To the editor:
Identification of interacting proteins essential for oncogenic functions of leukemia-associated transcription factors is important for understanding the underlying transformation mechanisms and designing effective cancer therapeutics.1 We have recently found that homo-oligomerization property, but not its interaction with core-binding factor beta subunit (CBFβ), is critical for AML1-ETO–mediated transformation of primary hematopoietic cells.2 Strikingly, the conclusion on CBFβ requirement contradicts another study by Roudaia et al, which reported an essential function of CBFβ interaction for AML1-ETO activity based on an AML1-ETO double-point mutant (Y113A/T161A).3
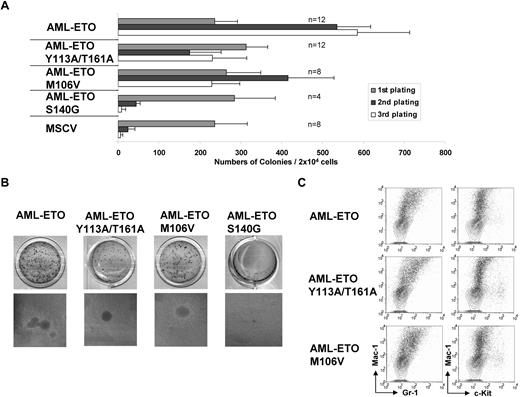
One possible explanation for these discrepancies is the use of different point mutants in these studies.4 To this end, we have generated an identical AML1-ETO Y113A/T161A mutation used in the study of Roudaia et al and compared it with our M106V point mutant in the transformation assay. In contrast to cells transduced with empty vector or the AML1-ETO DNA binding mutant that rapidly lost their proliferative capacity, cells transduced with the CBFβ defective mutants including Y113A/T161A could still form significant numbers of third and subsequent rounds of colonies in the serial replating assay (Figure 1A). Despite reduced number (an average of 12 different experiments), the resultant colonies exhibited very similar morphology and immunophenotypes as wild-type AML1-ETO transformed cells (Figure 1B-C and supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). This is in stark contrast to the results by Roudaia et al, where Y113A/T161A mutant when transduced into 5′FU-treated bone marrow cells failed to give third-round colonies. It is possible that 5′FU treatment may have depleted certain AML1-ETO target cells that are responsible for the observed phenotype in the assay using positively selected c-kit cells.5 We also note that the transformation data in Roudaia and colleagues' study had not been normalized with the number of plated cells; that is, colony number in the second and third platings were derived from 10 times more cells (104) compared with the first plating (103).3 If presented as normalized data, the results would look quite significantly different, and the difference between the wild-type and Y113A/T161A mutant would be more modest.
Analysis of transformation ability of various AML1-ETO point mutants in murine primary hematopoietic cells. (A) AML1-ETO point mutants used in the retroviral transduction/transformation assay are indicated on the left. The bar chart (right) represents the numbers of colonies after each plating in methylcellulose. Error bars show standard deviations of indicated numbers (n) of independent experiments. (B) Typical third-round colony morphology of primary transduced bone marrow cells transduced with indicated constructs. The top panel shows colonies stained with INT and the bottom panel shows unstained colonies (×10 magnification). (C) Phenotypical analysis of cells transformed by indicated constructs. Dot plots represent stainings obtained with antibodies specific for the indicated surface markers. Contour plots show unstained controls.
Analysis of transformation ability of various AML1-ETO point mutants in murine primary hematopoietic cells. (A) AML1-ETO point mutants used in the retroviral transduction/transformation assay are indicated on the left. The bar chart (right) represents the numbers of colonies after each plating in methylcellulose. Error bars show standard deviations of indicated numbers (n) of independent experiments. (B) Typical third-round colony morphology of primary transduced bone marrow cells transduced with indicated constructs. The top panel shows colonies stained with INT and the bottom panel shows unstained colonies (×10 magnification). (C) Phenotypical analysis of cells transformed by indicated constructs. Dot plots represent stainings obtained with antibodies specific for the indicated surface markers. Contour plots show unstained controls.
In addition, one must be cautious when interpreting mutagenesis data, as it is almost impossible to engineer absolutely specific mutations that will affect only a single property of the mutated protein. Thus it is critical to have an alternative approach targeting CBFβ expression without altering the structure of AML1-ETO. Consistent with our point mutant data, we further demonstrated that 2 independent shRNAs that effectively knocked down more than 95% CBFβ expression at protein level in primary hematopoietic cells did not compromise AML1-ETO–mediated transformation.2 Together, these results indicate that a significant reduction of CBFβ activity has only modest effect on AML1-ETO–mediated transformation. However, a decisive experiment to determine whether there is an absolute CBFβ dependence is to assess the behavior of AML1-ETO in a complete absence of CBFβ using genetic approaches such as conditional knockout mice that will be instrumental to this issue.
Authorship
The online version of this letter contains a data supplement.
Acknowledgments: We specially thank Mel Greaves and Jon Wilson for critical reading of the manuscript. C.W.E.S. is an Association for International Cancer Research (AICR) fellow and a European Molecular Biology Organization (EMBO) young investigator, supported in part by Leukaemia and Lymphoma Research, the Kay Kendall Leukaemia Fund, and the Childwick Trust.
Contribution: C.K. and B.B.Z. designed and performed the experiments and analyzed data; S.D. analyzed data; and C.W.E.S. designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Eric So, King's College London, Rayne Institute, 123 Coldharbour La, Denmark Hill, London SE5 9NU, United Kingdom; e-mail: eric.so@kcl.ac.uk.
References
Author notes
C.K. and B.B.Z. contributed equally to this article.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal