The results demonstrated that 18 of the 41 virus-specific CD8 and CD4 T-cell clones were alloreactive, as shown by recognition of at least 1 of the EBV-LCLs from the panel. Most alloreactive T-cell clones exhibited cross-reactivity against EBV-LCLs that shared an HLA molecule, suggesting allo-HLA reactivity. To confirm that the reactivity of a T-cell clone was directed against a specific allo-HLA molecule, the virus-specific T-cell clones were tested against HLA-negative K562 cell line or EBV-LCLs negative for the recognized allo-HLA molecules, which were transduced with the particular allo-HLA molecule. The allo-HLA reactivity of 7 representative T-cell clones is shown in Figure 2A-G. The EBV-EBNA3A/HLA-A3–specific CD8 T-cell clone exhibited alloreactivity against all HLA-A*3101–expressing EBV-LCLs within the panel. The reactivity directed against allo–HLA-A*3101 was confirmed by transfection of K562 with HLA-A*3101 and subsequent recognition by this T-cell clone (Figure 2A). The EBV-EBNA3A/HLA-A3 clone also recognized the HLA-A*3101–negative EBV-LCL RSW, which however expressed HLA-A*3001. HLA-A*3101 and HLA-A*3001 are very similar in sequence and therefore we hypothesized that the molecules exhibit strong similarity in structure and peptide presentation, explaining recognition by this T-cell clone. To analyze whether the EBV-EBNA3A/HLA-A3 clone recognized HLA-A*3001, the clone was tested against 3 HLA-A*3001+ EBV-LCLs, of which 1 is shown in Figure 2A, and 1 HLA-A*3101+ EBV-LCL with or without blocking mAbs directed against HLA class I, HLA-A30/A31, and HLA-A2. All HLA-A*3001+ EBV-LCLs and the HLA-A*3101+ EBV-LCLs were recognized, and this recognition was blocked by anti–HLA class I and anti–HLA-A30/A31 mAbs and not by anti–HLA-A2 mAb, indicating that the clone indeed also recognized HLA-A*3001. The CMV-pp50/HLA-A1–specific T-cell clone exhibited alloreactivity against all HLA-A*1101–expressing EBV-LCLs (Figure 2B). The allo–HLA-A*1101 reactivity could be confirmed by specific recognition of K562 transduced with HLA-A*1101 by this T-cell clone. In addition to reactivity against HLA-A*1101, the T-cell clone also exhibited low IFNγ production upon stimulation with a few EBV-LCLs negative for HLA-A*1101. Because these less recognized EBV-LCLs did not share one HLA molecule, we did not determine whether this alloreactivity was also based on allo-HLA cross-reactivity. The EBV-BRLF1–specific HLA-A3–restricted clone, shown in Figure 2C, exerted alloreactivity against all HLA-A*0201+ EBV-LCLs. The CD8 T-cell clone did not recognize K562 transduced with HLA-A*0201, however it showed recognition of EBV-LCLs transduced with HLA-A*0201, suggesting that the peptide recognized by the clone in the context of HLA-A*0201 is not presented by K562 cells. The clone also showed recognition of HLA-A*0201+ PHA-stimulated T-cell blasts (Figure 2C right panel), excluding the possibility that the clone recognized an EBV-derived peptide presented in HLA-A*0201. Next to allo–HLA-A*0201 reactivity, this T-cell clone also exhibited low cross-reactivity against EBV-LCLs negative for HLA-A*0201. Because these less recognized EBV-LCLs did not share 1 HLA molecule, we did not determine the alloreactivity in detail. The CMV-pp65/HLA-B7–specific CD8 T-cell clone exhibited alloreactivity against 2 HLA-DRB1*0801– and 1 HLA-DRB1*0806–expressing EBV-LCL present in the panel. The cross-reactivity exerted by this CD8 T-cell clone could be blocked with antibodies directed against HLA class II and HLA-DR and not by HLA class I, HLA-DQ, or HLA-DP antibodies, confirming that the cross-reactivity of this virus-specific CD8 T-cell clone was directed against HLA-DR8. The ability of CD8 T cells to cross-react against allo-HLA class II molecules was also demonstrated by the CMV-pp65–specific HLA-B35–restricted CD8 T-cell clone that was shown to be cross-reactive against HLA-DRB1*0401 (Table 3). As previously described, we observed that the EBV-EBNA3A/HLA-B8–specific T-cell clone exhibited alloreactivity against all EBV-LCLs expressing HLA-B*4402.22 Furthermore, we observed alloreactivity against EBV-LCLs expressing HLA-B*5501 as was recently described by us.33 Allo-HLA reactivity against HLA-B*4402 and HLA-B*5501 was confirmed by recognition of K562 transfected with either HLA-B*4402 or HLA-B*5501 by this T-cell clone (Figure 2E).
