Abstract
Humoral alloimmunization to red blood cell (RBC) antigens is a clinically significant problem that can lead to transfusion reactions and difficulty in locating future compatible blood for transfusion. However, factors regulating responder/nonresponder status are only partially understood. Herein, we identify a series of microbes with 100% identity in 8– to 9–amino acid peptides containing the variant amino acids in Kell, Kidd, and Duffy antigens. To test the hypothesis that infection with such a microbe could predispose to RBC alloimmunization, a mouse model was developed using murine polyoma virus expressing a defined CD4+ T-cell epitope ovalbumin323-339 ((OVA)323-339) and subsequent transfusion with RBCs expressing a B-cell epitope (hen egg lysozyme [HEL]) fused to (OVA)323-339. Whereas infection alone induced no detectable anti-HEL, subsequent RBC transfusion induced 100- to 1000-fold more anti-HEL in mice that had been previously infected compared with control mice. This effect did not occur with wild-type polyoma virus or RBCs expressing HEL alone. Together, these data indicate that prior exposure to a pathogen with small peptide homology to RBC antigens can lead to an enhanced primary alloantibody response. As such priming is not detectable by current clinical tests, it is unknown to what extent this occurs in human alloimmunization.
Introduction
Humoral immunization to red blood cell (RBC) alloantigens can occur as a result of transfusion or pregnancy. Antibodies against clinically significant blood group alloantigens (ie, RhD, Kell, Kidd, etc) can lead to both hemolysis of transfused RBCs and/or to hemolytic disease of the newborn.1,2 However, unlike humoral immunization to microbial infection, which approaches 100% in immunocompetent persons, exposure to RBC alloantigens induces a measurable antibody response in only a subset of recipients. Alloimmunization to the RhD antigen on RBCs ranges from 20% to 80%, with only 3% to 10% of recipients becoming immunized to the remaining RBC antigens (eg, Kell, Duffy, Kidd), despite chronic transfusion.3-5 The reason why some transfused patients but not others become alloimmunized is unclear, and factors influencing alloimmunization have been only partially defined. Immunogenetics plays some role in variability of alloimmunization to blood products, as antibody responses to certain alloantigens are confined to distinct recipient human leukocyte antigen (HLA) types.6-8 In addition, genetic variants outside of HLA may regulate RBC alloimmunization.9 Environmental differences between recipients also likely affect alloimmunization, as genetically identical animals still have variable alloantibody responses to transfused RBCs.10,11 One such environmental variable may be the inflammatory status of the recipient, which has been shown to have a substantial effect upon alloimmunization to transfused RBCs in mice,10,12,13 and potentially in humans.14 In the current report, we hypothesize that an additional potential factor is small peptide homology between microbial-derived peptides and blood group antigens.
It has long been appreciated that alloimmunity can be induced through exposure to microbial antigens that mimic the 3-dimensional structure and epitopes of alloantigens (molecular mimicry); thus, antibodies generated against the microbial antigen can cross-react with alloantigens. This is a well-documented event with anti-ABO antibodies, where humans make antibodies against “non–self-” ABO antigens without any prior exposure through transfusion, because of immunization with gut microbes that express A and/or B antigens.15 In the context of immunoglobulin G (IgG) responses to protein alloantigens, limited cross-reactivity of anti-RBC antibodies and microbes has been observed for K and Jkb antigens.16,17 However, the possibility of significant molecular mimicry inducing antibodies to non-ABO RBC antigens has been largely rejected, because alloantibodies against other alloantigens (eg, RhD, RhCc, RhEe, Kell, Duffy, Kidd) are rarely if ever detected in the absence of prior exposure to alloantigen through transfusion or pregnancy.2,18
A second potential mechanism of molecular mimicry, which has not been thoroughly evaluated in the context of humoral alloimmunization, is similarity at the level of CD4+ T-cell epitopes in the absence of mimicry of the 3-dimensional blood group antigen recognized by antibodies. In this case, mimicry would be restricted to homology of short peptide sequences presented by major histocompatibility complex class II (MHC II). Herein, we report a series of microbial peptides with substantial similarity or identity to peptides containing the polymorphisms responsible for 3 pairs of clinically significant antithetical human RBC alloantigens (K/k, Fya/Fyb, and Jka/Jkb). Based on these findings, we hypothesize that CD4+ T-cell responses to some microbes cross-react with CD4+ T-cell epitopes of RBC alloantigens. Alloantibody binding to blood group alloantigens typically requires precise 3-dimensional structure of the antigen. Because the identified CD4+ T-cell epitopes represent only small linear peptides surrounded by nonhomologous amino acids, it is predicted that exposure to the microbes would result in isolated activation of CD4+ T cells but not an induction of an alloantibody. The potential significance of this is that if the authentic RBC alloantigen were encountered during transfusion, the patient would have preformed CD4+ helper T-cell immunity, which could then provide the required help to allow B cells to make alloantibodies. As generation of CD4+ helper T cells is often a key regulatory event in humoral immunization, which may not occur in response to the weak stimulus of RBCs alone, microbial mimicry at the CD4+ T-cell level represents an additional hypothesis to explain why some but not other transfusion recipients make an anti-RBC antibody.
Current clinical methodologies in transfusion biology are limited to serologic analysis, and do not evaluate CD4+ T-cell immunity. Therefore, evaluating this hypothesis in humans with existing clinical data is not technically feasible. To allow a rigorous testing of the hypothesis in an ethical fashion, we engineered a murine model by generating a novel recombinant virus with insertion of a known CD4+ T-cell epitope (ovalbumin323-339 [(OVA)323-339]) in frame with a viral protein. We also used a transgenic mouse expressing an RBC-specific antigen containing a distinct B-cell epitope fused in frame with (OVA)323-339.19 This model recapitulates the observed scenario in humans, by providing a microbe with homology limited to a small peptide, and not the entire blood group antigen. Using this system, we report that an antecedent microbial infection substantially enhances the alloantibody response upon subsequent exposure to the whole antigen on a transfused RBC, yet does not itself induce detectable antibodies in the absence of exposure to the relevant RBC antigen.
Methods
Mice
C57BL/6, B10.BR, and C57BL/6.PL-Thy1.1 mice were purchased from The Jackson Laboratory. HOD, FVB, OT-II, OT-II Thy1.1, membrane-bound hen egg lysozyme (mHEL), and C57BL/6 × B10.BR mice were bred by the Emory University Department of Animal Resources. Mice were maintained on standard rodent chow and water in a temperature- and light-controlled environment and used at 8 to 12 weeks of age. All experiments were performed according to approved Institutional Animal Care and Use Committee procedures.
Viruses
These studies used 2 viruses: wild-type polyoma (PyV.WT) and polyoma virus expressing (OVA)323-339 (PyV.OVA-II), a model CD4+ T-cell ovalbumin epitope. PyV.OVA-II was generated by inserting the I-Ab–restricted (OVA)323-339 (ISQAVHAAHAEINEAGR) epitope from chicken ovalbumin at a unique BlpI site in the genome of mouse polyoma virus (PyV) strain A2 in frame with middle T-antigen. A fragment containing sequence for (OVA)323-339 was generated by polymerase chain reaction (PCR) using high-fidelity Taq polymerase (Invitrogen) with an overhanging forward primer encoding (OVA)323-339 and the BlpI recognition sequence (F: 5′-GTGTTGCTGAGCATCTCACAAGCTGTTCATGCAGCACACGCGGAAACAACGAAGCGGGAAGA AGCCCGATGACACGATATCC-3′, BlpI site underlined) and a reverse primer that included an EcoRI recognition site (R: 5′-TCAGAATTCGGGCCTGAACTTCC-3′, EcoRI site underlined). Genomic DNA from mouse polyoma virus (PyV) strain A2 was digested by BamHI and EcoRI to yield a small fragment and a large fragment, which were subcloned into pUC19. The PCR product encoding (OVA)323-339 was digested with BlpI and EcoRI then cloned into the BlpI/EcoRI region of small fragment in place of the original sequence. Insertion of (OVA)323-339 was confirmed by DNA sequencing. The large fragment and the recombinant small fragment were excised from pUC19 and ligated to form full-length recombinant PyV.OVA-II DNA. PyV.OVA-II DNA was transfected into primary baby mouse kidney cells by Lipofectamine 2000 (Invitrogen). Virus stocks were prepared on baby mouse kidney cells and titered by plaque assay using BALB/3T3 clone A31 cells (ATCC), as previously described.20 Full-length PyV.OVA-II DNA was sequenced to confirm retention of the (OVA)323-339 insert.
Treatment of mice and transfusion of blood
C57BL/6 or C57BL/6 × B10.BR recipient mice were infected with 2 × 105 pfu of PyV.WT or PyV.OVA-II via footpad injection. Two weeks after infection, recipients were transfused with 70 μL of packed mHEL, FVB, or HOD RBCs, as previously described.10 Given the ubiquitous expression of mHEL on all cells, mHEL RBCs were leukoreduced with Pall neonatal leukoreduction filters prior to transfusion, as previously described.10 Sera were obtained before infection, before transfusion, and 7 and 14 days after transfusion. Intracellular cytokine staining and quantitative real-time PCR (qRT-PCR) were performed 7 to 10 weeks after infection.
Blast homology search with Kell, Kidd, and Duffy antigens
On October 5, 2009, a blast search was carried out using peptides containing the polymorphisms that constitute the antithetical RBC antigens K/k, Fya/Fyb, and Jka/Jkb. Twenty amino acid inputs were entered into the BLAST tblastn database,21 and the genome search was restricted to microbial sequences. To focus on short, nearly exact matches, the following search parameters were used: word size = 2, expect threshold = 30 000, Matrix = PAM30, Gap Costs = Existence:9 Extension 1; low complexity filter was turned off; and compositional adjustment was set to “no adjustment.”
ELISA for anti-HEL and anti-OVA humoral response
Enzyme-linked immunosorbent assays (ELISAs) for anti-HEL IgG and anti-OVA IgG were performed on sera run in triplicate at dilutions from 1:50 to 1:500 000, as previously described.10 Fold difference in antibody production was determined in the linear range of optical density.
Flow cytometric cross-matching
Sera from experimental mice were diluted 1:10 and incubated with HOD, mHEL, or FVB RBCs in fluorescence activated cell sorter (FACS) buffer (phosphate-buffered saline + 0.2 mg/mL bovine serum albumin [Sigma-Aldrich] + 0.9 mg/mL ethylenediaminetetraacetic acid [Sigma-Aldrich] + 2% fetal bovine serum [Hyclone]). Goat anti–mouse immunoglobulins conjugated to allophycocyanin (Becton Dickinson) were used as a secondary antibody.
Intracellular cytokine staining and flow cytometry
Spleens were digested with collagenase (Sigma-Aldrich) and resuspended in complete media (RPMI 1640 + 1% l-glutamine + 1% penicillin/streptomycin + 1% 1M N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid + 1% sodium pyruvate + 1% nonessential amino acids [all from Cellgro] + 2-beta mercaptoethanol [EM Science] + 10% fetal bovine serum [Hyclone]). Splenocytes were restimulated in vitro using the BD Cytofix/Cytoperm plus Fixation/Permeabilization kit with BD GolgiPlug (BD Pharmingen), per the manufacturer's instructions. Briefly, 2 × 106 splenocytes were restimulated with 1μM LT359-368, the dominant Db-restricted CD8+ T-cell epitope for polyoma, in the presence of GolgiPlug. After 5 hours of stimulation, cells were treated with Fc block (2.4G2; BD PharMingen) and stained with a directly conjugated antibody against surface CD8 (clone 53-6.7; eBioscience). Cells were then permeabilized, fixed, and stained intracellularly with a directly conjugated antibody against interferon-γ (IFNγ, clone XMG1.2; BD PharMingen). All staining was performed in FACS buffer. Samples were acquired using a 6-color BD FACSCanto flow cytometer.
