Abstract
HIV “controllers” are persons infected with human immunodeficiency virus, type I (HIV) who maintain long-term control of viremia without antiviral therapy and who usually do not develop the acquired immune deficiency syndrome (AIDS). In this study, we have correlated results from polychromatic flow cytometry and oligonucleotide expression arrays to characterize the mucosal immune responses of these subjects in relation to untreated HIV+ persons with high viral loads and progressive disease (“noncontrollers”). Paired peripheral blood and rectosigmoid biopsies were analyzed from 9 controllers and 11 noncontrollers. Several cellular immune parameters were found to be concordant between the 2 compartments. Compared with noncontrollers, the mucosal tissues of controllers had similar levels of effector T cells and fewer regulatory T cells (Tregs). Using principal component analysis to correlate immunologic parameters with gene expression profiles, transcripts were identified that accurately distinguished between controllers and noncontrollers. Direct 2-way comparison also revealed genes that are significantly different in their expression between controllers and noncontrollers, all of which had reduced expression in controllers. In addition to providing an approach that integrates flow cytometry datasets with transcriptional profiling analysis, these results underscore the importance of the sustained inflammatory response that attends progressive HIV disease.
Introduction
Although most untreated HIV-infected persons will eventually experience a progressive loss of CD4+ T cells, some maintain undetectable or low viral loads in the absence of antiretroviral medication and do not progress to AIDS.1 These HIV controllers represent less than 5% of infected persons and are defined by their ability to maintain circulating virus loads at undetectable or very low levels.2 Usually, but not always, controllers are also long-term nonprogressors, as defined by their ability to maintain normal CD4+ T-cell counts and to remain clinically healthy for more than 10 years.3,4 At the opposite end of the spectrum, HIV-infected noncontrollers have sustained high viral loads (> 10000 copies/mL) and usually experience disease progression, as defined by the loss of circulating CD4+ T cells and the development of AIDS.
The viral-host interactions and the correlates of immunity that prevent disease progression in most HIV controllers are poorly understood. Recent reports have shown that these persons (1) have a higher frequency of polyfunctional HIV-specific CD4+ and CD8+ T-cell responses in the peripheral blood and in gut-associated mucosal lymphoid tissues (GALT),5-7 (2) present highly potent granzyme B–mediated cytolytic CD8+ T-cell activity against HIV-infected CD4+ T cells,8 (3) display decreased expression of immunoregulatory receptors that negatively impact upon CD4+ T-cell function,9 (4) are enriched for protective class HLA haplotypes,6,10 and/or (5) harbor HIV virions with reduced fitness.11 There is, however, no individual factor that appears to be common to all controllers.
Mucosal tissues including GALT undergo severe depletion of CD4+ T cells during primary lentiviral infection.12-14 Even with viral loads suppressed by highly active antiretroviral therapy (HAART), CD4+ T cells are restored only slowly15 and often incompletely,16 and low-level HIV production can persist for years.17,18 Hence, the immune responses against HIV in the intestinal mucosal tissues could be a crucial determinant of disease progression, both at the time of acute infection and after the initiation of HAART. If so, characterizing the cellular and molecular features of the intestinal mucosa response in HIV controllers may provide insight into mechanisms of protection, pathogenesis, and treatment. Because comprehensive analysis of mucosal T-cell function of HIV subjects is logistically difficult, it would also be useful if accurate surrogate markers of such responses could be measured in the peripheral blood. The relationship between responses that can be observed in the peripheral blood and in the mucosal tissues of such patients, however, remains unclear.
To better characterize the relationship between immune responses to HIV infection in the context of nonprogressive versus progressive disease, we analyzed multiple immune parameters in paired peripheral blood and rectosigmoid biopsy samples as well as whole genome expression profiles of rectosigmoid biopsy samples from controllers and noncontrollers. Whereas transcriptional profiling has been conducted on GALT tissue from long-term nonprogressors19 and flow cytometric studies have been conducted on controllers,6 this is the first time that gene expression patterns have been correlated with immune parameters in peripheral blood and in GALT of controllers and noncontrollers. By reducing the dimensions of our flow cytometry data through principal component analysis (PCA),20 we were able to associate immune parameters with specific molecular markers that distinguish these 2 groups of subjects. This is an approach that could be applied to other disease conditions where correlating complex cellular and molecular signatures of immunity may provide new insights in the mechanisms of pathogenesis.
Methods
Research subjects
Peripheral blood mononuclear cells (PBMCs) and rectosigmoid biopsies were obtained from HIV-infected adults enrolled in the University of California, San Francisco Study on the Consequences of the Protease Inhibitor Era (SCOPE) cohort,21 an ongoing prospective cohort study aimed at investigating the long-term clinical and immunologic consequences of treated and untreated HIV disease. Twenty participants from the SCOPE cohort were selected in 2 well-defined groups of “controllers” (n = 9) and “noncontrollers” (n = 11; Table 1). Controllers are defined as untreated persons with undetectable plasma HIV RNA levels (< 75 copies/mL; n = 4, “elite controllers”) or with very low but detectable plasma HIV RNA levels (< 2000 copies/mL; n = 5, “viremic controllers”). In this group, median age was 47 years (range, 42-54 years); median CD4+ T-cell counts, 701 cells/μL (range, 518-1507 cells/μL); median plasma viral load, 77 RNA copies/mL (range, < 50 to 1957 RNA copies/mL). Noncontrollers are defined as untreated persons with plasma HIV RNA levels of more than 10000 copies/mL and CD4+ T-cell count of 200 cells/μL or more. In this group, median age was 42 years (range, 36-59 years); median CD4+ T-cell counts, 253 cells/μL (range, 196-673 cells/μL); median plasma viral load, 24 734 copies RNA/mL (range, 15 534-91 199 copies RNA/mL; Table 1). This study was approved by the University of California at San Francisco (UCSF) Committee on Human Research and all subjects gave written informed consent in accordance with the Declaration of Helsinki.
Summary of study participant characteristics
| Group/patient ID . | Age, y . | Sex . | Viral load, copies/mL . | CD4+ T-cell count, cells/μL (PB) . | RB %CD4 (relative to CD3+) . |
|---|---|---|---|---|---|
| Controllers | |||||
| 1554 | 53.9 | Male | 77 | 823 | 45.3 |
| 1139 | 46.8 | Male | 50 | 518 | 41.1 |
| 1545 | 48.4 | Male | 249 | 625 | 27.9 |
| 1518 | 47.3 | Transgender | 75 | 1507 | 59.2 |
| 1256 | 44.7 | Male | 820 | 597 | 26.2 |
| 1560 | 41.8 | Male | 1864 | 701 | 29.7 |
| 1504 | 45.8 | Male | 1957 | 633 | 25.7 |
| 1198 | 51.9 | Male | 50 | 1311 | 57.3 |
| 1529 | 47.3 | Male | 50 | 1000 | 65.5 |
| Noncontrollers | |||||
| 1292 | 38.7 | Male | 24 644 | 673 | 44.5 |
| 1246 | 58.5 | Male | 32 944 | 221 | 18 |
| 1515 | 40.8 | Male | 15 534 | 196 | 8.35 |
| 1254 | 46.4 | Transgender | 32 236 | 211 | 10.8 |
| 1217 | 37.8 | Male | 16 212 | 389 | 14.9 |
| 1265 | 45.6 | Intersex | 20 420 | 643 | 32.2 |
| 1259 | 42.2 | Male | 56 299 | 206 | 19.5 |
| 1298 | 49.8 | Male | 91 199 | 208 | 20.7 |
| 1190 | 48.8 | Male | 21 523 | 415 | 13 |
| 1094 | 41.1 | Male | 24 734 | 253 | 14.2 |
| 1080 | 36.2 | Male | 33 904 | 354 | 22.9 |
| Group/patient ID . | Age, y . | Sex . | Viral load, copies/mL . | CD4+ T-cell count, cells/μL (PB) . | RB %CD4 (relative to CD3+) . |
|---|---|---|---|---|---|
| Controllers | |||||
| 1554 | 53.9 | Male | 77 | 823 | 45.3 |
| 1139 | 46.8 | Male | 50 | 518 | 41.1 |
| 1545 | 48.4 | Male | 249 | 625 | 27.9 |
| 1518 | 47.3 | Transgender | 75 | 1507 | 59.2 |
| 1256 | 44.7 | Male | 820 | 597 | 26.2 |
| 1560 | 41.8 | Male | 1864 | 701 | 29.7 |
| 1504 | 45.8 | Male | 1957 | 633 | 25.7 |
| 1198 | 51.9 | Male | 50 | 1311 | 57.3 |
| 1529 | 47.3 | Male | 50 | 1000 | 65.5 |
| Noncontrollers | |||||
| 1292 | 38.7 | Male | 24 644 | 673 | 44.5 |
| 1246 | 58.5 | Male | 32 944 | 221 | 18 |
| 1515 | 40.8 | Male | 15 534 | 196 | 8.35 |
| 1254 | 46.4 | Transgender | 32 236 | 211 | 10.8 |
| 1217 | 37.8 | Male | 16 212 | 389 | 14.9 |
| 1265 | 45.6 | Intersex | 20 420 | 643 | 32.2 |
| 1259 | 42.2 | Male | 56 299 | 206 | 19.5 |
| 1298 | 49.8 | Male | 91 199 | 208 | 20.7 |
| 1190 | 48.8 | Male | 21 523 | 415 | 13 |
| 1094 | 41.1 | Male | 24 734 | 253 | 14.2 |
| 1080 | 36.2 | Male | 33 904 | 354 | 22.9 |
PB indicates peripheral blood; and RB, rectosigmoid biopsies.