Allo-HLA reactivity of virus-specific T-cell clones
| Specificity . | Donor . | TCR Vβ . | EBV panel . | HLA trans/block . | Figure . |
|---|---|---|---|---|---|
| pp50/A1 | MBX | * | UD | ||
| pp50/A1 | MBX | 1 | |||
| pp50/A1 | MBX | 5.1 | |||
| pp50/A1 | MBX | 3 | A*1101 | A*1101 | 2B |
| pp65/A2 | MRJ | 2 | |||
| pp65/A2 | MRJ | 13 | |||
| pp65/A2 | HRN | 8 | UD | 2G | |
| pp65/A2 | HRN | 2 | |||
| pp65/A2 | AMJ | 3 | |||
| BMLF1/A2 | GFS | * | |||
| LMP2/A2 | JVW | * | |||
| FLU/A2 | FKR | 17 | B*6401 | ||
| FLU/A2 | FKR | 17 | |||
| VZV/A2 | PKN | 14 | B*5501 | B*5501 | 3A |
| VZV/A2 | PKN | * | B*5701 | B*5701 | 3B |
| VZV/A2 | PKN | 21,3 | A*0205 | A*0205 | 3C |
| EBNA3A/A3 | HRN | * | A*3101 | A*3101 | 2A |
| BRLF1/A3 | AKO | 7.1 | |||
| BRLF1/A3 | AKO | 14 | |||
| BRLF1/A3 | AKO | 17 | |||
| BRLF1/A3 | AKO | 1 | |||
| BRLF1/A3 | DVO | 7.1 | UD | ||
| BRLF1/A3 | DVO | 8 | |||
| BRLF1/A3 | DVO | 14 | UD | ||
| BRLF1/A3 | DVO | 17 | UD | ||
| BRLF1/A3 | DVO | * | |||
| BRLF1/A3 | DVO | 7.2 | A*0201 | A*0201 | 2C |
| pp65/B7 | BDV | 7.2 | |||
| pp65/B7 | BDV | 7.2 | DRB1*0801 | DRB1*0801 | 2D |
| EBNA3A/B8 | LDO | * | B*4402, B*5501 | B*4402, B*5501 | 2E |
| BZLF/B8 | AVK | 7.1 | |||
| BZLF/B8 | AVK | * | |||
| BZLF/B8 | AVK | 5.1 | |||
| pp65/B35 | MED | 3 | |||
| pp65/B35 | MED | 5.1 | DRB1*0401 | DRB1*0401 | |
| pp65/B35 | MED | * | |||
| pp65/DR1 | CBH | 2 | |||
| pp65/DR1 | CBH | * | |||
| pp65/DR1 | MSV | 8 | DRB1*0901 | ||
| pp65/DR1 | MSV | * | DRB3*0101 | DRB3*0101 | |
| FLU/DR4 | VKY | 3 | DRB1*1301 | DRB1*1301 | 2F |
| Specificity . | Donor . | TCR Vβ . | EBV panel . | HLA trans/block . | Figure . |
|---|---|---|---|---|---|
| pp50/A1 | MBX | * | UD | ||
| pp50/A1 | MBX | 1 | |||
| pp50/A1 | MBX | 5.1 | |||
| pp50/A1 | MBX | 3 | A*1101 | A*1101 | 2B |
| pp65/A2 | MRJ | 2 | |||
| pp65/A2 | MRJ | 13 | |||
| pp65/A2 | HRN | 8 | UD | 2G | |
| pp65/A2 | HRN | 2 | |||
| pp65/A2 | AMJ | 3 | |||
| BMLF1/A2 | GFS | * | |||
| LMP2/A2 | JVW | * | |||
| FLU/A2 | FKR | 17 | B*6401 | ||
| FLU/A2 | FKR | 17 | |||
| VZV/A2 | PKN | 14 | B*5501 | B*5501 | 3A |
| VZV/A2 | PKN | * | B*5701 | B*5701 | 3B |
| VZV/A2 | PKN | 21,3 | A*0205 | A*0205 | 3C |
| EBNA3A/A3 | HRN | * | A*3101 | A*3101 | 2A |
| BRLF1/A3 | AKO | 7.1 | |||
| BRLF1/A3 | AKO | 14 | |||
| BRLF1/A3 | AKO | 17 | |||
| BRLF1/A3 | AKO | 1 | |||
| BRLF1/A3 | DVO | 7.1 | UD | ||
| BRLF1/A3 | DVO | 8 | |||
| BRLF1/A3 | DVO | 14 | UD | ||
| BRLF1/A3 | DVO | 17 | UD | ||
| BRLF1/A3 | DVO | * | |||
| BRLF1/A3 | DVO | 7.2 | A*0201 | A*0201 | 2C |
| pp65/B7 | BDV | 7.2 | |||
| pp65/B7 | BDV | 7.2 | DRB1*0801 | DRB1*0801 | 2D |
| EBNA3A/B8 | LDO | * | B*4402, B*5501 | B*4402, B*5501 | 2E |
| BZLF/B8 | AVK | 7.1 | |||
| BZLF/B8 | AVK | * | |||
| BZLF/B8 | AVK | 5.1 | |||
| pp65/B35 | MED | 3 | |||
| pp65/B35 | MED | 5.1 | DRB1*0401 | DRB1*0401 | |
| pp65/B35 | MED | * | |||
| pp65/DR1 | CBH | 2 | |||
| pp65/DR1 | CBH | * | |||
| pp65/DR1 | MSV | 8 | DRB1*0901 | ||
| pp65/DR1 | MSV | * | DRB3*0101 | DRB3*0101 | |
| FLU/DR4 | VKY | 3 | DRB1*1301 | DRB1*1301 | 2F |
TCR indicates T-cell receptor; and UD, undetermined allo-HLA reactivity, which could not be characterized because the recognized EBV-LCLs did not share one particular allo-HLA molecule.
The TCR Vβ of the clone could not be determined with the TCR Vβ kit.