Quantitative real-time PCR for viral genome copy level
DNA was extracted from sections of spleen using QIAamp DNA mini kit (QIAGEN). Quantitative real-time PCR (qRT-PCR) was set up as previously described.22 The TaqMan probe used was 5′-AGA CGA AAT CCT TGT GTT GCT GAG C-3′. The forward primer was 5′-CGC ACA TAC TGC TGG AAG AAG A-3′, and the reverse primer was 5′-TCT TGG TCG CTT TCT GGA TAC AG-3′. A standard curve of known polyoma genome copy numbers was used to determine the threshold cycle of detection values for experimental samples, which were used to calculate genome copies per milligram of splenic tissue. The detection limit of this assay is 10 copies of viral genomic DNA.
Adoptive transfer of cells
OT-II spleens were digested in collagenase and resuspended in complete RPMI. Cells were washed twice, counted, and resuspended at 20 × 106 splenocytes/mL in phosphate-buffered saline. C57BL/6.PL-Thy1.1 recipients were injected via lateral tail vein with 10 × 106 total splenocytes. For studies characterizing viral responses, C57BL/6 recipient mice were injected via lateral tail vein with 1 × 105 OT-II splenocytes.
Statistical analysis
Statistical significance was determined using GraphPad Prism software (GraphPad Software; www.graphpad.com) and performing a Student t test for comparison of 2 samples or 2-way repeated measure analysis of variance with a Bonferroni posttest for 3 or more samples with multiple conditions. The measure of statistical significance was set at a P value less than .05.
Results
Homology of microbial peptides and polymorphisms that define human blood group antigens
To investigate potential homology at the peptide level between existing microbial sequence data and clinically relevant human blood group antigens, the BLAST database was searched using peptides from blood group antigens that contain allelic polymorphisms known to give rise to antithetical antigens. Peptides from Kell (K/k), Duffy (Fya/Fyb), and Kidd (Jka/Jkb) antigens were analyzed (Table 1) An 8–amino acid peptide was identified from Haemophilus influenzae with 100% identity to the Cellano (k) antigen. Likewise, a 9–amino acid peptide was found in Bacteroides fragilis that had 100% identity to a rare form of Kell (K) and varied from Cellano only in the amino acid constituting the K/k polymorphism. Finally, an 8–amino acid peptide from Vibrio species with 100% identity to Cellano (k) was observed. For Duffy, a common 8–amino acid identity to Fyb was found in Vibrio cholerae, Yersinia pestis, and Salmonella enterica. An additional 9–amino acid sequence was found that varied from Fya by 1 amino acid. Less significant homology was observed for Kidd antigens, but homologous sequences were observed from Bordetella parapertussis, and species of Bacteroides and Clostridium. For each of the observed sequences, identity/homology was restricted to 8 to 9 amino acids, and no significant homology was noted on flanking amino acids on either N or C ends.
Peptide homology of microbes and polymorphisms that define human blood group antigens
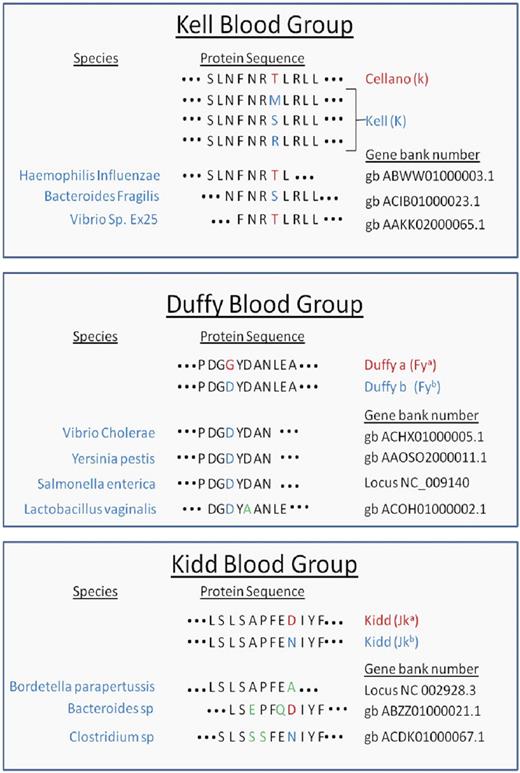
Peptide sequences from Kell, Duffy, and Kidd antigens were entered into the BLAST database as described in “Blast homology search with Kell, Kidd, and Duffy antigens.” Peptide identity and/or homology were detected for the indicated microbes. Red letters denote the predominant amino acids, whereas blue indicate polymorphisms. Green letters highlight amino acid variants between the blood group antigen and microbial amino acid sequences.
Antecedent viral infection enhances initial humoral alloimmunization to subsequent RBC transfusion
The homology data presented in Table 1 suggest that in at least some humans (depending upon HLA), certain microbial peptides may mimic CD4+ T-cell epitopes from blood group antigens, without themselves constituting the antibody binding epitope of humoral blood group antigens. The significance of such homology in small peptide epitopes depends upon the capacity of variant peptides on RBC antigens to display linked recognition characteristics with associated humoral alloantigens. We assessed this capacity by engineering a murine model system to test the hypothesis that a microbe expressing a CD4+ T-cell epitope from a blood group antigen can prime a recipient mouse for a strong humoral response to a subsequent RBC transfusion. Based upon this hypothesis, 2 outcomes can be predicted: (1) the microbial infection itself will not result in an immunization event that is detectable by serology (ie, no anti-RBC antigen antibodies), and (2) the microbial infection will convert an animal that would have otherwise had no response or a very weak response to a given RBC antigen into a strong responder.
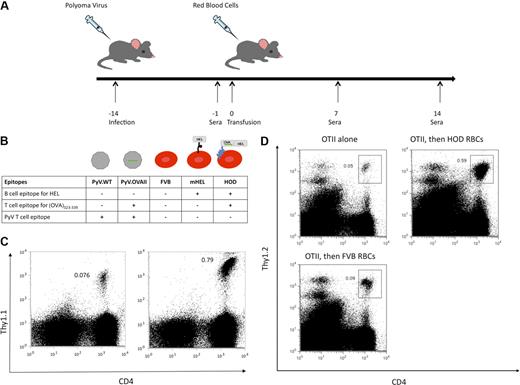
The experimental design to test this hypothesis consisted of infection of recipient mice with either polyoma virus encoding (OVA)323-339 (PyV.OVA-II) or control strain wild-type polyoma virus (PyV.WT). At 2 weeks after infection, recipient mice were then transfused with RBCs from transgenic HOD or control FVB donors (Figure 1A). The HOD mouse expresses a model antigen consisting of a well-characterized B-cell epitope, hen egg lysozyme (HEL), fused in frame with a portion of ovalbumin containing the (OVA)323-339 T-cell epitope, and anchored to the RBC membrane by a human blood group antigen (Duffy; Figure 1B). Thus, the humoral HEL epitope is linked to (OVA)323-339 on the same molecule; HOD is expressed selectively on RBCs and is not detectable on leukocytes or platelets.19
Experimental design and presentation of (OVA)323-339 peptide after exposure to either PyV.OVA-II or HOD RBCs. Recipient mice were infected with wild-type polyoma virus (PyV.WT) or polyoma virus expressing a known T-cell epitope (ovalbumin (OVA)323-339), PyV.OVA-II. Two weeks after infection, recipient mice were transfused with RBCs from control FVB donors (expressing no HEL epitope), mHEL donors (expressing HEL on RBCs), or HOD donors (expressing RBC-specific HEL fused to OVA). (A) Diagram of experimental design: Sera were collected prior to transfusion and at 7 and 14 days after transfusion. (B) Diagram of B- and T-cell epitopes expressed in PyV.WT and PyV.OVA-II, as well as in FVB RBCs, mHEL RBCs, and HOD RBCs. Presence of the (OVA)323-339 epitope is indicated by the green line. (C) OT-II Thy1.1 splenocytes were adoptively transferred into C56BL/6 mice followed by infection with either PyV.WT (C left) or PyV.OVA-II (C right). At 8 days after infection, splenocytes were harvested and stained for CD4 and Thy1.1. (D) OT-II splenocytes were adoptively transferred into C56BL/6.PL-Thy1.1 mice followed 24 hours later by transfusion with either HOD RBCs or FVB RBCs. At 4 days after transfusion, splenocytes were harvested and stained for CD4 and Thy1.2. Groups included mice receiving OT-II cells alone (left), OT-II cells and HOD transfusion (right), or OT-II cells and FVB transfusion (bottom).
Experimental design and presentation of (OVA)323-339 peptide after exposure to either PyV.OVA-II or HOD RBCs. Recipient mice were infected with wild-type polyoma virus (PyV.WT) or polyoma virus expressing a known T-cell epitope (ovalbumin (OVA)323-339), PyV.OVA-II. Two weeks after infection, recipient mice were transfused with RBCs from control FVB donors (expressing no HEL epitope), mHEL donors (expressing HEL on RBCs), or HOD donors (expressing RBC-specific HEL fused to OVA). (A) Diagram of experimental design: Sera were collected prior to transfusion and at 7 and 14 days after transfusion. (B) Diagram of B- and T-cell epitopes expressed in PyV.WT and PyV.OVA-II, as well as in FVB RBCs, mHEL RBCs, and HOD RBCs. Presence of the (OVA)323-339 epitope is indicated by the green line. (C) OT-II Thy1.1 splenocytes were adoptively transferred into C56BL/6 mice followed by infection with either PyV.WT (C left) or PyV.OVA-II (C right). At 8 days after infection, splenocytes were harvested and stained for CD4 and Thy1.1. (D) OT-II splenocytes were adoptively transferred into C56BL/6.PL-Thy1.1 mice followed 24 hours later by transfusion with either HOD RBCs or FVB RBCs. At 4 days after transfusion, splenocytes were harvested and stained for CD4 and Thy1.2. Groups included mice receiving OT-II cells alone (left), OT-II cells and HOD transfusion (right), or OT-II cells and FVB transfusion (bottom).
The model reagents used in the experimental design were tested to determine whether the (OVA)323-339 peptide was processed and presented on MHC II. CD4+ T cells were harvested from mice that express a TCR transgene specific for (OVA)323-339 presented by I-Ab (OT-II mice).24,25 Splenocytes from OT-II mice on a Thy1.1 background were adoptively transferred into C57BL/6 mice followed by PyV.WT or PyV.OVA-II infection. After 8 days, splenocytes were harvested and stained for CD4 and Thy1.1. OT-II cells expanded 10-fold in mice infected with PyV.OVA-II compared with mice infected with PyV.WT (Figure 1C). Similarly, to test whether HOD protein on RBCs can be processed such that (OVA)323-339 is presented on MHC II, OT-II cells were adoptively transferred into C57BL/6.PL-Thy1.1 mice followed by a HOD RBC or control FVB RBC transfusion 24 hours later. After 4 days, splenocytes were harvested and stained for CD4 and Thy1.2. OT-II cells expanded 10-fold in mice that received a HOD RBC transfusion compared with mice that received OT-II cells alone. OT-II expansion was specific to peptides presented from the HOD antigen as there was a lack of expansion in mice that received FVB RBC transfusion (Figure 1D). Thus, the (OVA)323-339 peptide from both the HOD antigen and the PyV.OVA-II virus can be efficiently processed and presented on MHC II of antigen-presenting cells.