Tissue collection and processing
All HIV-infected participants except one (1139) provided a 20-mL blood draw for purification of PBMCs. All HIV-infected participants underwent flexible sigmoidoscopy. The sigmoidoscope was advanced to the rectosigmoid region, and 30 mucosal biopsies were obtained in a circumferential fashion at approximately 15 to 20 cm from the anal verge. Focal areas with visible evidence of inflammation were avoided. For each participant, 4 biopsy pieces were placed in RNAlater (Ambion) for RNA extraction, and 20 biopsy specimens were placed in R-10 medium and transported within 2 hours to the laboratory for immediate processing. A suspension of mucosal cells from rectosigmoid biopsies was obtained after 2 successive 0.25-mg/mL collagenase type II treatments (Sigma-Aldrich), as previously described.22
Measurement of cellular parameters by flow cytometry
Panels of antibodies used for definition of phenotype, intracellular cytokine responses, and proliferation are described in supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article. Intracellular cytokine responses were performed in vitro after stimulation for 8 to 12 hours in the presence of media (mock), Staphylococcus aureus enterotoxin B (SEB, 1 μg/mL), phorbol myristyl acetate (PMA, 10 ng/mL), and ionomycin (1 μg/mL), or 15-mer overlapping peptide pools from HIV Gag, Env, and Nef (at 2 μg/mL) and from cytomegalovirus (CMV) IE-1 and pp65 (at 0.5 μg/mL), or heat-killed Candida albicans (105 cells per well). Proliferative responses were tracked in vitro with the intracellular dye, carboxyfluorescein succinimidyl ester (CFSE; Molecular Probes) after 8 days in culture. HIV peptide pools were from the National Institutes of Health (NIH) AIDS Repository and included HIV-1 Consensus Subtype B 15-mer peptides from Env, Nef, and Gag. CMV peptide pools (originated from JPT Peptide Technologies GmbH) included PepMix IE-1 and PepMix pp65. Results in CD4+ or CD8+ T cells from PBMCs or from rectosigmoid biopsies are reported as background-subtracted values from the unstimulated cell population from each patient and for each stimulus. Fluorescence-activated cell sorting analysis was performed on a 4-laser BD LSR-II flow cytometer (BD Biosciences). Data were analyzed using FlowJo software Version 6-8 (Treestar), and then transferred into analysis and graphic software including Excel (Windows; Microsoft), StatView (Abacus Concepts), SPICE (NIH), and/or Graphpad Prism4 (GraphPad Software).
Expression profiling of rectosigmoid biopsy samples
Total RNA was extracted from the rectosigmoid biopsies using the TRIzol Reagent (Invitrogen) and the Purelink RNA purification system (Invitrogen), and then amplified for microarray analysis using the Amino Allyl MessageAmp II aRNA Amplification Kit (Ambion). An RNA reference consisting of an equal quantity of amplified RNA from the colon of an uninfected healthy person (obtained during surgery) was labeled with cyanin 3 dye and hybridized against the cyanin 5–labeled experimental samples on Human Exonic Evidence-Based Oligonucleotide (HEEBO, http://www.microarray.org/sfgf/heebo.do; Invitrogen) microarrays printed in-house at the UCSF Center for Advanced Technologies. Arrays were scanned using a GenePix 4000B scanner and GenePix PRO Version 4.1 (Axon Instruments/Molecular Devices). The Spotreader program (Niles Scientific) was used for array gridding and image analysis. The resulting files were uploaded to Acuity Version 4.0 (Molecular Devices), where the raw data were log transformed and filtered for retention of spots that were of high quality and for removal of nonhuman control spots. All microarray data can be found at the Gene Expression Omnibus (GEO) database under accession number GSE20075.23
Multiparameter data analysis
Using the program Cluster (http://rana.lbl.gov/EisenSoftware.htm), cellular parameters and cytokine patterns measured by flow cytometry were organized using a hierarchic clustering algorithm based on Pearson correlative metric, with average linkage clustering. The values for these parameters were median-centered and normalized before average linkage clustering analysis for the parameters and persons. The results were then visualized using the program Treeview (http://rana.lbl.gov/EisenSoftware.htm). PCA was performed on the cellular parameters using the program Cluster to generate eigenvalues, and the resulting datasets were further compiled and plotted on Excel. The filtered expression profiling data (microarray spots with data in at least 80% of the arrays) from the HEEBO arrays underwent unsupervised hierarchic clustering analysis and were then visualized using Treeview. Filtered datasets were also analyzed for statistically significant genes using the Significance Analysis of Microarrays (SAM) software Version 2.23A (http://www-stat.stanford.edu/∼tibs/SAM/). Using SAM, quantitative regression analysis with PC1 eigenvalues or pairwise comparisons was performed between HIV controllers and noncontrollers. SAM was used separately with a false discovery rate (FDR) cutoff of 0% and less than 5%. Gene ontology and pathway analyses were performed using Protein Analysis Through Evolutionary Relationships (PANTHER; http://www.pantherdb.org/).24 Gene lists were input into PANTHER along with a background gene list representing all of the genes present in the human genome. For real-time polymerase chain reaction (PCR) analysis, 1 μg of RNA from each sample was reverse transcribed and the resulting cDNA was used in quantitative real-time PCR reactions, with SYBR green labeling using primers designed in-house that span introns to avoid amplification of genomic DNA. All values were normalized to β-actin values.
Results
Identification of immunologic parameters and transcriptional profiles from the peripheral blood and rectosigmoid mucosal biopsies of controllers and noncontrollers
A comprehensive analysis of immunologic parameters was performed using polychromatic flow cytometry on cells from the peripheral blood and from rectosigmoid biopsy tissue of 9 controllers and 11 noncontrollers (Table 1). The flow cytometry panels used for this analysis are shown in supplemental Table 1, and representative gating strategies are presented in Figure 1. Gating on live CD3+CD4+ and CD3+CD8+ T cells from both peripheral blood and rectosigmoid biopsy tissue (Figure 1A), 36 independent parameters were measured, including CD3+CD4+ and CD3+CD8+ T-cell immune activation markers (Ki67+ and/or CD38+; Figure 1B left); the frequency of CD3+CD4+FoxP3+ regulatory T cells (Tregs; Figure 1B right); proliferative (Figure 1C) and cytokine response profiles (Figure 1D) of peripheral blood CD4+ and CD8+ T cells after stimulation with CMV, HIV, Candida albicans, and SEB; and the frequency of “effector” CD4+ T helper subsets producing interferon-γ–positive (IFNγ)+ (Th1), interleukin-17–positive (IL-17+; Th17), IL-2+, and/or tumor necrosis factor-α–positive (TNFα+) after in vitro stimulation with PMA and ionomycin (Figure 1E).
Flow cytometry–based immunologic profiling: representative flow plots and gating strategy. (A) Gating strategy by flow cytometry for the detection of CD4+ and CD8+ T cells (singlet [forward scatter A{FSC-A} × FSC-H diagonal] live [Aqua] CD3+ lymphocytes) extracted from rectosigmoid biopsy tissue (RB, top panels) and from peripheral blood (PB, bottom panels). (B) Detection of CD38 and Ki67 expression in CD8+ T cells (left) as well as FoxP3 and Ki67 in CD8+ and CD4+ T cells from rectosigmoid biopsy tissue (top) and from peripheral blood (bottom). (C-D) Antigen-specific CD8+ (top) CD4+ (bottom) T-cell responses in peripheral blood after stimulation with PBS (mock), CMV, HIV, Candida albicans (C alb), or SEB antigens, as measured by CFSElow proliferative responses (C) or IFNγ intracellular cytokine detection (D). (E) Detection of intracellular cytokines (IFNγ, IL-2, IL-17, or TNFα) in CD4+ T cells from rectosigmoid biopsy tissue (top) and from peripheral blood (bottom), after PMA/ionomycin stimulation in vitro. All of these analyses were carried out using cells from a representative noncontroller (1256).