Allo-HLA reactivity of virus-specific T-cell clones. Forty-one virus-specific T-cell clones, of which 7 are shown in this figure, were stimulated with a panel of EBV-LCLs for 18 hours, and IFNγ production was measured by ELISA. (A) The EBV-EBNA3A/HLA-A3–specific T-cell clone 1 exhibited alloreactivity against all EBV-LCLs expressing HLA-A*3101 and 1 EBV-LCL expressing HLA-A*3001. To confirm allo–HLA-A*3101 reactivity, clone 1 was tested against K562 cells (K562), K562 cells transduced with HLA-A*0201 (K562+A2), K562 cells transfected with HLA-A*3101 (K562+A31), and HLA-A*3101–negative (A31neg LCL) and HLA-A*3101–positive (A31+LCL) EBV-LCLs. To confirm the reactivity against HLA-A*3001, the clone was tested against 3 HLA-A*3001+ EBV-LCLs, of which 1 is shown, and 1 HLA-A*3101+ EBV-LCLs with or without blocking mAbs directed against HLA class I, HLA-A30/A31, and HLA-A2. (B) The CMV-pp50/HLA-A1–specific T-cell clone 24 exhibited alloreactivity against all HLA-A*1101–expressing EBV-LCLs. To confirm allo–HLA-A*1101 reactivity, clone 24 was tested against K562 cells (K562), K562 cells transduced with HLA-A*0201 (K562+A2), K562 cells transduced with HLA-A*1101 (K562+A11), and HLA-A*1101–positive EBV-LCLs (A11+LCL). (C) The EBV-BRLF1/HLA-A3–specific clone 19 exerted alloreactivity against all HLA-A0201+ EBV-LCLs. This T-cell clone did not recognize K562 transduced with HLA-A*0201 (K562+A2). To confirm allo–HLA-A*0201 reactivity, clone 19 was tested against untransduced HLA-A*0201–negative EBV-LCLs (HLA-A2neg LCL) or transduced with HLA-A*0201 (A2 trans), and HLA-A*0201–positive EBV-LCLs (A2+LCL). To investigate whether this clone recognized an EBV-derived peptide in the context of HLA-A*0201, the clone was tested against HLA-A*0201+ PHA blasts and HLA-A*0201–positive and –negative EBV-LCLs as controls. (D) The CMV-pp65/HLA-B7–specific T-cell clone 11 exhibited reactivity against all 3 HLA-DRB1*0801+ EBV-LCLs. To confirm allo–HLA-DRB1*0801 reactivity, clone 11 was tested against the 3 HLA-DRB1*0801+ EBV-LCLs, of which 1 is shown, in the presence of either no blocking mAbs (no block) or anti–HLA class I (class I), anti–HLA class II (class II), anti–HLA-DR (DR), anti–HLA-DR (DQ), and anti–HLA-DR (DP) blocking mAbs. (E) The EBV-EBNA3A/HLA-B8–specific T-cell clone 9 exhibited alloreactivity against all EBV-LCLs expressing either HLA-B*4402 or HLA-B*5501. To confirm HLA-B*4402 and HLA-B*5501 cross-reactivity, clone 9 was tested against K562 cells (K562), or K562 cells transfected with HLA-B*4402 (K562+B4402) or HLA-B*5501 (K562+B5501). As controls, clone 9 was tested against HLA-B*4402– and HLA-B*5501–negative EBV-LCLs (neg LCL), HLA-B*4402+ EBV-LCLs (B4402+LCL), HLA-B*5501+ EBV-LCLs (B5501+LCL), or HLA-B*0801+ K562 loaded with viral peptide (K562+B8+pept). (F) The Flu-HA/HLA-DR4–specific clone 5 recognized all HLA-DRB1*1301+ EBV-LCLs. To confirm allo–HLA-DRB1*1301 reactivity, clone 5 was tested against HLA-DRB1*1301+ EBV-LCLs (FAQ DR13+ and IZA DR13+) as well as HLA-DR13–negative EBV-LCLs nontransduced (DR13 neg) or transduced with HLA-DRB1*1301 (DR13 trans). (G) The CMV-pp65/HLA-A2–specific clone HRN 3 recognized EBV-LCLs that did not share 1 particular allo-HLA molecule. To investigate whether this reactivity was based on allo-HLA recognition, the clone was tested against 4 of the recognized EBV-LCLs with and without blocking mAb directed against HLA class I. Experiments were performed in duplicate, mean values are shown ± SD.
Allo-HLA reactivity of virus-specific T-cell clones. Forty-one virus-specific T-cell clones, of which 7 are shown in this figure, were stimulated with a panel of EBV-LCLs for 18 hours, and IFNγ production was measured by ELISA. (A) The EBV-EBNA3A/HLA-A3–specific T-cell clone 1 exhibited alloreactivity against all EBV-LCLs expressing HLA-A*3101 and 1 EBV-LCL expressing HLA-A*3001. To confirm allo–HLA-A*3101 reactivity, clone 1 was tested against K562 cells (K562), K562 cells transduced with HLA-A*0201 (K562+A2), K562 cells transfected with HLA-A*3101 (K562+A31), and HLA-A*3101–negative (A31neg LCL) and HLA-A*3101–positive (A31+LCL) EBV-LCLs. To confirm the reactivity against HLA-A*3001, the clone was tested against 3 HLA-A*3001+ EBV-LCLs, of which 1 is shown, and 1 HLA-A*3101+ EBV-LCLs with or without blocking mAbs directed against HLA class I, HLA-A30/A31, and HLA-A2. (B) The CMV-pp50/HLA-A1–specific T-cell clone 24 exhibited alloreactivity against all HLA-A*1101–expressing EBV-LCLs. To confirm allo–HLA-A*1101 reactivity, clone 24 was tested against K562 cells (K562), K562 cells transduced with HLA-A*0201 (K562+A2), K562 cells transduced with HLA-A*1101 (K562+A11), and HLA-A*1101–positive EBV-LCLs (A11+LCL). (C) The EBV-BRLF1/HLA-A3–specific clone 19 exerted alloreactivity against all HLA-A0201+ EBV-LCLs. This T-cell clone did not recognize K562 transduced with HLA-A*0201 (K562+A2). To confirm allo–HLA-A*0201 reactivity, clone 19 was tested against untransduced HLA-A*0201–negative EBV-LCLs (HLA-A2neg LCL) or transduced with HLA-A*0201 (A2 trans), and HLA-A*0201–positive EBV-LCLs (A2+LCL). To investigate whether this clone recognized an EBV-derived peptide in the context of HLA-A*0201, the clone was tested against HLA-A*0201+ PHA blasts and HLA-A*0201–positive and –negative EBV-LCLs as controls. (D) The CMV-pp65/HLA-B7–specific T-cell clone 11 exhibited reactivity against all 3 HLA-DRB1*0801+ EBV-LCLs. To confirm allo–HLA-DRB1*0801 reactivity, clone 11 was tested against the 3 HLA-DRB1*0801+ EBV-LCLs, of which 1 is shown, in the presence of either no blocking mAbs (no block) or anti–HLA class I (class I), anti–HLA class II (class II), anti–HLA-DR (DR), anti–HLA-DR (DQ), and anti–HLA-DR (DP) blocking mAbs. (E) The EBV-EBNA3A/HLA-B8–specific T-cell clone 9 exhibited alloreactivity against all EBV-LCLs expressing either HLA-B*4402 or HLA-B*5501. To confirm HLA-B*4402 and HLA-B*5501 cross-reactivity, clone 9 was tested against K562 cells (K562), or K562 cells transfected with HLA-B*4402 (K562+B4402) or HLA-B*5501 (K562+B5501). As controls, clone 9 was tested against HLA-B*4402– and HLA-B*5501–negative EBV-LCLs (neg LCL), HLA-B*4402+ EBV-LCLs (B4402+LCL), HLA-B*5501+ EBV-LCLs (B5501+LCL), or HLA-B*0801+ K562 loaded with viral peptide (K562+B8+pept). (F) The Flu-HA/HLA-DR4–specific clone 5 recognized all HLA-DRB1*1301+ EBV-LCLs. To confirm allo–HLA-DRB1*1301 reactivity, clone 5 was tested against HLA-DRB1*1301+ EBV-LCLs (FAQ DR13+ and IZA DR13+) as well as HLA-DR13–negative EBV-LCLs nontransduced (DR13 neg) or transduced with HLA-DRB1*1301 (DR13 trans). (G) The CMV-pp65/HLA-A2–specific clone HRN 3 recognized EBV-LCLs that did not share 1 particular allo-HLA molecule. To investigate whether this reactivity was based on allo-HLA recognition, the clone was tested against 4 of the recognized EBV-LCLs with and without blocking mAb directed against HLA class I. Experiments were performed in duplicate, mean values are shown ± SD.
In addition to CD8+ T-cell lines and clones, we also analyzed the cross-reactive potential of CD4+ T-cell clones to allo-HLA molecules. The alloreactivity of a Flu-HA/HLA-DR4–specific T-cell clone demonstrated to be directed against HLA-DRB1*1301. As shown in Figure 2F, both HLA-DRB1*1301–positive EBV-LCLs present in the panel were efficiently recognized by this T-cell clone. Allo–HLA-DRB1*1301 reactivity of the T-cell clone could be confirmed because the T cells recognized EBV-LCLs transduced with HLA-DRB1*1301, whereas nontransduced EBV-LCLs were not recognized. In addition, allo-HLA reactivity was demonstrated for 2 other CD4+ T-cell clones of the 5 CD4+ T-cell clones tested, indicating that virus-specific CD4+ T cells also exert allo-HLA reactivity (Table 3).
For 5 of the 18 allo-HLA–reactive virus-specific T-cell clones, the recognized HLA molecules could not be determined because the recognized EBV-LCLs did not share 1 particular allo-HLA molecule. The alloreactivity of 1 of these clones, CMV-pp65/HLA-A2–specific clone HRN 3, is shown in Figure 2G. We hypothesize that this recognition is mediated by the recognition of several HLA molecules because the reactivity could be blocked by mAb specific for HLA class I.
The results of the allo-HLA reactivity exerted by the virus-specific T-cell clones are summarized in Table 3, and demonstrate that approximately 45% of the virus-specific memory CD4 and CD8 T-cell clones exhibit cross-reactivity to allo-HLA molecules. The cross-reactivity of the CD8 and CD4 T-cell clones was directed primarily against HLA class I and II, respectively. However, cross-reactivity of CD8 T cells directed against HLA class II was also observed.
Different allo-HLA recognition by T-cell clones with the same specificity but different TCR usage
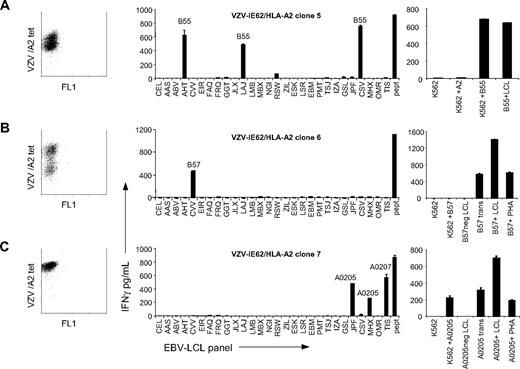
Burrows et al showed that EBV-EBNA3A–specific HLA-B8–restricted T-cell clones, derived from different HLA-B44–negative persons, were all alloreactive against HLA-B44.22,23 It could therefore be suggested that allo-HLA reactivity of virus-specific T cells can be predicted. However, the EBV-EBNA3A response in HLA-B8+, HLA-B44− persons is a very homogeneous response in which all T cells express an almost identical public TCR.34 This is in contrast to most virus responses, which are usually oligoclonal and different among persons16,35 (Table 3). To assess whether virus-specific T cells sharing the same antigen specificity but expressing different TCRs exert the same allo-HLA reactivity, we tested the alloreactivity of 3 T-cell clones derived from the same person, all specific for a peptide of the IE62 protein of VZV presented in HLA-A*0201 but with different TCR usage. As demonstrated in Figure 3A-C, all 3 VZV-specific T-cell clones recognized different allo-HLA molecules. Clone 5 showed alloreactivity against HLA-B*5501, clone 6 was alloreactive against HLA-B*5701, and clone 7 exhibited allo–HLA-A*0205 and allo–HLA-A*0207 reactivity. The recognition of the 3 clones together was comparable with the recognition exerted by the VZV-IE62–specific line derived from the same person. This confirms that alloreactivity exerted by the virus-specific lines shown in Figure 1 is the sum of the alloreactivity of the various clonal populations present within the lines. The allo-HLA reactivities of the T-cell clones were confirmed by transduction of the allo-HLA molecules in K562 or in nonrecognized EBV-LCLs. Clones 5 and 7 recognized K562 transduced with HLA-B*5501 and HLA-*0205, respectively. Clone 6 was unable to recognize K562 transduced with HLA-B*5701, whereas EBV-LCLs transduced with HLA-B*5701 were recognized. Because this clone also recognized HLA-B*5701–expressing PHA-stimulated T-cell blasts, specificity against EBV-derived peptide in the context of allo–HLA-B*5701 is excluded. These results demonstrate that virus-specific T cells with the same antigen specificity, but with different TCR complexes, can exert alloreactivity against different HLA molecules. Because T-cell responses against viruses are usually oligoclonal and different among people, these results indicate that allo-HLA reactivity cannot be predicted.