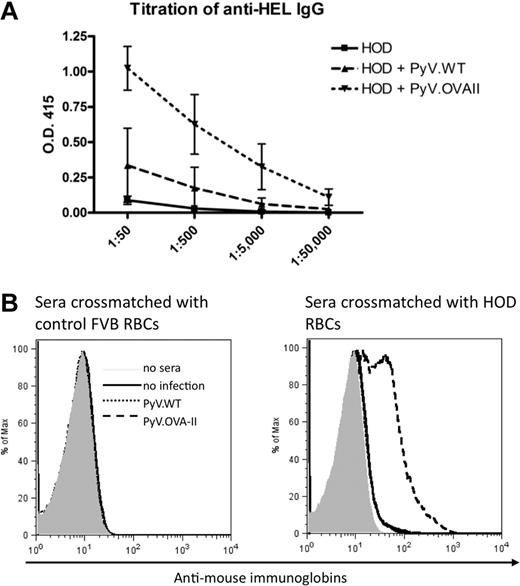
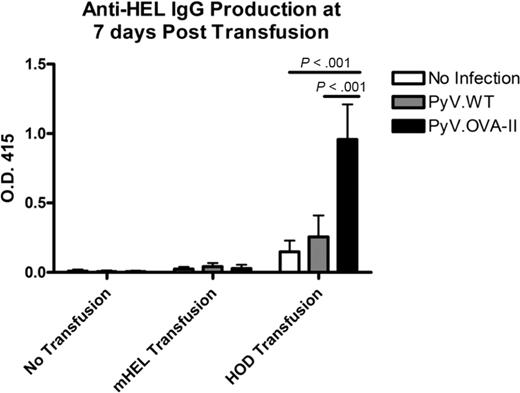
The experimental design shown in Figure 1A was carried out using the characterized reagents. In 3 of 3 experiments (5 mice/group per experiment), mice infected with PyV.OVA-II prior to HOD RBC transfusion had a dramatic increase in anti-HEL IgG compared with control mice that received PyV.WT prior to HOD RBCs or HOD RBCs alone (P < .05 at 1:50 dilution; Figure 2A). Titration of sera from 1:50 to 1:50 000 and extrapolation on the linear part of the curve indicated a 1000-fold higher level of anti-HEL IgG in mice infected with PyV.OVA-II prior to HOD RBC transfusion compared with mice receiving HOD RBCs alone. No anti-OVA IgG was detected in any group, indicating that HEL is a highly immunodominant antibody epitope for the HOD antigen (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Consistent with our prior observation that inflammation in and of itself can enhance RBC alloimmunization,10 infection with PyV.WT resulted in a small, reproducible, but statistically insignificant degree of enhancement (Figure 2A). Taking into account this inflammatory effect, PyV.OVA-II still had a 100-fold greater enhancement of anti-HEL IgG than infection with PyV.WT. To assess the ability of the antibody response to bind native HOD antigen on the RBC surface, flow cytometric cross-match was performed using indirect immunofluorescence. Antibodies that bound HOD RBCs were detected in sera from mice infected with PyV.OVA-II prior to HOD RBC transfusion (Figure 2B). This binding was specific to the HOD antigen, as no signal was observed with RBCs from wild-type FVB donors. Likewise, no anti-HOD RBC antibodies were detected by flow cytometric cross-match in mice that were uninfected or infected with PyV.WT prior to HOD RBC transfusion. Lack of detectable antibody in these latter 2 conditions is consistent with our previous report that the ELISA is substantially more sensitive than flow cross-match.10
Prior infection with PyV.OVA-II significantly enhances alloimmunization to a subsequent HOD RBC transfusion. C57BL/6 mice were infected with wild-type polyoma virus (PyV.WT) or polyoma virus expressing (OVA)323-339 (PyV.OVA-II). Additional control mice were uninfected. All groups received subsequent transfusion with HOD RBCs. Alloimmunization was assessed 2 weeks later by anti-HEL ELISA. (A) A representative experiment with 5 mice/group is shown, with mean ± SEM shown at sera dilutions of 1:50 to 1:50 000. (B) Enhancement was also evident by flow cytometric cross-matching with FVB or HOD RBC targets. These experiments have been reproduced 3 times (5 mice/group per experiment) with similar results.
Prior infection with PyV.OVA-II significantly enhances alloimmunization to a subsequent HOD RBC transfusion. C57BL/6 mice were infected with wild-type polyoma virus (PyV.WT) or polyoma virus expressing (OVA)323-339 (PyV.OVA-II). Additional control mice were uninfected. All groups received subsequent transfusion with HOD RBCs. Alloimmunization was assessed 2 weeks later by anti-HEL ELISA. (A) A representative experiment with 5 mice/group is shown, with mean ± SEM shown at sera dilutions of 1:50 to 1:50 000. (B) Enhancement was also evident by flow cytometric cross-matching with FVB or HOD RBC targets. These experiments have been reproduced 3 times (5 mice/group per experiment) with similar results.
To test the possibility that PyV.OVA-II infection induced anti-HEL IgG on its own (and transfusion with HOD RBCs is just boosting a secondary antibody response), specimens collected after infection but prior to transfusion were analyzed; no anti-HEL antibodies were detected in any group (supplemental Figure 2). Mice infected with the virus but not transfused were tested as late as 18 weeks after infection and no anti-HEL IgG was detected (data not shown). Together, these data demonstrate that a combination of infection with PyV.OVA-II and subsequent transfusion with HOD RBCs is required for the observed enhancement of anti-HEL IgG.
Analysis of immune responses to PyV.WT and PyV.OVA-II
Based on the presented data, we hypothesized that PyV.OVA-II induced a CD4+ T-cell response, which was then able to provide T-cell help to B cells that encountered the HOD antigen after RBC transfusion. However, it was also possible that the addition of (OVA)323-339 to PyV.OVA-II created a more virulent strain that could serve as a stronger adjuvant to any antigen encountered. Alternatively, it was possible that failure of Py.WT to enhance anti-HEL IgG was due to lack of infectivity. To assess these possibilities, viral infection and cellular immune response were assessed.
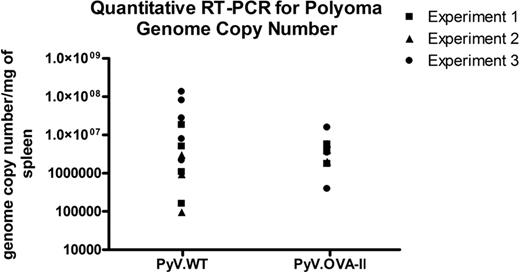
Quantitative RT-PCR was performed to enumerate polyoma genome copy number. DNA was amplified using polyoma-specific primers and probes that recognized both PyV.WT and PyV.OVA-II. The threshold cycle values were used to calculate the genome copy number per milligram of spleen. In 3 of 3 experiments (n = 3-5 mice/group per experiment), there was no statistically significant difference between polyoma genome copy number in recipient mice of PyV.WT or PyV.OVA-II infection (Figure 3). RT-PCR was specific for viral infection, as mice that received HOD RBCs alone (uninfected) had no detectable polyoma DNA (data not shown). These data indicate that the decreased enhancement of HOD alloimmunization by PyV.WT infection was not due to a lack of infectivity.
Quantitative real-time PCR enumeration of viral genome copy number. DNA was extracted from splenic tissue taken 7 to 10 weeks after infection from recipient mice infected with wild-type polyoma virus (PyV.WT) or polyoma virus expressing (OVA)323-339 (PyV.OVA-II) and then transfused with HOD RBCs. Quantitative RT-PCR was used to determine the polyoma virus genome copy number. No statistically significant difference in genome copy number between the 2 groups was seen. A compilation of data from 3 individual experiments (3-5 mice/group per experiment) is shown; uninfected mice had an undetectable viral genome copy number (data not shown).
Quantitative real-time PCR enumeration of viral genome copy number. DNA was extracted from splenic tissue taken 7 to 10 weeks after infection from recipient mice infected with wild-type polyoma virus (PyV.WT) or polyoma virus expressing (OVA)323-339 (PyV.OVA-II) and then transfused with HOD RBCs. Quantitative RT-PCR was used to determine the polyoma virus genome copy number. No statistically significant difference in genome copy number between the 2 groups was seen. A compilation of data from 3 individual experiments (3-5 mice/group per experiment) is shown; uninfected mice had an undetectable viral genome copy number (data not shown).
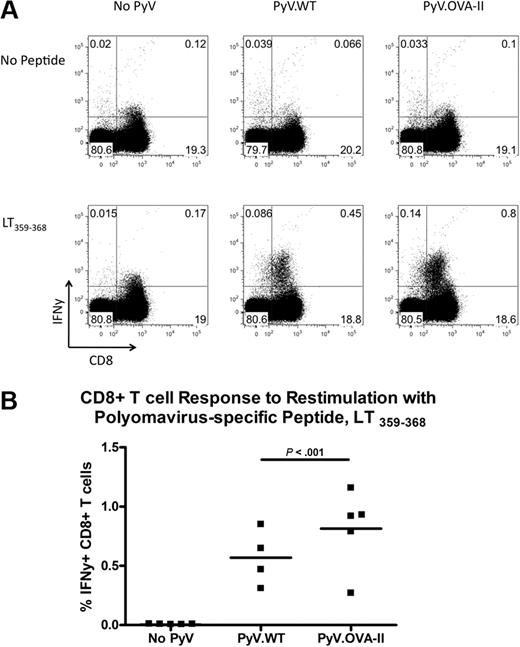
The similar viral genome copy number suggested a similar immunologic clearance of either strain of virus. However, to quantify the extent of antiviral immunity, intracellular cytokine staining was performed. Splenocytes were restimulated in vitro with the immunodominant Db-restricted PyV CD8+ T-cell epitope (LT359-368), and then surface stained for CD8 and intracellularly stained for IFNγ. Infection with either strain resulted in an increase in CD8+ T cells that expressed IFNγ compared with background levels in uninfected mice (Figure 4A). However, in 2 of 3 experiments, there was a significant increase in the percentage of viral-specific CD8+ T cells in PyV.OVA-II–infected mice compared with PyV.WT-infected mice (Figure 4B; representative experiment). Together, these data indicate that both PyV strains were infectious and induced a detectable CD8+ T-cell antiviral response, excluding the possibility that enhancement of anti-HEL IgG by PyV.OVA-II and not PyV.WT was due to a lack of viral infectivity by PyV.WT.
Enumeration of CD8+ T-cell antiviral responses to both PyV.WT and PyV.OVA-II. Intracellular cytokine staining was performed on splenocytes 7 to 10 weeks after infection with either wild-type PyV.WT or PyV.OVA-II; uninfected controls were also analyzed. Splenocytes were restimulated with and without a dominant MHC class I–restricted PyV peptide (LT359-368) and stained with anti-IFNγ. Representative flow plots are shown (A), and compiled data are presented from a representative experiment (B). This experiment has been repeated 3 times (3-5 mice/group per experiment). In all cases, CD8+ T cells from infected mice expressed IFNγ upon peptide stimulation. In 2 of 3 experiments, a greater percentage of IFNγ-producing CD8+T cells was seen in PyV.OVA-II–infected mice compared with PyV.WT-infected mice; the data shown are from a representative experiment displaying this difference.
Enumeration of CD8+ T-cell antiviral responses to both PyV.WT and PyV.OVA-II. Intracellular cytokine staining was performed on splenocytes 7 to 10 weeks after infection with either wild-type PyV.WT or PyV.OVA-II; uninfected controls were also analyzed. Splenocytes were restimulated with and without a dominant MHC class I–restricted PyV peptide (LT359-368) and stained with anti-IFNγ. Representative flow plots are shown (A), and compiled data are presented from a representative experiment (B). This experiment has been repeated 3 times (3-5 mice/group per experiment). In all cases, CD8+ T cells from infected mice expressed IFNγ upon peptide stimulation. In 2 of 3 experiments, a greater percentage of IFNγ-producing CD8+T cells was seen in PyV.OVA-II–infected mice compared with PyV.WT-infected mice; the data shown are from a representative experiment displaying this difference.