Flow cytometry–based immunologic profiling: representative flow plots and gating strategy. (A) Gating strategy by flow cytometry for the detection of CD4+ and CD8+ T cells (singlet [forward scatter A{FSC-A} × FSC-H diagonal] live [Aqua] CD3+ lymphocytes) extracted from rectosigmoid biopsy tissue (RB, top panels) and from peripheral blood (PB, bottom panels). (B) Detection of CD38 and Ki67 expression in CD8+ T cells (left) as well as FoxP3 and Ki67 in CD8+ and CD4+ T cells from rectosigmoid biopsy tissue (top) and from peripheral blood (bottom). (C-D) Antigen-specific CD8+ (top) CD4+ (bottom) T-cell responses in peripheral blood after stimulation with PBS (mock), CMV, HIV, Candida albicans (C alb), or SEB antigens, as measured by CFSElow proliferative responses (C) or IFNγ intracellular cytokine detection (D). (E) Detection of intracellular cytokines (IFNγ, IL-2, IL-17, or TNFα) in CD4+ T cells from rectosigmoid biopsy tissue (top) and from peripheral blood (bottom), after PMA/ionomycin stimulation in vitro. All of these analyses were carried out using cells from a representative noncontroller (1256).
Data analysis techniques typically used for the characterization of gene expression profiles were applied to this complex set of immunologic parameters. To understand the relationship between the distinct immunologic parameters and the control of HIV replication, unsupervised hierarchic clustering analysis (with Pearson correlations) was performed comparing the median-centered, normalized parameters across all controllers and noncontrollers (Figure 2). With the exception of subject 1292 (a documented controller for approximately 30 months before losing viral control), this analysis segregated the HIV controllers and noncontrollers into 2 distinct groups based on their immune and clinical parameters (Figure 2A). High expression of immune activation markers (Ki67 and CD38) on CD4+ and CD8+ T cells from peripheral blood (PB CD4+Ki67+ and PB CD38+ CD8+) and from rectosigmoid biopsies (RB CD4+Ki67+ and RB CD38+ CD8+) clustered together with viral load (“VL num”), underscoring the known strong association between viral replication and T-cell immune activation.1,25,26
Patterns of the immune response in peripheral blood and mucosal tissue samples from HIV-infected persons. (A) Cellular parameters measured by flow cytometry and individual samples were organized using hierarchic clustering. Each row represents a single cellular parameter and each column represents a single person. Black indicates the median values of each parameter; red, a value greater than the median; green, a value lower than the median; and gray, missing data. Horizontal bars (in blue for controllers and red for noncontrollers) at the bottom of the figure indicate that the samples from controllers and noncontrollers cluster together. Vertical brackets (at the right) indicate cellular parameters that cluster together, reflecting values that are closely related to each other. (B) Representative scatter plots with linear regression analysis to confirm the association between cellular parameters measured in the peripheral blood (PB) relative to those measured in rectosigmoid biopsies (RB).
Patterns of the immune response in peripheral blood and mucosal tissue samples from HIV-infected persons. (A) Cellular parameters measured by flow cytometry and individual samples were organized using hierarchic clustering. Each row represents a single cellular parameter and each column represents a single person. Black indicates the median values of each parameter; red, a value greater than the median; green, a value lower than the median; and gray, missing data. Horizontal bars (in blue for controllers and red for noncontrollers) at the bottom of the figure indicate that the samples from controllers and noncontrollers cluster together. Vertical brackets (at the right) indicate cellular parameters that cluster together, reflecting values that are closely related to each other. (B) Representative scatter plots with linear regression analysis to confirm the association between cellular parameters measured in the peripheral blood (PB) relative to those measured in rectosigmoid biopsies (RB).
Viral load was closely related to the frequency of FoxP3+ Tregs, both in the rectosigmoid biopsies (Spearman ρ = 0.6340, P = .003) as well as in peripheral blood (Spearman ρ = 0.6544, P = .002). The strong positive relationship between the frequency of Tregs and the frequency of activated T cells suggests that Tregs can be viewed as yet another sign of generalized immune activation.
The peripheral CD4+ T-cell count was most consistently related to the frequency of cytokine-producing CD4+ T cells in rectal biopsy tissue, including those producing IFNγ+ (Th1), IL-17+ (Th17), IL-2+, and TNFα+ (“RB PI CD4 IFNγ+,” “RB PI CD4 IL-17+,” etc; Figure 2A). In contrast, the peripheral CD4+ T-cell count did not show any consistent association with the frequency of cytokine-producing CD4+ T cells in the peripheral blood. This finding suggests that the frequency of effector CD4+ T cells in GALT is a better reflection of CD4 counts during HIV infection than is the frequency of effector CD4+ T cells in the peripheral blood. These results also show that, although HIV controllers have lower frequencies of Tregs in GALT, they have higher frequencies of effector CD4+ T cells in this tissue.
We also noted that the frequencies of effector CD4+ T cells from rectal biopsies (as defined by IFNγ production [“PB HIV IFNγ CD4+ and CD8+”[) clustered with HIV antigen–specific CD4+ and CD8+ T-cell responses in peripheral blood, and that both of these were associated with high peripheral CD4+ T-cell counts. These data support and extend prior observations1,5,26,27 that stronger systemic (ie, peripheral blood) responses against HIV are positively associated with good outcomes, including high CD4+ T-cell numbers and the preservation of cytokine-producing CD4+ T cells in the mucosa. Because viral loads and CD4+ T-cell counts are used to define the patient groups, we repeated the clustering analysis without these parameters and found that the controllers and noncontrollers still segregated into 2 distinct groups (with the exception of subject 1292) based on their immunologic parameters alone (data not shown).
Because it is more difficult to analyze immune responses in the GALT than in the peripheral blood, it would be useful to identify peripheral blood responses that could serve as reliable surrogate markers of mucosal responses. To this end, a strong relationship was observed between markers of immune activation in the peripheral blood relative to those measured in rectosigmoid biopsy samples, including the percentage of CD4+FoxP3+ Tregs as well as the fractions of CD8+CD38+ and of CD8+Ki67+ T cells (Figure 2B). By contrast, cytokine-producing CD4+ T cells in rectosigmoid biopsies and in peripheral blood did not cluster together (Figure 2A). These results indicate that some but not all parameters measured in peripheral blood will be reflective of (and might serve as surrogates for) analogous parameters found in rectosigmoid biopsy tissue.
Transcriptional profiling of rectosigmoid biopsies
To determine whether transcriptional profiling can distinguish the mucosal responses of controllers and noncontrollers, we analyzed gene expression patterns from rectosigmoid biopsy samples of the 20 persons on the whole genome HEEBO oligonucleotide array platform. Using “unsupervised” hierarchic clustering, the expression levels of 9324 individual spots (representing 7349 genes) present in more than 80% of the arrays for further analysis segregated the 20 samples into 2 main clusters, each of which was composed of a mixture of controller and noncontroller samples. Hence, the mucosal immune responses of controllers and noncontrollers could not be separated simply on the basis of expression patterns alone.
To add another layer of dimensionality to this dataset, we next correlated immune parameters (as detected by flow cytometry) with the gene expression patterns. To reduce the complexity of the 36 available immunologic parameters, we performed PCA, an approach toward large datasets that can help explain the variance in the data by reducing the number of dimensions, thereby revealing the internal structure of the data.20 The variance of the principal components when PCA is applied to all 36 parameters is shown in Figure 3A. The first 2 components, PC1 and PC2, retain 27.4% of the original variance. When data from the 20 individual samples were projected onto these 2 components (Figure 3B), controllers and noncontrollers could be separated along the axis defined by PC1 but not by PC2. This indicates that PC1 can explain an important part of the variance that separates controllers from noncontrollers. Consistent with the unsupervised hierarchic clustering analysis, factors loading on PC1 from the PCA analysis were found to have a positive relationship with immune parameters associated with immune activation and with the level of viremia, and a negative association with CD4+ T-cell counts and the number of cytokine-producing effector CD4+ T cells in rectosigmoid biopsies (Figure 3C).
PCA of cellular parameters determined by flow cytometry. (A) Plot of the eigenvalues that reflect the variance of the principal components when PCA is applied to all 36 cellular parameters with values from all 20 HIV-infected persons; 27.36% of the variance in this matrix of cellular parameters is contained in the first 2 principal components. (B) Each dot represents an HIV-infected person plotted in 2 dimensions using their projections onto the first 2 principal components. (C) The factors loading on the eigenvalues for PC1 reflect the amount of variance shared by the parameter with the PC1 values. Vertical brackets indicate the cellular parameters that are most notable for either their positive or negative association with the PC1 values.
PCA of cellular parameters determined by flow cytometry. (A) Plot of the eigenvalues that reflect the variance of the principal components when PCA is applied to all 36 cellular parameters with values from all 20 HIV-infected persons; 27.36% of the variance in this matrix of cellular parameters is contained in the first 2 principal components. (B) Each dot represents an HIV-infected person plotted in 2 dimensions using their projections onto the first 2 principal components. (C) The factors loading on the eigenvalues for PC1 reflect the amount of variance shared by the parameter with the PC1 values. Vertical brackets indicate the cellular parameters that are most notable for either their positive or negative association with the PC1 values.
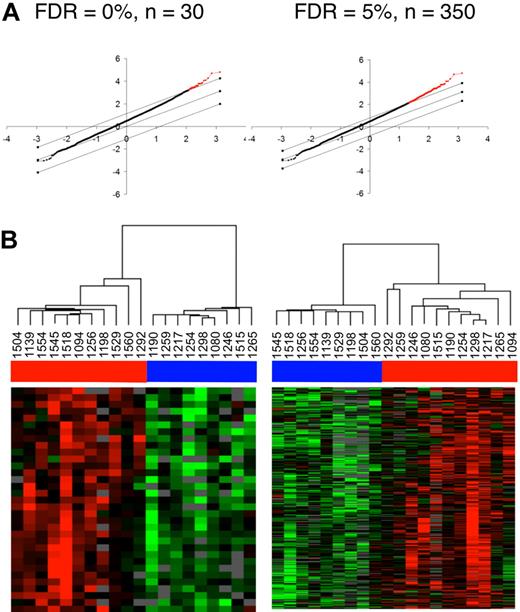
Quantitative regression analysis using the Significance Analysis of Microarrays (SAM) was performed to identify genes that have expression patterns significantly correlated with the eigenvalues calculated from the first component of the PCA analysis (PC1). A total of 99 genes were significantly correlated with the eigenvalues from first major component of the PCA analysis at an FDR of 0% (Table 2 and Figure 4B), whereas 535 genes were correlated with an FDR of less than 5% (Figure 4C and supplemental Table 2). Thus, by reducing the dimensions of the 36 parameters into principal components, the first major principal component could be effectively correlated with expression patterns from the transcriptional profiling analysis. When, however, regression analysis was conducted with individual parameters alone (eg, CD4+ T-cell counts or viral load), many fewer significant genes were identified (data not shown). Quantitative regression analysis of the second and third components of the PCA analysis with the transcriptional profiling dataset also did not reveal any significant hits (data not shown). This analysis indicates that the parameters that contribute toward the variance explained by the first major component (PC1; Figure 3C) represent the primary drivers of important differences in gene expression patterns in the rectosigmoid biopsies of these infected persons.
Ninety-nine genes significantly correlated (at an FDR of 0%) with eigenvalues from the PCA of immunologic parameters in the 20 HIV-infected persons
| Symbol . | Score . | Gene name . |
|---|---|---|
| IRF9 | 7.63969424 | Interferon-stimulated transcription factor 3, gamma 48 kDa |
| IFI44 | 6.463064537 | Interferon-induced protein 44 |
| PARP9 | 6.331458105 | Poly (ADP-ribose) polymerase family, member 9 |
| OAS2 | 6.080896015 | 2′-5′-Oligoadenylate synthetase 2, 69/71kDa |
| ISG15 | 5.653887207 | Interferon, alpha-inducible protein (clone IFI-15K) |
| LAP3 | 5.562707184 | Leucine aminopeptidase 3 |
| IFITM1 | 5.309911759 | Interferon-induced transmembrane protein 1 (9-27) |
| SERPING1 | 5.295948182 | Serpin peptidase inhibitor, clade G (C1 inhibitor), member 1, (angioedema, hereditary) |
| RNF213 | 5.268344777 | Chromosome 17 open reading frame 27 |
| SP110 | 5.256872524 | SP110 nuclear body protein |
| TRIM22 | 5.161555429 | Tripartite motif–containing 22 |
| STAT1 | 5.160281208 | Signal transducer and activator of transcription 1, 91 kDa |
| FGL2 | 5.059764921 | Fibrinogen-like 2 |
| MX1 | 5.052279793 | Myxovirus (influenza virus) resistance 1, interferon-inducible protein P78 (mouse) |
| HERC6 | 4.969209215 | HECT domain and RLD 6 |
| TAP1 | 4.925673126 | Transporter 1, ATP-binding cassette, subfamily B (MDR/TAP) |
| IFIT2 | 4.851202317 | Interferon-induced protein with tetratricopeptide repeats 2 |
| TYROBP | 4.643418344 | Tyro protein tyrosine kinase binding protein |
| PSMB9 | 4.61684985 | Proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) |
| ACOT7 | 4.474890582 | ACYL-COA thioesterase 7 |
| CCL4L2 | 4.435460418 | Chemokine (C-C motif) ligand 4 |
| GIMAP7 | 4.387779639 | GTPASE, IMAP family member 7 |
| IFIT3 | 4.377142549 | Interferon-induced protein with tetratricopeptide repeats 3 |
| STAT2 | 4.360968495 | Signal transducer and activator of transcription 2, 113 kDa |
| C1S | 4.356075207 | Complement component 1, S subcomponent |
| IFI35 | 4.297139761 | Interferon-induced protein 35 |
| IFITM4P | 4.23435714 | Interferon induced transmembrane protein 4 pseudogene |
| PNCK | 4.23295043 | Pregnancy up-regulated nonubiquitously expressed CAM kinase |
| ARHGAP25 | 4.190769686 | RHO GTPase activating protein 25 |
| GIMAP5 | 4.184925108 | GTPase, IMAP family member 5 |
| CXCL10 | 4.165976731 | Chemokine (C-X-C motif) ligand 10 |
| IL8RB | 4.152775449 | Interleukin 8 receptor, BETA |
| APOL3 | 4.134889167 | TNF-inducible protein CG12-1 |
| IFITM2 | 4.071757562 | Interferon induced transmembrane protein 2 (1-8D) |
| IL15 | 4.056626891 | Interleukin 15 |
| ZNFX1 | 3.992991172 | ZINC finger, NFX1-type containing 1 |
| CTSK | 3.987687377 | Cathepsin K (pycnodysostosis) |
| OASL | 3.972065117 | 2′-5′-Oligoadenylate synthetase-like |
| TRAalpha | 3.95014906 | T-cell receptor alpha locus |
| SRGN | 3.909639447 | Proteoglycan 1, secretory granule |
| CCL4L2 | 3.880280761 | Chemokine (C-C motif) ligand 4 |
| BTN3A3 | 3.862827621 | Butyrophilin, subfamily 3, member A3 |
| DTX3L | 3.834935982 | Deltex 3-like (drosophila) |
| NA | 3.812138625 | NA |
| CCL19 | 3.782911188 | Chemokine (C-C MOTIF) ligand 19 |
| ADAR | 3.779240486 | Adenosine deaminase, RNA specific |
| REC8 | 3.777102872 | REC8-like 1 (yeast) |
| KIAA1949 | 3.745843871 | KIAA1949 |
| ARL4C | 3.727007283 | ADP-ribosylation factor-like 4C |
| ARHGAP9 | 3.725234545 | RHO GTPase activating protein 9 |
| CORO1A | 3.715238075 | Coronin, actin binding protein, 1A |
| INDO | 3.67830532 | Indoleamine-pyrrole 2,3 dioxygenase |
| RAB31 | 3.67336941 | RAB31, member RAS oncogene family |
| COTL1 | 3.659705292 | Coactosin-like 1 (dictyostelium) |
| GMFG | 3.658192955 | GLIA maturation factor, GAMMA |
| SLA | 3.656310796 | SRC-like adaptor |
| BUB1 | 3.648122755 | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) |
| C1QA | 3.639355563 | Complement component 1, Q subcomponent, A chain |
| RAC2 | 3.632653776 | RAS-related C3 botulinum toxin substrate 2 (RHO family, small GTP binding protein RAC2) |
| CDC2 | 3.631566177 | Cell division cycle 2, G1 TO S and G2 to M |
| CLU | 3.626085643 | Clusterin |
| CCL3 | 3.621733073 | Chemokine (C-C MOTIF) ligand 3 |
| ECGF1 | 3.603371522 | Endothelial cell growth factor 1 (platelet-derived) |
| C1ORF54 | 3.599083135 | Chromosome 1 open reading frame 54 |
| HCLS1 | 3.598945235 | Hematopoietic cell-specific lyn substrate 1 |
| NA | 3.587802729 | NA |
| CCL4L2 | 3.584485049 | Chemokine (C-C MOTIF) ligand 4 |
| GBP2 | 3.584220194 | Guanylate binding protein 2, interferon-inducible |
| CXORF9 | 3.569357482 | Chromosome X open reading frame 9 |
| FAM14B | 3.552293235 | Family with sequence similarity 14, member B |
| EMILIN1 | 3.549670469 | Elastin microfibril interfacer 1 |
| PRPS2 | 3.539994216 | Phosphoribosyl pyrophosphate synthetase 1 |
| KIAA0082 | 3.530559508 | KIAA0082 |
| TSPAN14 | 3.527596769 | Tetraspanin 14 |
| FCER1G | 3.52450567 | Fc fragment of ige, high affinity I, receptor for gamma polypeptide |
| TREX1 | 3.514961993 | Three prime repair exonuclease 1 |
| VAMP1 | 3.510867551 | Vesicle-associated membrane protein 1 (synaptobrevin 1) |
| ALAS2 | 3.504662969 | Aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) |
| CD3D | 3.495196273 | CD3D antigen, delta polypeptide (TIT3 complex) |
| SLC7A7 | 3.490283635 | Solute carrier family 7 (cationic amino acid transporter, Y+ system), member 7 |
| CTSH | 3.489681294 | Cathepsin H |
| GRB2 | 3.47560278 | Growth factor receptor-bound protein 2 |
| SELL | 3.462829854 | Selectin L (lymphocyte adhesion molecule 1) |
| CCL3L3 | 3.452677466 | Chemokine (C-C MOTIF) ligand 3-like 1 |
| NA | 3.448890739 | NA |
| PHF11 | 3.446650391 | PHD finger protein 11 |
| DOCK8 | 3.446173435 | Dedicator of cytokinesis 8 |
| TRAC | 3.438194233 | T-cell receptor alpha constant |
| DAPP1 | 3.415611 | Dual adaptor of phosphotyrosine and 3-phosphoinositides |
| MPEG1 | 3.404638568 | Macrophage expressed gene 1 |
| NLRC5 | 3.399119856 | Nucleotide-binding oligomerization domains 27 |
| UGT1A10 | 3.374779661 | UDP glucuronosyltransferase 1 family, polypeptide A10 |
| LMO2 | 3.36809907 | LIM domain only 2 (rhombotin-like 1) |
| WARS | 3.349658421 | Interferon-induced protein 53 |
| CD74 | 3.340708593 | CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| TRIM56 | 3.339288432 | Tripartite motif-containing 56 |
| CD53 | 3.330644072 | CD53 antigen |
| MGC39372 | 3.329207645 | Hypothetical protein MGC39372 |
| RPS6KB2 | 3.316673927 | Ribosomal protein S6 kinase, 70 kDa, polypeptide 2 |
| Symbol . | Score . | Gene name . |
|---|---|---|
| IRF9 | 7.63969424 | Interferon-stimulated transcription factor 3, gamma 48 kDa |
| IFI44 | 6.463064537 | Interferon-induced protein 44 |
| PARP9 | 6.331458105 | Poly (ADP-ribose) polymerase family, member 9 |
| OAS2 | 6.080896015 | 2′-5′-Oligoadenylate synthetase 2, 69/71kDa |
| ISG15 | 5.653887207 | Interferon, alpha-inducible protein (clone IFI-15K) |
| LAP3 | 5.562707184 | Leucine aminopeptidase 3 |
| IFITM1 | 5.309911759 | Interferon-induced transmembrane protein 1 (9-27) |
| SERPING1 | 5.295948182 | Serpin peptidase inhibitor, clade G (C1 inhibitor), member 1, (angioedema, hereditary) |
| RNF213 | 5.268344777 | Chromosome 17 open reading frame 27 |
| SP110 | 5.256872524 | SP110 nuclear body protein |
| TRIM22 | 5.161555429 | Tripartite motif–containing 22 |
| STAT1 | 5.160281208 | Signal transducer and activator of transcription 1, 91 kDa |
| FGL2 | 5.059764921 | Fibrinogen-like 2 |
| MX1 | 5.052279793 | Myxovirus (influenza virus) resistance 1, interferon-inducible protein P78 (mouse) |
| HERC6 | 4.969209215 | HECT domain and RLD 6 |
| TAP1 | 4.925673126 | Transporter 1, ATP-binding cassette, subfamily B (MDR/TAP) |
| IFIT2 | 4.851202317 | Interferon-induced protein with tetratricopeptide repeats 2 |
| TYROBP | 4.643418344 | Tyro protein tyrosine kinase binding protein |
| PSMB9 | 4.61684985 | Proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) |
| ACOT7 | 4.474890582 | ACYL-COA thioesterase 7 |
| CCL4L2 | 4.435460418 | Chemokine (C-C motif) ligand 4 |
| GIMAP7 | 4.387779639 | GTPASE, IMAP family member 7 |
| IFIT3 | 4.377142549 | Interferon-induced protein with tetratricopeptide repeats 3 |
| STAT2 | 4.360968495 | Signal transducer and activator of transcription 2, 113 kDa |
| C1S | 4.356075207 | Complement component 1, S subcomponent |
| IFI35 | 4.297139761 | Interferon-induced protein 35 |
| IFITM4P | 4.23435714 | Interferon induced transmembrane protein 4 pseudogene |
| PNCK | 4.23295043 | Pregnancy up-regulated nonubiquitously expressed CAM kinase |
| ARHGAP25 | 4.190769686 | RHO GTPase activating protein 25 |
| GIMAP5 | 4.184925108 | GTPase, IMAP family member 5 |
| CXCL10 | 4.165976731 | Chemokine (C-X-C motif) ligand 10 |
| IL8RB | 4.152775449 | Interleukin 8 receptor, BETA |
| APOL3 | 4.134889167 | TNF-inducible protein CG12-1 |
| IFITM2 | 4.071757562 | Interferon induced transmembrane protein 2 (1-8D) |
| IL15 | 4.056626891 | Interleukin 15 |
| ZNFX1 | 3.992991172 | ZINC finger, NFX1-type containing 1 |
| CTSK | 3.987687377 | Cathepsin K (pycnodysostosis) |
| OASL | 3.972065117 | 2′-5′-Oligoadenylate synthetase-like |
| TRAalpha | 3.95014906 | T-cell receptor alpha locus |
| SRGN | 3.909639447 | Proteoglycan 1, secretory granule |
| CCL4L2 | 3.880280761 | Chemokine (C-C motif) ligand 4 |
| BTN3A3 | 3.862827621 | Butyrophilin, subfamily 3, member A3 |
| DTX3L | 3.834935982 | Deltex 3-like (drosophila) |
| NA | 3.812138625 | NA |
| CCL19 | 3.782911188 | Chemokine (C-C MOTIF) ligand 19 |
| ADAR | 3.779240486 | Adenosine deaminase, RNA specific |
| REC8 | 3.777102872 | REC8-like 1 (yeast) |
| KIAA1949 | 3.745843871 | KIAA1949 |
| ARL4C | 3.727007283 | ADP-ribosylation factor-like 4C |
| ARHGAP9 | 3.725234545 | RHO GTPase activating protein 9 |
| CORO1A | 3.715238075 | Coronin, actin binding protein, 1A |
| INDO | 3.67830532 | Indoleamine-pyrrole 2,3 dioxygenase |
| RAB31 | 3.67336941 | RAB31, member RAS oncogene family |
| COTL1 | 3.659705292 | Coactosin-like 1 (dictyostelium) |
| GMFG | 3.658192955 | GLIA maturation factor, GAMMA |
| SLA | 3.656310796 | SRC-like adaptor |
| BUB1 | 3.648122755 | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) |
| C1QA | 3.639355563 | Complement component 1, Q subcomponent, A chain |
| RAC2 | 3.632653776 | RAS-related C3 botulinum toxin substrate 2 (RHO family, small GTP binding protein RAC2) |
| CDC2 | 3.631566177 | Cell division cycle 2, G1 TO S and G2 to M |
| CLU | 3.626085643 | Clusterin |
| CCL3 | 3.621733073 | Chemokine (C-C MOTIF) ligand 3 |
| ECGF1 | 3.603371522 | Endothelial cell growth factor 1 (platelet-derived) |
| C1ORF54 | 3.599083135 | Chromosome 1 open reading frame 54 |
| HCLS1 | 3.598945235 | Hematopoietic cell-specific lyn substrate 1 |
| NA | 3.587802729 | NA |
| CCL4L2 | 3.584485049 | Chemokine (C-C MOTIF) ligand 4 |
| GBP2 | 3.584220194 | Guanylate binding protein 2, interferon-inducible |
| CXORF9 | 3.569357482 | Chromosome X open reading frame 9 |
| FAM14B | 3.552293235 | Family with sequence similarity 14, member B |
| EMILIN1 | 3.549670469 | Elastin microfibril interfacer 1 |
| PRPS2 | 3.539994216 | Phosphoribosyl pyrophosphate synthetase 1 |
| KIAA0082 | 3.530559508 | KIAA0082 |
| TSPAN14 | 3.527596769 | Tetraspanin 14 |
| FCER1G | 3.52450567 | Fc fragment of ige, high affinity I, receptor for gamma polypeptide |
| TREX1 | 3.514961993 | Three prime repair exonuclease 1 |
| VAMP1 | 3.510867551 | Vesicle-associated membrane protein 1 (synaptobrevin 1) |
| ALAS2 | 3.504662969 | Aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) |
| CD3D | 3.495196273 | CD3D antigen, delta polypeptide (TIT3 complex) |
| SLC7A7 | 3.490283635 | Solute carrier family 7 (cationic amino acid transporter, Y+ system), member 7 |
| CTSH | 3.489681294 | Cathepsin H |
| GRB2 | 3.47560278 | Growth factor receptor-bound protein 2 |
| SELL | 3.462829854 | Selectin L (lymphocyte adhesion molecule 1) |
| CCL3L3 | 3.452677466 | Chemokine (C-C MOTIF) ligand 3-like 1 |
| NA | 3.448890739 | NA |
| PHF11 | 3.446650391 | PHD finger protein 11 |
| DOCK8 | 3.446173435 | Dedicator of cytokinesis 8 |
| TRAC | 3.438194233 | T-cell receptor alpha constant |
| DAPP1 | 3.415611 | Dual adaptor of phosphotyrosine and 3-phosphoinositides |
| MPEG1 | 3.404638568 | Macrophage expressed gene 1 |
| NLRC5 | 3.399119856 | Nucleotide-binding oligomerization domains 27 |
| UGT1A10 | 3.374779661 | UDP glucuronosyltransferase 1 family, polypeptide A10 |
| LMO2 | 3.36809907 | LIM domain only 2 (rhombotin-like 1) |
| WARS | 3.349658421 | Interferon-induced protein 53 |
| CD74 | 3.340708593 | CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| TRIM56 | 3.339288432 | Tripartite motif-containing 56 |
| CD53 | 3.330644072 | CD53 antigen |
| MGC39372 | 3.329207645 | Hypothetical protein MGC39372 |
| RPS6KB2 | 3.316673927 | Ribosomal protein S6 kinase, 70 kDa, polypeptide 2 |
FDR indicates false discovery rate; and PCA, principal component analysis.
Transcriptional profiling analysis of rectosigmoid biopsies and the relationship with cellular parameters. (A) Gene expression patterns in the rectosigmoid biopsy samples from controllers and noncontrollers. Hierarchic clustering analysis was used to organize genes and samples. Each row represents an individual gene and each column, an individual patient. Black indicates the median level of expression; red, greater than median expression, green, less than median expression; and gray, missing data. Horizontal bars (in blue for controllers and red for noncontrollers) at the top of the figure indicate the dispersal of controller and noncontroller samples. (B-C) Clustering of the controller and noncontroller samples on the basis of gene expression patterns associated with cellular parameters identified through regression analysis with PC1 eigenvalues. (B) Quantitative analysis by Significance Analysis of Microarrays (SAM) was used to identify data from elements of the 99 genes that were positively associated with PC1 eigenvalues at a false discovery rate (FDR) of 0%. The graph at the top displays the positively associated genes in red, with the x-axis value denoting the expected expression and the y-axis denoting the observed expression values. The threshold lines indicate the FDR cutoff rate of 0%. The heat map shows the 20 patient samples after being reorganized using the expression values of these 99 genes. Horizontal bars (in blue for controllers and red for noncontrollers) at the top of the figure indicate the clustering of controller and noncontroller samples. (C) As described in panel B, 535 genes were identified by SAM analysis at an FDR of < 5%, and the 20 patient samples were reorganized using the expression values of these 535 genes. Horizontal bars at the top of the figure indicate the clustering of controller (blue) and noncontroller (red) samples.
Transcriptional profiling analysis of rectosigmoid biopsies and the relationship with cellular parameters. (A) Gene expression patterns in the rectosigmoid biopsy samples from controllers and noncontrollers. Hierarchic clustering analysis was used to organize genes and samples. Each row represents an individual gene and each column, an individual patient. Black indicates the median level of expression; red, greater than median expression, green, less than median expression; and gray, missing data. Horizontal bars (in blue for controllers and red for noncontrollers) at the top of the figure indicate the dispersal of controller and noncontroller samples. (B-C) Clustering of the controller and noncontroller samples on the basis of gene expression patterns associated with cellular parameters identified through regression analysis with PC1 eigenvalues. (B) Quantitative analysis by Significance Analysis of Microarrays (SAM) was used to identify data from elements of the 99 genes that were positively associated with PC1 eigenvalues at a false discovery rate (FDR) of 0%. The graph at the top displays the positively associated genes in red, with the x-axis value denoting the expected expression and the y-axis denoting the observed expression values. The threshold lines indicate the FDR cutoff rate of 0%. The heat map shows the 20 patient samples after being reorganized using the expression values of these 99 genes. Horizontal bars (in blue for controllers and red for noncontrollers) at the top of the figure indicate the clustering of controller and noncontroller samples. (C) As described in panel B, 535 genes were identified by SAM analysis at an FDR of < 5%, and the 20 patient samples were reorganized using the expression values of these 535 genes. Horizontal bars at the top of the figure indicate the clustering of controller (blue) and noncontroller (red) samples.
When data from the 20 HIV-infected persons were reclustered using unsupervised hierarchic clustering of the gene expression values of only these 99 or 535 genes (Figure 4B-C), the controllers and the noncontrollers (with the exception of 1292 and 1094) were found to segregate quite accurately into 2 main groups. Many of the notable genes on this list (Table 2) are related to IFN signaling and antiviral responses (eg, IRF3, STAT1, OAS2, IFI44, and IFIT3).28 Two members of the HERC family of ubiquitin ligases (HERC5 and HERC6) were also included, which is particularly interesting because HERC5 conjugates ISG15 to proteins in a process known as ISGylation.29 ISG15 is an IFN-inducible, ubiquitin-like protein that has 2 ubiquitin-like domains and a wide range of target proteins.28 Notably, ISG15 could be involved in IFN-mediated inhibition of viral budding and release.30
“Supervised” 2-way comparisons using SAM were then conducted to identify genes that are differentially expressed between controllers and noncontrollers. The 2 groups were distinguishable by 33 genes at a false discovery rate (FDR) of 0% and 387 genes at an FDR rate of less than 5% (Figure 5A). As expected, when the 20 samples were reclustered using either of these 33 or 387 genes, the controllers and noncontrollers segregated perfectly into 2 distinct subgroups (Figure 5B). Many of the more notable transcripts from these lists (supplemental Figure 1) are also associated with interferon signaling and antiviral responses (eg, STAT1, OAS1, OASL, IFI27, ISG20, and IFI30). Interestingly, APOBEC3G, IDO, RIGI (or RARRES3), TAP1, CCL4, CCL5, and CCL19 were also down-regulated in the controllers (supplemental Figure 1). The reduced expression of IDO in the controllers is of particular interest because this gene is also down-regulated in the nonpathogenic model of simian immunodeficiency virus infection of African green monkeys, relative to the pathogenic model of pigtailed macaques.31,32
Identification of differentially expressed genes between HIV controllers and noncontrollers by 2-way supervised comparisons. (A) By predefining controllers and noncontrollers on the basis of viral load, 2-way comparisons between the groups were conducted using SAM analysis to identify 30 genes at an FDR of 0% and 350 genes at an FDR of 5%. The graphs display the significantly different genes in red, with the x-axis value denoting the expected expression and the y-axis denoting the observed expression values. The threshold lines indicate the FDR cutoff rate of 0% and 5%. (B) The 20 patient samples were then reorganized using the expression values of either the 30 genes (FDR = 0%) or the 350 genes (FDR = 5%). Horizontal bars (in blue for controllers and red for noncontrollers) at the top of the figure indicate the clustering of controller and noncontroller samples.
Identification of differentially expressed genes between HIV controllers and noncontrollers by 2-way supervised comparisons. (A) By predefining controllers and noncontrollers on the basis of viral load, 2-way comparisons between the groups were conducted using SAM analysis to identify 30 genes at an FDR of 0% and 350 genes at an FDR of 5%. The graphs display the significantly different genes in red, with the x-axis value denoting the expected expression and the y-axis denoting the observed expression values. The threshold lines indicate the FDR cutoff rate of 0% and 5%. (B) The 20 patient samples were then reorganized using the expression values of either the 30 genes (FDR = 0%) or the 350 genes (FDR = 5%). Horizontal bars (in blue for controllers and red for noncontrollers) at the top of the figure indicate the clustering of controller and noncontroller samples.
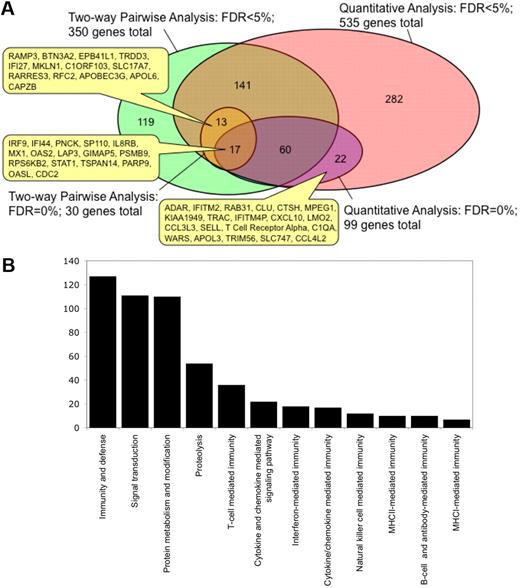
We then compared the lists of genes that were identified using 2-way supervised comparison with unsupervised correlations with PC1, and found that the distinct analysis strategies identified different sets of genes. Thus, Figure 6A shows a Venn diagram representing the overlap between these different types of analysis. At the FDR of 0%, 17 genes (including STAT1, IRF9, PARP9, MX1, OAS2, and OASL) were found in common by 2-way comparison and by quantitative analysis with PC1, whereas 13 genes (including APOBEC3G, IFI27, and RARRES3) were identified by 2-way comparison alone and 82 genes (including CXCL10, CCL3L3, and RAB31) were identified by quantitative analysis alone. However, whereas all of 13 genes identified by 2-way comparison at an FDR of 0% were included among the 535 genes identified by quantitative analysis at an FDR of 5%, there were 22 genes identified by quantitative analysis at an FDR of 0% that were not among the 350 genes identified by 2-way comparison at an FDR of 5%. This shows that the use of several strategies of identifying candidate transcriptional markers may provide a more complete picture of the observed responses than a 2-way based supervised comparison alone. In total, we identified 608 genes of significant interest in this study (supplemental Table 2).
Integrating transcriptional profiling results from 2-way supervised and unsupervised analyses. (A) Relationship between genes that were identified from the expression profiling dataset by 2-way supervised pairwise comparisons or through unsupervised quantitative analysis with cellular parameters using PC1 at an FDR of 0% or < 5%. The numbers in the Venn diagram show the number unique to that set or in the overlap between sets. The positions of certain genes in these overlapping gene sets are indicated. (B) Classification of the combined gene list into biologic process categories by gene ontology analysis using the PANTHER tool. Only categories that are significantly enriched (P < .05) relative to all the genes in the human genome are shown. The y-axis denotes the number of genes that belong to each biologic process category that is significantly enriched.
Integrating transcriptional profiling results from 2-way supervised and unsupervised analyses. (A) Relationship between genes that were identified from the expression profiling dataset by 2-way supervised pairwise comparisons or through unsupervised quantitative analysis with cellular parameters using PC1 at an FDR of 0% or < 5%. The numbers in the Venn diagram show the number unique to that set or in the overlap between sets. The positions of certain genes in these overlapping gene sets are indicated. (B) Classification of the combined gene list into biologic process categories by gene ontology analysis using the PANTHER tool. Only categories that are significantly enriched (P < .05) relative to all the genes in the human genome are shown. The y-axis denotes the number of genes that belong to each biologic process category that is significantly enriched.
To better understand the function of the identified genes, gene ontology analysis was conducted to identify associated biologic pathways (Figure 6B). Of the 608 genes, 127 were involved in the “immunity and defense” category (enrichment P = 1.38 × 10−39), 36 genes in “T-cell mediated immunity” (P = 1.87 × 10−18), 18 genes in “interferon-mediated immunity” (P = 7.01 × 10−12), 54 genes in “proteolysis” (P = 1.53 × 10−6), 17 genes in “cytokine/chemokine mediated immunity,” 10 genes in “MHCII-mediated immunity,” 12 genes in “NK-cell mediated immunity,” 7 genes in “MHCI-mediated immunity,” and 10 genes in “B-cell and antibody mediated immunity.” This analysis indicates that the differential expression of genes involved in inflammation and in immunity is a major distinguishing factor between the intestinal mucosa of controllers and the noncontrollers and is also the major correlate with our flow cytometry parameters. Notably, all of these genes were more highly expressed in noncontrollers than in controllers.
Verification of gene expression by real-time PCR analysis
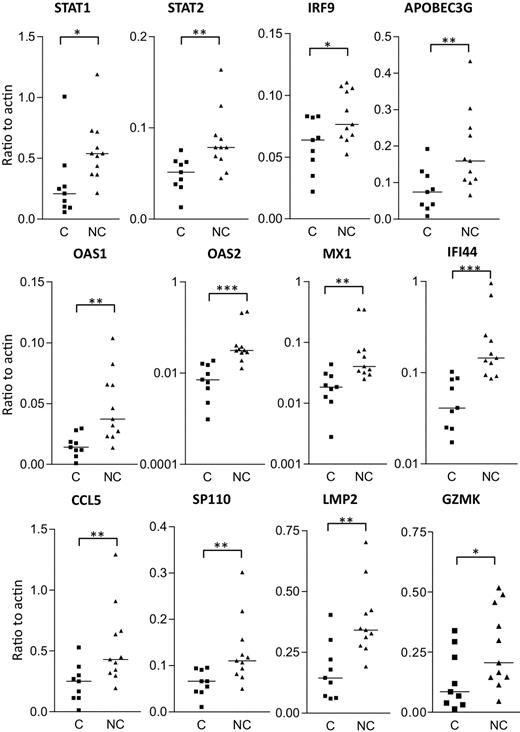
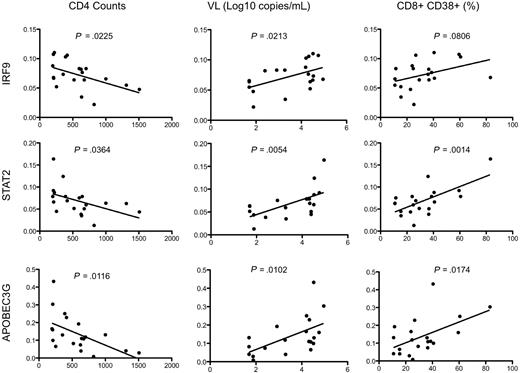
Real-time PCR analysis was used to independently confirm the differential expression of some of the more interesting transcripts that distinguish HIV controllers from noncontrollers (Figure 7). Of the 18 genes selected for real-time PCR analysis, 14 had a P value less than .05 (and 12 of these are shown in Figure 7). Type I IFN signaling induces formation of a heterotrimeric transcription complex that contains STAT1, STAT2, and IRF9. We confirmed that all 3 genes were significantly up-regulated in the rectosigmoid biopsies of HIV noncontrollers. Expression levels of these genes were found to be strongly associated with reduced CD4+ T-cell counts, increased viral loads, and higher levels of immune activation (as shown for IRF9 and STAT2 in Figure 8). Several antiviral or antimicrobial mechanisms are up-regulated in the HIV noncontrollers relative to the controllers (Figure 7). These include APOBEC3G (also shown in Figure 8 to be associated with CD4 counts, viral loads, and immune activation), a cytidine deaminase that can degrade HIV DNA when it becomes packaged into HIV virions33-35 ; MX1, an antiviral GTPase that can trap viral nucleocapsid-like structures by altering regular vesicle trafficking36 ; the OAS genes (OAS1 and OAS2) that encode enzymes contributing to innate responses that result in the degradation of viral RNA and the inhibition of viral replication28,37 ; CCL5 (or RANTES),38 an antagonist of the CCR5 coreceptor for HIV39 ; SP110, a nuclear body protein40 that has been associated with resistance and susceptibility to Mycobacterium tuberculosis infection in mice41 and humans42 ; and IFI44, an IFNα/β-induced microtubular aggregate protein associated with hepatitis C virus infection of chimpanzees.43,44 Increased expression of LMP2 was also confirmed in noncontrollers (Figure 7), suggesting increased antigen processing. LMP2 is a component of an IFNγ-induced proteasome that could favor the production of peptides that preferentially bind major histocompatibility complex (MHC) class I molecules.45 Although not confirmed by real-time PCR, TAP1 was also up-regulated in the noncontrollers in the array analysis. Surprisingly, although having a more potent granzyme B–mediated cytolytic CD8+ T-cell activity against HIV-infected CD4+ T cells is thought to be a feature of viral controllers,8 expression levels of granzyme K were significantly higher in the noncontrollers. In aggregate, these results suggest that, even though HIV-specific CTL activity may be enhanced in viral controllers,8 the mucosal microenvironment of noncontrollers displays broad activation of host antiviral mechanisms.
Verification of gene expression by real-time PCR analysis of rectosigmoid biopsy samples. Transcript levels of selected genes were measured in controllers (C) and noncontrollers (NC) and normalized to β-actin transcript levels. Horizontal bars indicate median expression levels. Statistical significance between groups was determined by 2-tailed Mann-Whitney test. *P < .05; **P < .01;***P < .001.
Verification of gene expression by real-time PCR analysis of rectosigmoid biopsy samples. Transcript levels of selected genes were measured in controllers (C) and noncontrollers (NC) and normalized to β-actin transcript levels. Horizontal bars indicate median expression levels. Statistical significance between groups was determined by 2-tailed Mann-Whitney test. *P < .05; **P < .01;***P < .001.