Variable allo-HLA recognition by T-cell clones with the same specificity but different TCR Vβ usage. To investigate whether virus-specific T cells sharing the same antigen specificity but expressing different TCRs exert the same allo-HLA reactivity, 3 VZV-IE62/HLA-A2–specific T-cell clones expressing different TCRs were stimulated for 18 hours with a panel of EBV-LCLs and IFNγ production was measured by ELISA. (A) VZV clone 5 showed alloreactivity against all HLA-B*5501+ EBV-LCLs. To confirm allo–HLA-B*5501 reactivity, the clone was tested against K562 cells (K562), K562 cells transduced with HLA-A*0201 (K562+A2), K562 cells transduced with HLA-B*5501 (K562+B55), and HLA-B55+ EBV-LCLs (B55+LCL). (B) VZV clone 6 was alloreactive against the HLA-B*5701+ EBV-LCLs. Clone 6 did not show reactivity against K562 cells transduced with HLA-B*5701 (K562+B57). To confirm allo–HLA-B*5701 reactivity, clone 6 was tested against HLA-B*5701–negative EBV-LCLs (HLA-B57neg LCL) or transduced with HLA-B*5701 (B57 trans), HLA-B*5701+ EBV-LCLs (B57+LCL), and HLA-B*5701+ PHA blasts (B57+PHA). (C) VZV clone 7 exhibited cross-reactivity against all HLA-A*0205+ and HLA-A*0207+ EBV-LCLs. Allo–HLA-A*0205 reactivity was confirmed by testing the clone against K562 cells (K562), K562 transduced with HLA-HLA-A*0205 (K562+HLA-A0205), HLA-A*0205–negative EBV-LCLs (HLA-A0205neg LCL), or these EBV-LCLs transduced with HLA-A*0205 (A0205 trans), HLA-A*0205+ EBV-LCLs (A0205+LCL), and HLA-A*0205+ PHA blasts (A0205+PHA). The results demonstrate that virus-specific T cells with the same antigen specificity, but with different TCR usage, exert alloreactivity against different HLA molecules. Experiments were performed in duplicate, mean values are shown ± SD.
Variable allo-HLA recognition by T-cell clones with the same specificity but different TCR Vβ usage. To investigate whether virus-specific T cells sharing the same antigen specificity but expressing different TCRs exert the same allo-HLA reactivity, 3 VZV-IE62/HLA-A2–specific T-cell clones expressing different TCRs were stimulated for 18 hours with a panel of EBV-LCLs and IFNγ production was measured by ELISA. (A) VZV clone 5 showed alloreactivity against all HLA-B*5501+ EBV-LCLs. To confirm allo–HLA-B*5501 reactivity, the clone was tested against K562 cells (K562), K562 cells transduced with HLA-A*0201 (K562+A2), K562 cells transduced with HLA-B*5501 (K562+B55), and HLA-B55+ EBV-LCLs (B55+LCL). (B) VZV clone 6 was alloreactive against the HLA-B*5701+ EBV-LCLs. Clone 6 did not show reactivity against K562 cells transduced with HLA-B*5701 (K562+B57). To confirm allo–HLA-B*5701 reactivity, clone 6 was tested against HLA-B*5701–negative EBV-LCLs (HLA-B57neg LCL) or transduced with HLA-B*5701 (B57 trans), HLA-B*5701+ EBV-LCLs (B57+LCL), and HLA-B*5701+ PHA blasts (B57+PHA). (C) VZV clone 7 exhibited cross-reactivity against all HLA-A*0205+ and HLA-A*0207+ EBV-LCLs. Allo–HLA-A*0205 reactivity was confirmed by testing the clone against K562 cells (K562), K562 transduced with HLA-HLA-A*0205 (K562+HLA-A0205), HLA-A*0205–negative EBV-LCLs (HLA-A0205neg LCL), or these EBV-LCLs transduced with HLA-A*0205 (A0205 trans), HLA-A*0205+ EBV-LCLs (A0205+LCL), and HLA-A*0205+ PHA blasts (A0205+PHA). The results demonstrate that virus-specific T cells with the same antigen specificity, but with different TCR usage, exert alloreactivity against different HLA molecules. Experiments were performed in duplicate, mean values are shown ± SD.
The cytotoxic potential and affinity of the alloreactivity exerted by virus-specific T cells
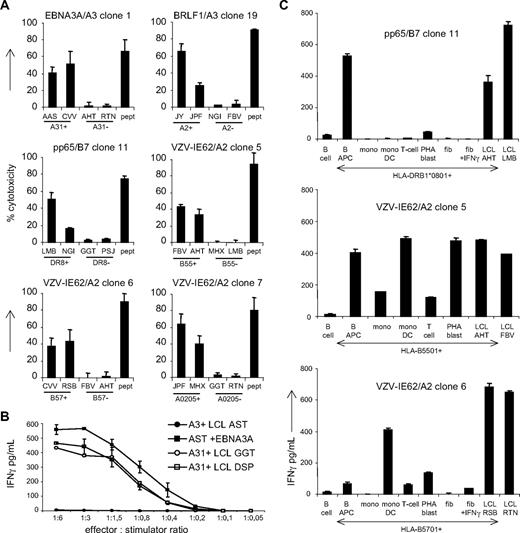
Because cytotoxicity may be a relevant measure to predict the potency of the virus-specific T cells to induce GVHD or graft rejection in vivo, we investigated the allo-HLA–reactive cytotoxic capacity of the virus-specific T cells. Six virus-specific T-cell clones were tested against a panel of EBV-LCLs positive and negative for the recognized allo-HLA molecules. As shown in Figure 4A, all 6 T-cell clones tested showed cytotoxicity against the allo-HLA–expressing EBV-LCLs.