Enhancement of alloimmunization to HOD transfusion by PyV.OVA-II requires linkage of (OVA)323-339 to HEL
We hypothesized that PyV.OVA-II induced CD4+ T cells specific for (OVA)323-339 presented by MHC II, resulting in enhanced anti-HEL IgG responses to HOD RBC transfusion due to antigen-specific linked recognition. However, the increased CD8+ T-cell response to infection with PyV.OVA-II compared with PyV.WT (Figure 4) raised the possibility that increased anti-HEL IgG in response to HOD after PyV.OVA-II was simply an adjuvant effect. To directly test the requirements for linked recognition, a cohort of mice was transfused with mHEL RBCs instead of HOD RBCs. mHEL RBCs express the same HEL epitope on their surface fused to a transmembrane domain, but lack fusion to OVA, and thus are not linked to the (OVA)323-339 peptide (Figure 1B).26 In contrast to what was observed with HOD RBCs, in 2 of 2 experiments (5 mice/group per experiment, using both C57BL/6 as well as C57BL/6 × B10.BR recipients), no enhancement of anti-HEL IgG in response to mHEL RBC transfusion was observed in mice previously infected with PyV.OVA-II, compared with uninfected or PyV.WT-infected controls at 7 days (Figure 5) or 14 days (data not shown) after transfusion. In aggregate, these findings demonstrate that anti-HEL IgG responses to HEL on transfused RBCs are not enhanced by antecedent infection with PyV.OVA-II when (OVA)323-339 is not linked to the HEL.
Enhanced alloimmunization to HOD transfusion by PyV.OVA-II requires linkage of (OVA)323-339 to HEL on the antigen. C57BL/6 mice were infected with PyV.WT or PyV.OVA-II and transfused with mHEL or HOD RBCs (uninfected and/or untransfused control groups were also included). One week after transfusion, sera were analyzed for anti-HEL IgG. Similar results were observed at 14 days (data not shown). A representative experiment with 5 mice/group is shown (mean ± SD depicted, with sera at 1:50 dilution); this experiment has been repeated twice (using C57BL/6 as well as C57BL/6 × B10.BR recipients) with similar results.
Enhanced alloimmunization to HOD transfusion by PyV.OVA-II requires linkage of (OVA)323-339 to HEL on the antigen. C57BL/6 mice were infected with PyV.WT or PyV.OVA-II and transfused with mHEL or HOD RBCs (uninfected and/or untransfused control groups were also included). One week after transfusion, sera were analyzed for anti-HEL IgG. Similar results were observed at 14 days (data not shown). A representative experiment with 5 mice/group is shown (mean ± SD depicted, with sera at 1:50 dilution); this experiment has been repeated twice (using C57BL/6 as well as C57BL/6 × B10.BR recipients) with similar results.
Discussion
Herein, we demonstrate homology between infectious microbes such as H influenzae, Y pestis, and B parapertussis and the RBC antigens Kell, Duffy, and Kidd, respectively. Although the homology was generally restricted to 8– to 9–amino acid stretches, the MHC II molecule has open ends to its presentation pocket (both in humans and mice), which allows substantial promiscuity in anchor residues. Peptides can be of variable lengths (often 13 amino acids), and obligate residues adjacent to the peptide recognized by the T-cell receptor are less constrained than MHC I and more difficult to predict.27 Thus, whether adjacent amino acids allow the peptides to be presented on MHC II is difficult to surmise based on known MHC II biology, and likely varies depending upon the particular HLA type of a given person.28 However, it is likely that at least certain HLA types will be capable of presenting microbial peptides containing the observed homologous sequences.
We hypothesize that exposure to these infectious agents results in activation of CD4+ T cells, which would not be detectable by current serology-based blood-banking methodologies. Subsequent transfusion with RBCs containing a peptide sequence shared with the microbe then leads to RBC alloimmunization, with help from the preformed CD4+ T cells. Thus, molecular mimicry at the CD4+ T-cell level may be a previously unappreciated factor influencing rates of RBC alloimmunization.
To formally test this hypothesis in a model system, we recapitulated in mice the scenario hypothesized to occur in humans. Using a virus containing a CD4+ T-cell epitope shared with a small peptide on the donor RBCs, we have shown that infection with PyV.OVA-II enhances alloimmunization to HOD RBCs (containing linked HEL and OVA peptides). This enhancement requires that both the virus and the RBC antigen share a CD4+ T-cell epitope; the effect is lost if the epitope is absent from the virus (PyV.WT infection followed by HOD transfusion) or if the epitope is absent from the RBC antigen (PyV.OVA-II infection followed by mHEL transfusion).
It is worth noting that the baseline responses to mHEL RBCs are substantially lower than to HOD RBCs (Figure 5). There are several possible reasons, including decreased levels of mHEL transgene expression compared with HOD (data not shown) and lack of additional CD4+ T-cell epitopes provided by the fusion partners in HOD. However, peptide(s) from HEL are known to be efficiently presented by MHC II encoded by the H-2k haplotype. Accordingly, these experiments were carried out in C57BL/6 × B10.BR recipients (H-2b × H-2k) as well as C57BL/6 recipients (H-2b), with similar results. We have previously reported that anti-HEL responses to mHEL transfusion can be significantly enhanced by inflammation (with poly (I:C))10 and are also markedly enhanced by increasing CD4+ T cells specific for a peptide contained in HEL itself.12 Thus, lack of increased anti-HEL in response to mHEL was not due to an inability to be enhanced by either inflammation or increased CD4+ helper T cells.
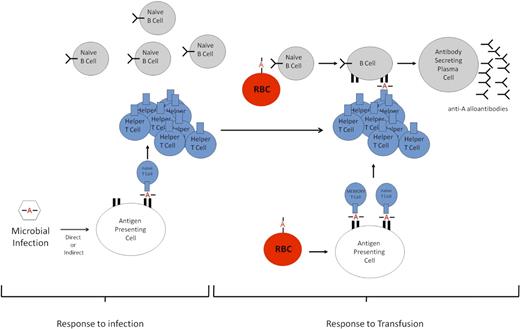
Our interpretation of these data is that after HOD transfusion, anti-HEL B cells selectively recognize and, through receptor-mediated endocytosis, internalize the HOD antigen (whole or partial RBC) via their HEL-specific B-cell receptor. Because the HEL antigen is linked to OVA, both antigens are endocytosed, broken down into peptides, and presented in the MHC II of B cells as well as professional antigen-presenting cells (ie, dendritic cells). We hypothesize that (OVA)323-339–specific CD4+ T cells that were previously generated in response to the PyV.OVA-II infection persist, recognizing OVA peptide presented by MHC II on anti-HEL B cells that have processed HOD antigen. Through this interaction, anti-HEL B cells are activated and differentiate into antibody-secreting plasma cells with the help of viral-induced (OVA)323-339-specific CD4+ T cells (Figure 6).
Schematic of proposed enhanced alloimmunization. Response to infection. Peptides containing a polymorphism (designated by A) from a microbe will be processed and presented by host antigen-presenting cells; peptides presented in MHC II will be recognized by CD4+ T cells. However, in the absence of a B-cell epitope, the B cells will not be able to receive CD4+ T-cell help to generate an antibody response. Response to transfusion. Upon a second antigenic exposure (transfusion) that contains the same polymorphism A, the polymorphism in the blood group constitutes not only a CD4+ T-cell epitope but also a de novo B-cell epitope. Through receptor-mediated endocytosis, naive B cells phagocytose the polymorphism-containing blood group molecule. The polymorphism is then presented on the MHC II of B cells to the preformed helper or memory CD4+ T cells generated against the microbial infection. The B cells are then stimulated to differentiate into plasma cells that secrete antibodies against the polymorphism.
Schematic of proposed enhanced alloimmunization. Response to infection. Peptides containing a polymorphism (designated by A) from a microbe will be processed and presented by host antigen-presenting cells; peptides presented in MHC II will be recognized by CD4+ T cells. However, in the absence of a B-cell epitope, the B cells will not be able to receive CD4+ T-cell help to generate an antibody response. Response to transfusion. Upon a second antigenic exposure (transfusion) that contains the same polymorphism A, the polymorphism in the blood group constitutes not only a CD4+ T-cell epitope but also a de novo B-cell epitope. Through receptor-mediated endocytosis, naive B cells phagocytose the polymorphism-containing blood group molecule. The polymorphism is then presented on the MHC II of B cells to the preformed helper or memory CD4+ T cells generated against the microbial infection. The B cells are then stimulated to differentiate into plasma cells that secrete antibodies against the polymorphism.
Additional considerations include the possibility that PyV.OVA-II had a B-cell epitope that mimicked HEL, such that PyV.OVA-II infection induced a primary anti-HEL antibody response and the enhancement of immunization with HOD RBC transfusion was actually due to boosting. However, we reject this hypothesis because no anti-HEL antibody was detected after PyV.OVA-II infection in the absence of transfusion (up to 18 weeks after inoculum). Moreover, there was no increase of anti-HEL in response to mHEL RBC transfusion, demonstrating that introducing the HEL B-cell epitope(s) was not sufficient for augmentation by PyV.OVA-II. Finally, we rule out that the failure of the PyV.WT to enhance anti-HEL was an artifact due to the viral stock not being infectious, as a strong CD8+ T-cell antiviral response was observed and viral genome copy number was consistent with an active infection.
As the current genomic databases for microbes are far from complete, discovery of additional homology with ongoing microbial sequence analysis is possible. Moreover, the likelihood of peptide mimicry is increased by the recent appreciation of additional polymorphisms in alloantigens outside of those traditionally detected by patient serology.29 However, cross-reactivity alone would not be sufficient, as the peptide in question would have to fit into the particular MHC encoded by a given recipient's HLA. Although monitoring of CD4+ T cell–based immunity is not currently available in the routine clinical setting, it may be possible to predict a response to a future alloantigen based on prior pathogen exposure or gut flora. Furthermore, identification of environmental antigens that cross-react with alloantigen CD4+ T-cell epitopes may lead to the development of specific diagnostic MHC II tetramers capable of detecting CD4+ T cell–based immunization to relevant peptides. Together, these advancements would help identify patients at high risk for humoral alloimmunization and provide peptide-based therapeutic interventions.
In summary, the current findings provide additional insight into factors that may regulate RBC alloimmunization. A patient may be primed to respond to an RBC antigen through prior exposure to a microbial infection with a shared CD4+ T-cell epitope, yet this initial priming would go undetected by current serology-based blood-banking methodologies. Although the described studies use an RBC transfusion model, the principle demonstrated could extend to platelet and HLA antigens, with potential relevancy to both solid organ and bone marrow transplantation. Future comprehensive genomic and proteomic studies, in the context of a growing database of known sequences, will be useful to investigate potential homology between microbial peptides and RBC, white blood cell, and platelet antigens.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Chantel Cadwell and Nicole Smith for excellent technical assistance.
This study was funded in part by grants from the National Institutes of Health (P01 HL086773 [J.C.Z.] and R01 CA071971 [A.E.L.]).
National Institutes of Health
Authorship
Contribution: K.E.H., J.E.H., and J.C.Z. designed experiments and wrote the paper; K.E.H. and J.E.H. performed experiments, analyzed data, and made figures; and E.L. and A.E.L. contributed vital reagents.
Conflict-of-interest disclosure: J.C.Z. has a research grant from Immucor Inc for a project unrelated to the research in this paper. The remaining authors declare no competing financial interests.
Correspondence: James C. Zimring, Center for Transfusion and Cellular Therapies, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Woodruff Memorial Bldg, Ste 7107, 101 Woodruff Cir, Atlanta, GA 30322; e-mail: jzimrin@emory.edu.