Association of gene expression by real-time PCR analysis of rectosigmoid biopsy samples with clinical and immune parameters. Representative scatterplots with linear regression analysis to confirm the association between expression levels of selected transcripts from rectosigmoid biopsy samples with CD4+ T-cell counts, viral load, and levels of immune activation (represented by the percentage of CD8+CD38+ cells).
Association of gene expression by real-time PCR analysis of rectosigmoid biopsy samples with clinical and immune parameters. Representative scatterplots with linear regression analysis to confirm the association between expression levels of selected transcripts from rectosigmoid biopsy samples with CD4+ T-cell counts, viral load, and levels of immune activation (represented by the percentage of CD8+CD38+ cells).
Discussion
This study presents a novel strategy for direct comparison of immunologic parameters in GALT and peripheral blood with whole genome transcriptional profiling data from rectosigmoid biopsy tissue in well-characterized cohorts of HIV-infected controllers and noncontrollers. The most notable observation pertaining to the immunologic measurements was a consistent association between markers of immune activation in the peripheral blood and the expression of IFN-related genes in the rectal mucosa. We also found a strong relationship between some markers in the peripheral blood and paired rectosigmoid biopsy samples from the same persons (eg, immune activation and Tregs were positively associated in both peripheral blood and mucosal samples). In the case of transcriptional profile analysis, there was a diminished IFN-related inflammatory response in the intestinal mucosa of HIV controllers relative to that found in HIV noncontrollers, although no transcripts were found to be more highly expressed in the HIV controllers relative to the noncontrollers. Although it is not possible to determine whether an increased IFN response causes disease progression or is simply the result of increased viral replication, these results are consistent with the hypothesis that a reduced IFN-mediated inflammatory response in HIV controllers enables preservation of functional CD4+ T cells and/or other host mechanisms of viral control. Alternatively, heightened IFN-mediated inflammatory responses may be caused by higher levels of viral replication in the mucosal tissues of the noncontrollers.
Although previous studies have closely examined CD8+ T-cell responses in the rectal mucosa of HIV controllers, the CD4+ T-cell response is less well understood. Among the HIV controllers studied here, the presence of mucosal CD4+ helper T cells capable of producing cytokines such as IL-2, IFNγ, TNFα, and IL-17 was strongly associated with controller status, whereas the presence of peripheral blood cytokine responses by CD4+ T cells was not correlated with protection. It is not clear whether these are HIV-specific T cells or whether this observation simply reflects an accelerated depletion of functional CD4+ T cells in the mucosal tissue of noncontrollers. It is also unclear whether these cytokine-producing CD4+ T cells may have any role in the protection of HIV controllers, although the presence of more functional CD4+ helper T cells in the mucosa of HIV controllers would predictably sustain the quality and frequency of CD8+ T cells against HIV and/or other mucosal pathogens.
One limitation of our study is that only 2 groups of HIV-infected persons (controllers and noncontrollers) were characterized. Although all of the transcripts that we identified were up-regulated in noncontrollers relative to controllers, it is not possible to state that HIV controllers have mucosal profiles that are similar to those found in uninfected seronegative persons. In a smaller expression profiling study of GALT tissue, long-term nonprogressors had higher expression levels of many immune response genes, although the levels of expression of these genes were even greater in the noncontrollers.19 It is possible that the controllers in our study have higher expression levels of immune response genes relative to HIV-negative healthy persons, but the levels are still relatively low with respect to the noncontrollers. Finally, it will be of interest to characterize the same set of parameters in HAART-suppressed patients who have undetectable viral loads as a result of drug-mediated viral suppression as opposed to natural resistance. Studies to analyze the immunologic and transcriptional parameters in these additional groups of patients are now ongoing.
In summary, this analysis confirms the observation that HIV disease progression is associated with immunologic and transcriptional signs of increased immune activation and inflammation.6,19,25 Our analysis has validated some parameters of such activation that are concordant between the peripheral blood and rectosigmoid biopsy tissue. Using several forms of analysis, we have also delineated transcripts in the rectosigmoid biopsy tissue of noncontrollers that are highly related to disease progression. The complete dataset should be valuable to the research community because it provides a detailed characterization of mucosal tissues from HIV controllers. In addition, we anticipate that the approach of relating genomic signatures to a complex set of immune parameters by PCA will be applicable to the evaluation of cellular and molecular signatures of viral control, disease progression, and vaccine protection in studies that include even more complex datasets. Further studies will be aimed at understanding whether the patterns of inflammation distinguishing HIV controllers from noncontrollers are induced by disease progression or if, alternatively, they underlie mechanisms that may drive disease progression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Suzanne Noble, Department of Microbiology and Immunology, UCSF, for the provision of Candida albicans; Dr Mario Roederer of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases (NIAID)/NIH, for SPICE software; Dr Keith A. Reimann of the Beth Israel Deaconess Medical Center, Harvard University, for providing antibodies through the NIH Reagent Resource Program; and Dr William P. Schecter of the UCSF Division of Surgery at San Francisco General Hospital for assistance in gathering tissue samples from patients. Invaluable advice on the Significance Analysis of Microarrays data were provided by Dr Charles C. Kim from the Department of Biochemistry and Biophysics, UCSF; Dr Mark R. Segal of the Center of Bioinformatics and Molecular Biostatistics; and James Bullard from the Department of Statistics at UC Berkeley.
These studies were supported by a grant from the Elizabeth Glaser Pediatric AIDS Foundation (77510-29; D.F.), a K23 grant from NIH (AI065244; P.W.H.), a grant from the Centers for AIDS Research at UCSF (AI27763, MH59037; S.G.D.), and NIH grants R37 AI40312 and DPI OD00329 (J.M.M.). Additional support was provided by NIAID (K24 AI069994), the UCSF Clinical and Translational Science Institute (UL1 RR024131-01), the American Foundation for AIDS Research (106710-40-RGRL), and the Harvey V. Berneking Living Trust. J.M.M. is a recipient of the NIH Director's Pioneer Award Program, part of the NIH Roadmap for Medical Research, through grant DPI OD00329.
National Institutes of Health
Authorship
Contribution: P.L. was involved in study design, protocol development, microarray analysis, and writing the paper; D.F. was involved in study design, obtaining samples, flow cytometric analysis, and writing the paper; P.W.H. was involved in subject recruitment and specimen collection; J.M.L. was involved in microarray analysis; B.K. was involved in obtaining samples, preparing rectal biopsies, and editing the paper; J.N.M. was involved in subject recruitment, specimen collection, and data management; S.G.D. was involved in study design, subject recruitment, specimen collection, and paper editing; and J.M.M. was involved in study design and writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joseph M. McCune, Division of Experimental Medicine, University of California at San Francisco, San Francisco, CA; e-mail: mike.mccune@ucsf.edu.
References
Author notes
P.L. and D.F. contributed equally to this study.

![Figure 1. Flow cytometry–based immunologic profiling: representative flow plots and gating strategy. (A) Gating strategy by flow cytometry for the detection of CD4+ and CD8+ T cells (singlet [forward scatter A{FSC-A} × FSC-H diagonal] live [Aqua] CD3+ lymphocytes) extracted from rectosigmoid biopsy tissue (RB, top panels) and from peripheral blood (PB, bottom panels). (B) Detection of CD38 and Ki67 expression in CD8+ T cells (left) as well as FoxP3 and Ki67 in CD8+ and CD4+ T cells from rectosigmoid biopsy tissue (top) and from peripheral blood (bottom). (C-D) Antigen-specific CD8+ (top) CD4+ (bottom) T-cell responses in peripheral blood after stimulation with PBS (mock), CMV, HIV, Candida albicans (C alb), or SEB antigens, as measured by CFSElow proliferative responses (C) or IFNγ intracellular cytokine detection (D). (E) Detection of intracellular cytokines (IFNγ, IL-2, IL-17, or TNFα) in CD4+ T cells from rectosigmoid biopsy tissue (top) and from peripheral blood (bottom), after PMA/ionomycin stimulation in vitro. All of these analyses were carried out using cells from a representative noncontroller (1256).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/15/10.1182_blood-2009-12-257451/4/m_zh89991051240001.jpeg?Expires=1767733747&Signature=38BJogDrCxI-fVI3uaw6Xrn2eTQdE-g-2FjQ6bCSM-XR~qHsjbUS02EHmENeNU4ZmKiVFGg9UaqtXtIwQ1CrDFMZyIKbAQ8wdh0ohqYA6IgQnyyqHfZxt1O2OjpTXrXkQPD9sMgo4ZAOePyxpcp0ji~3i5UiBkbW9MZ1x9tICIZ7T2lFysEJY-6YCYSOM5dvw~pVCp6UwU~WnPqzUNEOO4ehyOjVkjQdGmFZD0ORu88kGg3R6dMMvddZop~i~vnt0vJo2Xs~SSy8H-1BF0IG8FlAIqmGset1ECAIKRT-2GHBDpiNqxg917j~hj97QvlRqNd-utJUJNAac7P3B08PvA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)