The potency of the alloreactivity exerted by virus-specific T cells. (A) To investigate the allo-HLA–reactive cytotoxic capacity of virus-specific T cells, 6 virus-specific cell clones were tested in cytotoxicity assays against 2 EBV-LCLs expressing the recognized allo-HLA molecules and 2 EBV-LCLs negative for the allo-HLA molecules. EBV-LCLs expressing the virus-specific restriction molecule were loaded with the viral peptide and used as positive control for cytotoxicity. (B) To compare the affinities of the allo-HLA–reactive response and the virus-specific response, the allo–HLA-A30/A31–reactive EBV-EBNA3A/HLA-A3–specific clone 19 was tested against the HLA-A*0301+ EBV-LCL AST transduced with a retrovirus encoding for EBNA3A and against 2 HLA-A*3101+ EBV-LCLs GGT and DSP. To compare the kinetics of the 2 responses, the clone was tested against the EBV-LCLs in different effector-stimulator ratios. (C) To extrapolate the results obtained with the EBV-LCLs and K562 cells to the recognition of normal cell subsets in vivo, we tested virus-specific T-cell clones against allo-HLA–expressing B cells, CD40 ligand–activated B cells (B APC), T cells, PHA blasts, monocytes, monocyte-derived DCs, and fibroblasts with and without IFNγ pretreatment. Experiments were performed in duplicate, mean values are shown ± SD.
The potency of the alloreactivity exerted by virus-specific T cells. (A) To investigate the allo-HLA–reactive cytotoxic capacity of virus-specific T cells, 6 virus-specific cell clones were tested in cytotoxicity assays against 2 EBV-LCLs expressing the recognized allo-HLA molecules and 2 EBV-LCLs negative for the allo-HLA molecules. EBV-LCLs expressing the virus-specific restriction molecule were loaded with the viral peptide and used as positive control for cytotoxicity. (B) To compare the affinities of the allo-HLA–reactive response and the virus-specific response, the allo–HLA-A30/A31–reactive EBV-EBNA3A/HLA-A3–specific clone 19 was tested against the HLA-A*0301+ EBV-LCL AST transduced with a retrovirus encoding for EBNA3A and against 2 HLA-A*3101+ EBV-LCLs GGT and DSP. To compare the kinetics of the 2 responses, the clone was tested against the EBV-LCLs in different effector-stimulator ratios. (C) To extrapolate the results obtained with the EBV-LCLs and K562 cells to the recognition of normal cell subsets in vivo, we tested virus-specific T-cell clones against allo-HLA–expressing B cells, CD40 ligand–activated B cells (B APC), T cells, PHA blasts, monocytes, monocyte-derived DCs, and fibroblasts with and without IFNγ pretreatment. Experiments were performed in duplicate, mean values are shown ± SD.
To investigate whether the affinity of the allo-HLA–reactive response was comparable with the affinity of the virus-specific response, the kinetics of recognition and antigen threshold were tested for both specificities. For this purpose, the allo–HLA-A*3101/A*3001 reactive EBV-EBNA3A/HLA-A3–specific clone 19 was tested against HLA-A*0301+ EBV-LCL AST transduced with a retrovirus encoding for EBNA3A and against 2 HLA-A*3101+ EBV-LCLs, GGT and DSP at different effector-stimulator ratios. As shown in Figure 4B, the T-cell clone produced comparable amounts of IFNγ against the virus antigen–expressing EBV-LCLs as against the allo-HLA expressing–EBV-LCLs in the different effector-stimulator ratios, indicating that the kinetics of recognition and antigen threshold of the alloreactive response and the virus-specific T-cell response are comparable.
Normal cell subsets are recognized by virus-specific T cells
To extrapolate the results obtained with the EBV-LCLs and K562 cells to the recognition of normal cell subsets in vivo, we tested virus-specific T-cell clones against allo-HLA–expressing B cells, CD40L-activated B cells, T cells, PHA blasts, monocytes, monocyte-derived DCs, and fibroblasts with and without IFNγ pretreatment. The results shown in Figure 4C demonstrate the reactivity of 3 virus-specific T-cell clones directed against the different cell subsets. Pp65/HLA-B*0702–specific clone 11 showed high recognition of HLA-DRB1*0801–positive CD40L-activated B cells and low recognition of B cells and PHA blasts. VZV-IE62/HLA-A*0201–specific clone 5 showed high recognition of HLA-B*5501–positive CD40L-activated B cells, DCs, and PHA blasts and low recognition of monocytes and T cells. This clone could unfortunately not be tested against fibroblasts because HLA-B*5501–positive fibroblasts were not available. VZV-IE62/HLA-A*0201–specific clone 6 highly recognized HLA-B*5701–positive DCs and showed low reactivity against CD40L-activated B cells, T cells, PHA blasts, and IFNγ-stimulated fibroblasts. These results indicate that virus-specific T cells can also be reactive against in vivo relevant normal cell subsets.
One TCR complex mediates both virus specificity and allo-HLA reactivity
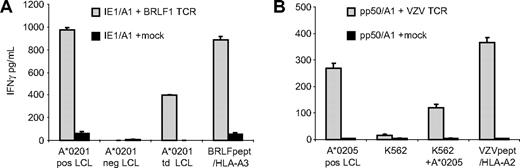
Because alloreactivity mediated by T cells may be explained by T cells expressing 2 TCR complexes at the cell surface,36,37 we wanted to exclude that the allo-HLA reactivity was mediated via another TCR than the virus-specific TCR. For this purpose, we determined the TCR usage of 2 representative allo-HLA–reactive virus-specific clones, the allo–HLA-A*0201–reactive BRLF1/HLA-A*0301–specific clone 19 (Figure 2C) and the allo–HLA-A*0205–reactive VZV-IE62/HLA-A*0201–specific clone 7 (Figure 3C). By reverse transcriptase–polymerase chain reaction, we established that the BRLF1/HLA-A*0301–specific clone 19 expressed 1 TCRβ gene transcript, BV7S2, and 2 TCRα transcripts, AV12S1 and AV18S1. However, 1 of the TCRα chains, AV12S1, contained a stop codon in the CDR3 region, indicating that this TCRα was not expressed. Flow cytometric analysis confirmed that 100% of the T cells expressed TCR BV7S2 at the cell surface (data not shown). No antibodies were available for analysis of the specific TCRα chain expression at the cell surface. To investigate whether the BV7S2 and the AV18S1 mediated the dual recognition, IE1/HLA-A1–specific T cells were transduced with retroviral vectors encoding for these TCRα and TCRβ chains. The results shown in Figure 5A demonstrate that the BRLF1-TCR–transduced T cells exerted reactivity against HLA-A*0201–expressing target cells as well as against EBV-BRFL1 peptide–loaded HLA-A*0301+ target cells. No reactivity directed against peptide-loaded or HLA-A*0201–expressing target cells was observed with mock-transduced T cells.
One TCR complex mediates both virus specificity and allo-HLA reactivity. To exclude that allo-HLA reactivity was mediated via another TCR than the virus-specific TCR, the TCR of 2 representative clones was transferred to T cells with a different specificity. (A) IE1/A1-specific T cells transduced with viral vectors encoding for the TCR of BRLF1/A3-specific clone 19 (IE1/A1+BRLF1 TCR) and IE1/A1-specific T cells transduced with a mock viral vector (IE1/A1+mock) were tested for allo–HLA-A*0201 reactivity against HLA-A*0201–positive EBV-LCLs (A*0201 pos LCLs), HLA-A*0201–negative EBV-LCLs (A*0201 neg LCLs), and HLA-A*0201–negative EBV-LCLs transduced with HLA-A*0201 (A*0201 td LCLs) and for BRLF1 specificity against BRLF1 peptide–loaded HLA-A*0301–positive EBV-LCLs (BRLF1pept /HLA-A3). (B) Pp50/A1-specific T cells transduced with viral vectors encoding for the TCR of VZV clone 7 (pp50/A1+VZV TCR) and pp50-specific T cells transduced with a mock viral vector (pp50/A1+mock) were tested for allo–HLA-A*0205 reactivity against HLA-A*0205–positive EBV-LCLs (A*0205 pos LCLs), K562 cells (K562), and K562 cells transduced with HLA-A*0205 (K562+A*0205) and for VZV specificity against VZV-IE62 peptide–loaded HLA-A*0201–positive EBV-LCLs (VZVpept/HLA-A2). The results demonstrate that virus specificity and allo-HLA reactivity exerted by virus-specific T cells were mediated by 1 TCR complex. Experiments are shown in duplicate, mean values are shown ± SD.
One TCR complex mediates both virus specificity and allo-HLA reactivity. To exclude that allo-HLA reactivity was mediated via another TCR than the virus-specific TCR, the TCR of 2 representative clones was transferred to T cells with a different specificity. (A) IE1/A1-specific T cells transduced with viral vectors encoding for the TCR of BRLF1/A3-specific clone 19 (IE1/A1+BRLF1 TCR) and IE1/A1-specific T cells transduced with a mock viral vector (IE1/A1+mock) were tested for allo–HLA-A*0201 reactivity against HLA-A*0201–positive EBV-LCLs (A*0201 pos LCLs), HLA-A*0201–negative EBV-LCLs (A*0201 neg LCLs), and HLA-A*0201–negative EBV-LCLs transduced with HLA-A*0201 (A*0201 td LCLs) and for BRLF1 specificity against BRLF1 peptide–loaded HLA-A*0301–positive EBV-LCLs (BRLF1pept /HLA-A3). (B) Pp50/A1-specific T cells transduced with viral vectors encoding for the TCR of VZV clone 7 (pp50/A1+VZV TCR) and pp50-specific T cells transduced with a mock viral vector (pp50/A1+mock) were tested for allo–HLA-A*0205 reactivity against HLA-A*0205–positive EBV-LCLs (A*0205 pos LCLs), K562 cells (K562), and K562 cells transduced with HLA-A*0205 (K562+A*0205) and for VZV specificity against VZV-IE62 peptide–loaded HLA-A*0201–positive EBV-LCLs (VZVpept/HLA-A2). The results demonstrate that virus specificity and allo-HLA reactivity exerted by virus-specific T cells were mediated by 1 TCR complex. Experiments are shown in duplicate, mean values are shown ± SD.
The VZV-IE62/HLA-A*0201 clone 7 expressed 1 TCRα transcript, AV6S1, and 1 TCRβ transcript, BV21S3. Pp50/A1-specific T cells were transduced with retroviral vectors encoding for the VZV TCR chains. The results shown in Figure 5B demonstrate that the VZV-TCR–transduced T cells exerted reactivity against HLA-A*0205–expressing target cells as well as against VZV-IE62 peptide–loaded HLA-A*0201+ target cells. No reactivity directed against peptide-loaded or HLA-A*0205–expressing target cells was observed with mock-transduced T cells. These results demonstrate that the virus specificity and the allo-HLA reactivity exerted by these virus-specific T cells were mediated via 1 TCR complex.
Discussion
In this study, we demonstrated that a high percentage of virus-specific memory T cells exhibits cross-reactivity against allogeneic HLA molecules. CD8 as well as CD4 virus-specific memory T cells were demonstrated to have allo-HLA–reactive potential. In addition, we determined that the alloreactivity exerted by CD8 T cells was directed against either HLA class I or HLA class II molecules, and that the alloreactivity of the T cells was mediated by cytotoxicity and cytokine production. Furthermore, we demonstrate that virus-specific T cells can exert allo-HLA reactivity against normal cell subsets, indicating the potential clinical relevance of the response. By TCR transfer, we confirmed that the allo-HLA reactivity and virus specificity were mediated via the same TCR.
Most virus-specific T-cell lines and 45% of virus-specific T-cell clones directed against EBV, CMV, VZV, and Flu exerted alloreactivity when tested against a panel of EBV-LCLs covering almost all common HLA molecules. The cross-reactivity exerted by the virus-specific T cells was confirmed to be based on allo-HLA recognition by testing the T-cell clones against K562 cells and EBV-LCLs transduced with single HLA molecules. Some of the alloreactive virus-specific T-cell clones did not recognize K562 cells transduced with the specific allo-HLA molecules, but showed reactivity against EBV-LCLs transduced with the allo-HLA molecules. These data support the previous findings that allo-HLA reactivity is dependent on endogenous peptide.38 Allo-HLA cross-reactivity was directed not only against EBV-LCLs but also against PHA-stimulated T cells (Figure 3B), indicating that the peptides responsible for allo-HLA reactivity were not EBV derived. Differential recognition of HLA-transduced K562 cells and EBV-LCLs may indicate recognition of tissue-specific peptides in allo-HLA molecules. Therefore, it may be possible that we even underestimated the allo-HLA–reactive repertoire of T cells by initially screening only against an EBV-LCL panel.
Burrows et al showed that EBV-EBNA3A–specific HLA-B8–restricted T cells derived from different HLA-B44–negative persons all exert cross-reactivity against allo–HLA-B44.22,23 Based on these findings, it could be suggested that the allo-HLA reactivity of virus-specific T cells can be predicted. The EBV-EBNA3A–specific T cells, however, express an almost identical public TCR in all HLA-B8+ HLA-B44− persons,34 whereas most other virus responses are oligoclonal and the TCR usage of T cells directed against the same viral epitope is variable between persons16,35 (Table 3). We have demonstrated that virus-specific T cells with the same antigen specificity, but expressing different TCRs, exhibit cross-reactivity against different HLA molecules (Figure 3). In addition, we have shown that 3 specific T-cell lines with the same specificity for CMV-pp50, but derived from different persons, exerted a very variable pattern of allo-HLA reactivity, ranging from no allo-HLA reactivity to very broad alloreactivity (Figure 1). These results together illustrate that the cross-reactive potential of antigen-specific T cells against allo-HLA molecules is difficult to predict.
The alloreactivity exerted by the virus-specific memory CD8 and CD4 T cells was directed primarily against allo-HLA class I and II molecules, respectively, suggesting that the coreceptors expressed by the T cells contributed to the affinity of the allo-HLA reactivity. However, we also demonstrated allo-HLA class II recognition by a small proportion of antigen-specific CD8 T cells, as was also shown recently by Rist et al.39 HLA class II allorecognition by CD8+ T cells could indicate an HLA class II–TCR interaction that is independent of CD8 coreceptor binding. It is, however, also possible that CD8 coreceptors bind to HLA class I molecules expressed on the target cell and thereby strengthen the TCR–HLA class II interaction, as was previously shown.27 Although we did not observe HLA class I cross-reactive CD4 T cells, only a limited number of CD4 T-cell clones were tested, and therefore we cannot exclude that CD4 T cells may also cross-react with HLA class I complexes.
The results of our study illustrate that approximately 45% of all T cells exert allo-HLA cross-reactivity. However, because T cells were analyzed only for allo-HLA cross-reactivity against an EBV-LCL panel expressing most common HLA molecules, missing all infrequent HLA molecules as well as all tissue-specific peptides presented in allo-HLA molecules, we speculate that virtually all T cells may be allo-HLA reactive. Based on this assumption, and the fact that the TCR repertoire of humans is highly diverse, after HLA-mismatched transplantations sufficient allo-mismatched HLA cross-reactive T cells are likely to be present to induce acute GVHD or graft rejection. However, HLA-mismatched stem cell transplantation or solid organ transplantations do not always lead to acute GVHD or graft rejection, indicating that other factors must be involved in these transplantation-related complications. A range of acute viral infections have been linked to initiating GVHD and graft rejection after transplantation,26 suggesting that virus-specific T cells may be mediators of GVHD and graft rejection. Because virus-specific T-cell responses usually have a restricted TCR usage, high numbers of T cells expressing an identical TCR can be found. In addition, herpes virus–specific T-cell populations can remain present at high percentages for long periods of time in healthy persons as well as in patients,18-21 and viral infections, leading to expansion of virus-specific T cells, are very common after HLA-mismatched SCT or solid organ transplantation.40-43 Therefore, given the high proportion of virus-specific T cells and the less stringent requirements for activation of memory T cells,12,13 it is tempting to speculate that if the HLA type of patient or the transplanted organ matches the cross-reactivity of the virus-specific T cells, these allo-HLA–reactive virus-specific memory T cells may easily induce GVHD or graft rejection.
The ability of virus-specific T cells to exert allo-HLA reactivity may also have implications for the clinical applicability of virus-specific T-cell lines. Because immune deficiency for viruses is a common complication after SCT, broad administration of virus-specific T cells lines over HLA barriers to SCT patients has been proposed.44,45 The results of this study demonstrate that administration of virus-specific T cells over HLA barriers may increase risk of GVHD, and indicate that virus-specific lines should be tested for alloreactivity against the patient before administration.
Based on our results, we postulate that virtually all antigen-specific T cells will be cross-reactive against allo-HLA class I or II molecules. The high alloreactive potential of particularly virus-specific memory T cells may have important clinical implications in transplantation settings.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by grant LSBR0611 from the Landsteiner Foundation for Blood Transfusion Research.
Authorship
Contribution: A.L.A. performed research and wrote the paper; L.J.A.D. performed research; D.L.R. contributed to research design; M.M.L. helped in generation of EBV-LCL panel; R.S.H. generated TCR gene constructs; R.B. helped in generation of EBV-LCL panel and transduced the TCR gene constructs; M.A.W.G.H. generated virus-specific clones; M.G.D.K. generated tetramers; I.I.N.D. contributed to research design; J.H.F.F. supervised writing; F.H.J.C. supervised research; and M.H.M.H. supervised research and writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: M. H. M. Heemskerk, Department of Hematology, Leiden University Medical Center, Albinusdreef 2, PO Box 9600, 2300 RC Leiden, The Netherlands; e-mail: m.h.m.heemskerk@lumc.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal