Abstract
Global hypomethylation and regional hypermethylation are well-known epigenetic features of cancer; however, in chronic lymphocytic leukemia (CLL), studies on genome-wide epigenetic modifications are limited. Here, we analyzed the global methylation profiles in CLL, by applying high-resolution methylation microarrays (27 578 CpG sites) to 23 CLL samples, belonging to the immunoglobulin heavy-chain variable (IGHV) mutated (favorable) and IGHV unmutated/IGHV3-21 (poor-prognostic) subsets. Overall, results demonstrated significant differences in methylation patterns between these subgroups. Specifically, in IGHV unmutated CLL, we identified methylation of 7 known or candidate tumor suppressor genes (eg, VHL, ABI3, and IGSF4) as well as 8 unmethylated genes involved in cell proliferation and tumor progression (eg, ADORA3 and PRF1 enhancing the nuclear factor-κB and mitogen-activated protein kinase pathways, respectively). In contrast, these latter genes were silenced by methylation in IGHV mutated patients. The array data were validated for selected genes using methylation-specific polymerase chain reaction, quantitative reverse transcriptase–polymerase chain reaction, and bisulfite sequencing. Finally, the significance of DNA methylation in regulating gene promoters was shown by reinducing 4 methylated tumor suppressor genes (eg, VHL and ABI3) in IGHV unmutated samples using the methyl-inhibitor 5-aza-2′-deoxycytidine. Taken together, our data for the first time reveal differences in global methylation profiles between prognostic subsets of CLL, which may unfold epigenetic silencing mechanisms involved in CLL pathogenesis.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by clonal proliferation and accumulation of long-lived neoplastic B cells, which arise due to altered control of apoptosis as well as proliferation. No common disease-causing mutation has yet been identified in CLL, although certain recurrent genomic aberrations (ie, 11q−, +12, 13q−, 17p−) provide important prognostic information.1 Besides these aberrations, the mutation status of the immunoglobulin heavy-chain variable (IGHV) genes segregates patients into 2 main prognostic subgroups, where patients with unmutated IGHV genes show inferior outcome compared with those with mutated IGHV genes.2,3 In addition, CLL patients using the IGHV3-21 gene display poor outcome independent of mutation status.4
Aberrant DNA methylation has been shown to play a strong role in tumorigenesis, where genome-wide hypomethylation and regional hypermethylation of tumor suppressor gene promoters are characteristic hallmarks of many cancers.5 In CLL, the epigenetic mechanism of gene regulation has thus far received limited attention. That notwithstanding, a strong correlation between promoter methylation and transcriptional silencing was shown for certain individual gene promoters in CLL, for example, DAPK1, TWIST2, ZAP70, and HoxA4.6-9 The methylation status of several of these genes has also been reported to correlate with IGHV mutational status. To date, only one study has been performed using genome-wide methylation analysis in CLL using the “Restriction Landmark Genomic Scanning” technique. They observed that 2% to 8% of CpG islands were aberrantly methylated compared with normal controls and that methylation events showed a nonrandom distribution.10 However, this technique spans only 3000 CpG islands and does not give a full coverage of the genome.
The main aim of this present investigation was to study and compare the genome-wide methylation profiles of different CLL prognostic subgroups to identify differentially methylated genes that can play a role in CLL pathogenesis. Recent studies have revealed that DNA hypermethylation in cancer is not always restricted to discrete CpG islands or single genes but can affect multiple adjacent CpG-rich regions resulting in gene silencing across large genomic distances.11 Unlike promoter CpG island methylation arrays that cover sites near the promoter region, we here applied an Illumina Infinium HumanMethylation27 array (27 578 methylated sites, covering 14 495 genes) to 23 CLL patient samples belonging to the IGHV mutated, IGHV unmutated, and IGHV3-21 CLL subgroups. We identified distinct methylation profiles between different subgroups and the array data were validated for selected genes using methylation-specific primer (MSP) polymerase chain reaction (MSP-PCR), real-time quantitative PCR (RQ-PCR) and bisulfite sequencing. Finally, methylated genes were also reexpressed in CLL patient samples using methyl inhibitors to ascertain the role of DNA methylation in regulating gene expression.
Methods
Patient samples and clinical data
In this study, 23 tumor samples from patients with CLL (6 IGHV mutated, 7 IGHV unmutated, and 10 IGHV3-21) were collected from the Biobank at the Department of Pathology, Uppsala University Hospital (Uppsala, Sweden). All samples were diagnosed according to recently revised criteria,12 showing typical CLL immunophenotype and 70% or more tumor cells. Clinical and molecular characteristics including IGHV mutation status and genomic aberrations are summarized in Table 1. Two normal healthy control samples (1 sample with peripheral blood mononuclear cells [PBMC]) and 1 sample with CD19 sorted B cells) and 1 negative control (whole-genome amplified DNA) were included for analysis. In addition, 2 CLL Epstein-Barr virus (EBV)–transformed cell lines (I83 derived from a mutated CLL patient and HG3 derived from an unmutated CLL patient) were included for analysis. Finally, 10 additional CLL samples (5 IGHV mutated and 5 IGHV unmutated) were included in the validation experiments using MSP-PCR and another 50 CLL samples (27 IGHV mutated and 23 IGHV unmutated) were included in RQ-PCR analysis. None of these 60 latter samples were included in methylation array analysis. Informed consent was obtained according to the Declaration of Helsinki and the study was approved by the ethical review committee of Uppsala University.
Clinical and molecular data for the CLL patients included
| CLL sample . | Percent identity* . | Age at diagnosis, y . | Sex . | Survival, mo† . | Binet stage . | Treated before sampling‡ . | Genomic aberrations§ . |
|---|---|---|---|---|---|---|---|
| IGHV mutated | |||||||
| M1 | 94.6 | 63 | M | 195¶ | A | No | del(13q)3 |
| M2 | 91.5 | 74 | M | 55 | A | No | del(13q)1 |
| M3‖ | 93.2 | 72 | M | 129¶ | A | ND | None3 |
| M4 | 94.1 | 70 | M | 134¶ | A | No | del(13q)3 |
| M5 | 91.3 | 65 | M | 104¶ | A | No | del(13q)1 |
| M6 | 91.1 | 64 | F | 94¶ | A | No | None3 |
| IGHV unmutated | |||||||
| UM1 | 100.0 | 49 | M | 99 | A | Yes | del(11q)3 |
| UM2 | 100.0 | 44 | M | 84 | B | ND | del(11q)3 |
| UM3 | 100.0 | 79 | M | 10 | B | No | ND |
| UM4 | 100.0 | 82 | F | 50 | B | No | ND |
| UM5 | 100.0 | 57 | M | 49 | B | No | del(13q)1 |
| UM6 | 100.0 | 69 | M | 24 | C | No | None3 |
| UM7 | 100.0 | 51 | M | 112¶ | A | No | None1 |
| IGHV3-21 | |||||||
| 3-21-1 | 100.0 | 45 | F | 58 | ND | ND | +122 |
| 3-21-2 | 100.0 | 63 | M | 58 | C | No | del(13q), del(11q)1 |
| 3-21-3 | 96.50# | 61 | M | 52 | C | No | del(13q),del(11q)1 |
| 3-21-4 | 96.9# | 62 | F | 67 | ND | No | del (13q)2 |
| 3-21-5 | 100.0 | 65 | F | 52 | ND | Yes | del(17p),+121 |
| 3-21-6 | 98.7# | 67 | M | ND | ND | ND | del(13q),del(11q)1 |
| 3-21-7 | 96.49# | 63 | M | ND | ND | ND | del(13q)3 |
| 3-21-8 | 95.5# | 60 | M | 72 | B | No | None1 |
| 3-21-9 | 96.8# | 62 | M | 111 | ND | ND | del(13q)2 |
| 3-21-10 | 97.4# | 76 | M | 82 | A | No | del(13q), del(11q)2 |
| CLL sample . | Percent identity* . | Age at diagnosis, y . | Sex . | Survival, mo† . | Binet stage . | Treated before sampling‡ . | Genomic aberrations§ . |
|---|---|---|---|---|---|---|---|
| IGHV mutated | |||||||
| M1 | 94.6 | 63 | M | 195¶ | A | No | del(13q)3 |
| M2 | 91.5 | 74 | M | 55 | A | No | del(13q)1 |
| M3‖ | 93.2 | 72 | M | 129¶ | A | ND | None3 |
| M4 | 94.1 | 70 | M | 134¶ | A | No | del(13q)3 |
| M5 | 91.3 | 65 | M | 104¶ | A | No | del(13q)1 |
| M6 | 91.1 | 64 | F | 94¶ | A | No | None3 |
| IGHV unmutated | |||||||
| UM1 | 100.0 | 49 | M | 99 | A | Yes | del(11q)3 |
| UM2 | 100.0 | 44 | M | 84 | B | ND | del(11q)3 |
| UM3 | 100.0 | 79 | M | 10 | B | No | ND |
| UM4 | 100.0 | 82 | F | 50 | B | No | ND |
| UM5 | 100.0 | 57 | M | 49 | B | No | del(13q)1 |
| UM6 | 100.0 | 69 | M | 24 | C | No | None3 |
| UM7 | 100.0 | 51 | M | 112¶ | A | No | None1 |
| IGHV3-21 | |||||||
| 3-21-1 | 100.0 | 45 | F | 58 | ND | ND | +122 |
| 3-21-2 | 100.0 | 63 | M | 58 | C | No | del(13q), del(11q)1 |
| 3-21-3 | 96.50# | 61 | M | 52 | C | No | del(13q),del(11q)1 |
| 3-21-4 | 96.9# | 62 | F | 67 | ND | No | del (13q)2 |
| 3-21-5 | 100.0 | 65 | F | 52 | ND | Yes | del(17p),+121 |
| 3-21-6 | 98.7# | 67 | M | ND | ND | ND | del(13q),del(11q)1 |
| 3-21-7 | 96.49# | 63 | M | ND | ND | ND | del(13q)3 |
| 3-21-8 | 95.5# | 60 | M | 72 | B | No | None1 |
| 3-21-9 | 96.8# | 62 | M | 111 | ND | ND | del(13q)2 |
| 3-21-10 | 97.4# | 76 | M | 82 | A | No | del(13q), del(11q)2 |
ND indicates not defined.
IGHV gene analysis was performed using PCR amplification and nucleotide sequencing as previously described.13 Cases with ≥ 98% identity to germline were classified as unmutated, whereas cases with < 98% identity were considered mutated. IGHV3-21 cases were classified as stereotyped (#) or nonstereotyped according to Murray et al.13
indicates months from diagnosis to last observation or death; and ¶ indicates that the patient is alive.
denotes treatment status before sampling; yes indicates that the patient was treated before sampling; and no indicates that the patient remained untreated.
Known recurrent aberrations, that is, del(13q), del(11q), del(17p) and trisomy 12, were determined using fluorescence in situ hybridization (FISH) analysis (1), SNP-array analysis (2) or quantitative PCR analysis (3). FISH analysis was performed using commercially available probes as decribed previously,14 whereas single nucleotide polymorphism (SNP) array analysis was performed using high-resolution Affymetrix 250K SNP arrays from which data on known recurrent genomic aberrations were extracted.14 Quantitative PCR was performed by applying specifically designed primers for each locus (primers available on request) using the SYBR Green master mix according to the manufacturer's protocol (Fermentas). Copy-number changes were normalized against a copy-number neutral internal control region and ratios relative to the DNA copy number from a pooled control (Promega) were tallied for each locus.
A second mutated, in-frame IGHV gene rearrangement (92% identity) was also detected in this case.
Methylation array analysis
We applied the genome-wide Illumina Infinium HumanMethylation27 BeadChip array (Illumina) which allows interrogation of 27 578 CpG dinucleotides, covering 14 495 genes. First, bisulfite conversion of genomic DNA was performed using the EZ DNA Methylation Kit (Zymo Research) according to the manufacturer's protocol. Briefly, 1 μg of DNA was sodium bisulfite treated, denatured at 95°C for 30 seconds, and bisulfite converted at 50°C for 12 hours. After conversion, samples were desulphonated and eluted using column preparation. Approximately 100 to 125 ng of bisulfite-converted DNA was processed according to the Illumina Infinium Methylation Assay protocol. This assay is based on the conversion of unmethylated C-nucleotides into U(T) nucleotides by the bisulfite treatment. The DNA was whole-genome amplified, enzymatically fragmented, precipitated, resuspended, and hybridized overnight at 48°C to locus-specific oligonucleotide primers on the BeadArray. After hybridization, the C or T nucleotides were detected by single-base primer extension. The fluorescence signals corresponding to the C or T nucleotides were measured from the BeadArrays using an Illumina BeadStation GX scanner. The fluorescence data were then analyzed using the BeadStudio software (Illumina), which assigns a quantitative measure of the methylation levels (β value or methylation index [MI] value) for each CpG site, that corresponds to the ratio between the fluorescence signal from the methylated allele (C) and the sum of the fluorescent signals of the methylated (C) and unmethylated (T) alleles. The methylation status for each detected CpG site ranged between 0.1 (completely unmethylated) to 1 (completely methylated). MI cutoffs of 0.7 or more and 0.4 or less (average of all MI values in each group) were considered methylated and unmethylated, respectively.
Further bioinformatic analysis of the methylation data were carried out in the freely available statistical computing language R (http://www.r-project.org). To search for the differentially methylated genes between the different prognostic subgroups (ie, IGHV mutated/unmutated and IGHV3-21 CLL patients), the data were arcsin transformed and an empirical Bayes moderated t test was then applied using the “limma” package. The P values were adjusted using the method of Benjamini and Hochberg15 and a level of P less than .05 was used as a cutoff. To identify differentially methylated genes of largest difference between subgroups, an additional filter incorporating the average geometric difference was applied to ensure only genes with large absolute differences remained. Consequently, an average difference in MI of 0.45 between the IGHV mutated and IGHV unmutated subgroups, 0.35 between the IGHV3-21 and IGHV mutated subgroups, and 0.35 between IGHV3-21 and IGHV unmutated subgroups was applied.
MSP-PCR analysis
To verify the findings from the methylation arrays, we selected 4 genes for MSP-PCR. MSP were designed according to MSP selection criteria as outlined by Li and Dahiya.16 Specifically, sequences covering approximately 250 bp flanking the identified CpG sites on either side were used for primer design in the Methprimer software (Bisearch). The unmethylated-specific primers (USP) were designed around 50 bp upstream and downstream of the MSP, as to cover the same CpG sites as those contained by the MSP region. Both MSP and USP are listed in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The PCRs contained 1.5mM MgCl2, 200μM dNTP mix (Invitrogen), 0.2μM primers (Sigma-Aldrich), 1× PCR AmpliTaq gold buffer, 1.25 U of AmpliTaq Gold (Applied Biosystems), 5% dimethyl sulfoxide (DMSO; Merck) and 50 ng of bisulfite-treated DNA. The PCR was performed as follows: denaturation at 95°C for 10 minutes; 35 cycles of 94°C for 45 seconds, 58°C for 45 seconds, 72°C for 45 seconds and a final extension step of 72°C for 5 minutes. The PCR products were visualized on 2% agarose gels containing ethidium bromide.
RQ-PCR analysis
To further analyze whether methylation status influences the gene expression of specific genes, we selected 4 genes for RQ-PCR. Total RNA was extracted using the RNA Extraction Kit (QIAGEN) according to the manufacturer's protocol. The reverse transcription (RT) reaction was performed using the MMLV-RT Kit (Invitrogen) and random hexamers (Fermentas) according to the manufacturer's protocol. RT-PCR primers were designed using the Primer3 software (Broad Institute). All primer sequences are listed in supplemental Table 1. Quantitative RT-PCR analysis was performed using 2× SYBR Green master mix according to manufacturer's protocol (Fermentas). Expression was analyzed using the Stratagene Mx 3005p (Stratagene) detection system and calculated with the Max Pro QPCR software (Stratagene) using beta-actin as a reference gene and the ΔΔ cycle threshold (Ct) method. The statistical significance of differences in expression was calculated by a t test and represented in box plot graphs using Statistica 8.0 software (Stat Soft).
Bisulfite sequencing
To confirm the methylation status of individual genes, we selected 2 genes for bisulfite sequencing. Bisulfite-sequencing primers (BSP) were designed according to BSP selection criteria as outlined by Li and Dahiya16 (supplemental Table 1). PCR was performed using 1.5mM MgCl2, 200μM dNTP mix, 0.2μM primers (Sigma-Aldrich), 1× PCR Taq buffer, 1 U of Platinum Taq (Invitrogen), and approximately 50 ng of bisulfite-treated DNA. Touchdown PCR cycling conditions included 5 cycles at 95°C for 30 seconds, 55°C for 45 seconds, and 72°C for 1 minute, followed by 5 cycles at 95°C for 30 seconds, 53°C for 45 seconds, and 72°C for 1 minute, and a further 30 cycles at 94°C for 30 seconds, 50°C for 45 seconds, and 72°C for 45 seconds, followed by a final extension step of 72°C for 5 minutes. The PCR product was cloned using the Invitrogen TOPO TA Cloning Kit 2.1 TOPO vector (Invitrogen), and 10 to 12 clones per sample were selected for verification of the correct-sized insert via PCR using vector-specific M13 forward and reverse primers (provided by the cloning kit). Verified PCR products were treated using 0.5 U of ExoI and 1.0 U of shrimp alkaline phosphatase (Fermentas) at 37°C for 45 minutes and 80°C for 15 minutes and sequenced with vector-specific primers using the BigDye Terminator Cycle Sequencing Reaction Kit (Perkin-Elmer). All sequence reactions were analyzed using an automated DNA sequencer (ABI 377; Applied Biosystems). Sequences were aligned and analyzed using the bisulfite sequencing web-based tool BiQ Analyzer software (Max-Planck Institut fur Informatik, Saarbrucken, Germany) and the degree of methylation represented as a lollipop grid.
DAC and TSA treatment
Frozen primary CLL cell samples and the CLL cell line HG3 were cultured in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich), 4mM glutamine, and 1× penicillin/streptomycin (Invitrogen) until confluent. Cells were then subdivided to contain approximately 1 million cells/mL per well 12 hours before treatment, to allow the cells to adjust to conditions. Cells were subsequently cultured over 3 days in supplemented RPMI media treated with one of the following treatments: (1) medium containing 5-aza-2′-deoxycytidine (DAC; 5μM/L, Sigma-Aldrich) for 72 hours whereby medium was changed every 24 hours; (2) medium containing trichostatin A (TSA, 500nM/L; Sigma-Aldrich) for the last 24 hours; and (3) medium containing DAC for 48 hours followed by addition of TSA for 24 hours. Control cells were cultured in similar way with no drugs added.
Results
Distribution of samples based on overall MI
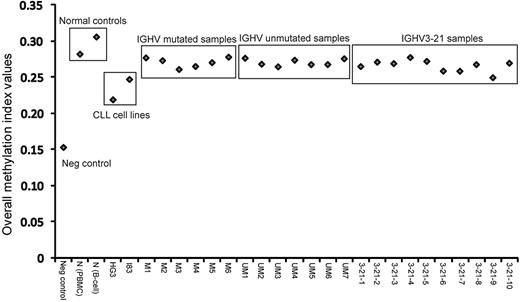
This study investigated a total of 27 samples, encompassing 6 IGHV-mutated, 7 IGHV-unmutated, and 10 IGHV3-21 CLL patient samples, as well as 2 healthy control samples (1 obtained from peripheral blood mononuclear cell [PBMC] and 1 from sorted B cells), 2 EBV-transformed CLL cell lines and 1 negative control (whole genome–amplified DNA). Figure 1 graphically represents the MI values obtained from all samples. As expected, the negative control sample showed lowest MI and the healthy control samples displayed the highest values. All CLL patient samples fell in the range between these controls, whereas the 2 CLL cell lines displayed lower MIs compared with the CLL samples. Hence, healthy controls had a higher degree of genome-wide methylation, whereas the CLL cell lines showed less methylation compared with the CLL patient samples.
Methylation profiling of different prognostic subgroups of CLL
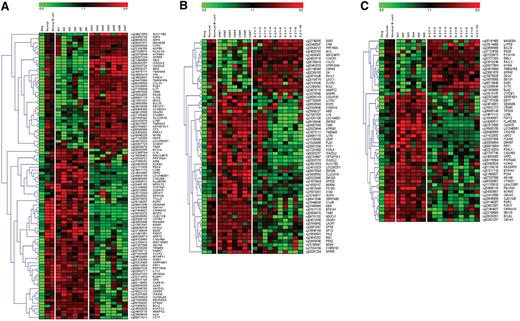
Genome-wide methylation profiles were compared between samples from 3 different CLL subgroups, that is, IGHV mutated, IGHV unmutated, and IGHV3-21 CLL. In addition, healthy controls were included for comparison. Because the CLL cell lines showed completely different methylation profiles compared with CLL patient samples, these were not included in the comparisons. Using highly stringent selection criteria (as detailed in “Methylation array analysis”), a total of 64 genes were identified as significantly differentially methylated between IGHV mutated and unmutated CLL (Figure 2A). Similarly, 60 and 31 genes were identified in the comparison between IGHV unmutated versus IGHV3-21 CLL and IGHV mutated versus IGHV3-21 CLL, respectively (Figure 2B-C). Based on the functionality of all these identified differentially methylated genes, they could be grouped into various categories, such as apoptotic-related genes (antiapoptotic or proapoptotic), known or candidate tumor suppressor genes, genes involved in high proliferative activity, genes enhancing tumorigenesis, drug-resistance genes, and genes associated with prognosis, as described in Table 2. Importantly, we identified known/candidate tumor suppressor genes that were preferentially methylated in the IGHV unmutated (7 genes, eg, VHL, ABI3), IGHV3-21 (1 gene, SLC22A18), and IGHV mutated (2 genes, PPP1R3A and WISP3) subgroups. We also identified 10 unmethylated and hence potentially expressed genes shown to be involved in activation of proliferative pathways such as the nuclear factor κB (NFκB) pathway (eg, ADORA3 and CARD15), and the mitogen-activated protein/extracellular signal–regulated kinase (MAPK/ERK) kinase pathway (eg, PRF1 and FABP7) in the IGHV unmutated and IGHV3-21 subgroups. Of the identified genes, most have previously not been associated with CLL; however, many were known to be involved in the tumorigenesis of several other cancers and leukemia.
Supervised hierarchical clustering of methylated/unmethylated genes between CLL subgroups. Three different comparisons were made: (A) IGHV mutated (samples M1 to M6) versus IGHV unmutated (samples UM1 to UM7), (B) IGHV unmutated versus IGHV3-21 (samples 3-21-1 to 3-21-10), and (C) IGHV mutated versus IGHV3-21. Two normal healthy control samples and 1 negative control were included for comparison. A gradient color scale ranging between green (completely unmethylated) and red (completely methylated) is included.
Supervised hierarchical clustering of methylated/unmethylated genes between CLL subgroups. Three different comparisons were made: (A) IGHV mutated (samples M1 to M6) versus IGHV unmutated (samples UM1 to UM7), (B) IGHV unmutated versus IGHV3-21 (samples 3-21-1 to 3-21-10), and (C) IGHV mutated versus IGHV3-21. Two normal healthy control samples and 1 negative control were included for comparison. A gradient color scale ranging between green (completely unmethylated) and red (completely methylated) is included.
Functional characterization of selected differentially methylated genes
| Functionality of genes/gene names . | IGHV mutated . | IGHV unmutated . | IGHV3-21 . | Implicated role in cancer . | Accession number . | Reference . |
|---|---|---|---|---|---|---|
| IGHV mutated and IGHV unmutated CLL | ||||||
| Tumor-suppressor genes | ||||||
| ABI3* | Unmethylated | Methylated | Lost in cancer cell lines, implicated in lung metastasis | NM_016428.3 | 17 | |
| SCGB2A1* | Unmethylated | Methylated | Methylated in lung cancer | NM_002407.1 | 18 | |
| VHL | Unmethylated | Methylated | Mutated in sporadic cell renal carcinoma and von Hippel-Lindau syndrome | NT_022517.17 | 19 | |
| PPP1R3A* | Methylated | Unmethylated | Deregulated in colorectal cancer | NM_002711.2 | 20 | |
| WISP3 | Methylated | Unmethylated | Loss of expression in inflammatory breast cancer | NM_003880.2 | 21 | |
| Genes enhancing proliferation | ||||||
| ADORA3 | Methylated | Unmethylated | Activation of the NFκB pathway | NM_020683.5 | 22 | |
| AIRE | Methylated | Unmethylated | Regulated by the PKC pathway | NM_000383.1 | 23 | |
| CARD15 | Methylated | Unmethylated | Activation of the NFκB pathway | NM_022162.1 | 24 | |
| FABP7 | Methylated | Unmethylated | Regulated by the MAPK/ERK pathway | NM_001446.3 | 25 | |
| LOC340061 | Methylated | Unmethylated | Activation of the NFκB pathway | NM_198282.1 | 26 | |
| PRF1 | Methylated | Unmethylated | Enhancing the MAPK pathway | NM_005041.3 | 27 | |
| UNC5CL | Methylated | Unmethylated | Activation of the NFκB pathway | NM_173561.1 | 28 | |
| Genes facilitating tumorigenesis | ||||||
| ANGPT2 | Methylated | Unmethylated | Preferentially expressed in unmutated CLL | NM_001147.1 | 29 | |
| IFNB1 | Methylated | Unmethylated | Shown to enhance B-cell proliferation | NM_002176.2 | 30 | |
| URP2 | Methylated | Unmethylated | Implicated in facilitating CLL tumorigenesis | NM_031471.4 | 31 | |
| Antiapoptotic genes | ||||||
| BCL2 | Methylated | Unmethylated | Known antiapoptotic gene, up-regulated in many cancers | NT_025028.13 | 32 | |
| PLD1 | Unmethylated | Methylated | Stimulates cell-cycle progression by repressing p53 regulation of p21 | NM_002662.2 | 33 | |
| Proapoptotic genes | ||||||
| BCL10 | Unmethylated | Methylated | Proapoptotic genes, involved in p53 mediated cell death | NM_003921.2 | 34 | |
| IL19 | Methylated | Unmethylated | Regulating apoptosis by activation of the STAT3 pathway | NM_013371.2 | 35 | |
| Genes involved in prognosis | ||||||
| NGFR | Unmethylated | Methylated | Highly expressed in favorable prognostic ALL patients | NM_002507.1 | 36 | |
| IGHV unmutated and IGHV3-21 CLL | ||||||
| Tumor-suppressor genes | ||||||
| ABI3* | Methylated | Unmethylated | Lost in cancer cell lines, implicated in lung metastasis | NM_016428.3 | 17 | |
| GPX3 | Methylated | Unmethylated | Deleted or methylated in prostate cancer | NM_002084.2 | 37 | |
| IGSF4* | Methylated | Unmethylated | Down-regulated by methylation in neuroblastoma | NM_014333.2 | 38 | |
| SERPIND5* | Methylated | Unmethylated | Tumor-suppressive properties in breast cancer | NM_002639.2 | 39 | |
| SLC22A18 | Unmethylated | Methylated | Deregulated in several cancer forms | NM_183233.1 | 40 | |
| Genes repressing proliferation | ||||||
| LDOC1 | Unmethylated | Methylated | Implicated in repressing the NFκB pathway | NM_012317.2 | 41 | |
| ZNF540 | Methylated | Unmethylated | Involved in down-regulating the MAPK pathway | NM_152606.2 | 42 | |
| Genes facilitating tumorigenesis | ||||||
| IL17RC | Unmethylated | Methylated | Shown to facilitate tumorigenesis in prostate cancer | NM_032732.2 | 43 | |
| ANGPT2 | Unmethylated | Methylated | Expressed in IGHV unmutated CLL | NM_001147.1 | 29 | |
| FHL2 | Methylated | Unmethylated | NM_201556.1 | |||
| S100A14 | Unmethylated | Methylated | NM_020672.1 | |||
| Antiapoptotic genes | ||||||
| BCL2 | Unmethylated | Methylated | Known antiapoptotic gene, up-regulated in many cancers | NT_025028.13 | 32 | |
| PLD1 | Methylated | Unmethylated | Stimulates cell-cycle progression by repressing p53 regulation of p21 | NM_002662.2 | 33 | |
| IGHV mutated and IGHV3-21 CLL | ||||||
| Genes enhancing proliferation | ||||||
| LOC340061 | Methylated | Unmethylated | Activation of the NFκB pathway | NM_198282.1 | 26 | |
| PRF1 | Methylated | Unmethylated | Enhancing the MAPK pathway | NM_005041.3 | 27 | |
| RASGRP3 | Methylated | Unmethylated | Regulated B-cell proliferation by facilitating B-cell receptor–RAS signaling | NM_170672.1 | 44 | |
| Genes facilitating tumorigenesis | ||||||
| LMO2 | Methylated | Unmethylated | Known proto-oncogene, expressed in human B-cell lymphomas | NM_005574.2 | 45 | |
| URP2 | Methylated | Unmethylated | Implicated in facilitating CLL tumorgenesis | NM_031471.4 | 31 | |
| NCRMS | Methylated | Unmethylated | XR_000219.2 | |||
| Proapoptotic genes | ||||||
| TP53I3 | Methylated | Unmethylated | Shown to induce p53-mediated cell death | NM_004881.2 | 46 | |
| BCL10 | Unmethylated | Methylated | Proapoptotic genes, involved in p53-mediated cell death | NM_003921.2 | 34 | |
| Genes involved in prognosis | ||||||
| DNTT | Methylated | Unmethylated | Associated with poor prognosis in acute myeloid leukemia | NM_004088.3 | 47 |
| Functionality of genes/gene names . | IGHV mutated . | IGHV unmutated . | IGHV3-21 . | Implicated role in cancer . | Accession number . | Reference . |
|---|---|---|---|---|---|---|
| IGHV mutated and IGHV unmutated CLL | ||||||
| Tumor-suppressor genes | ||||||
| ABI3* | Unmethylated | Methylated | Lost in cancer cell lines, implicated in lung metastasis | NM_016428.3 | 17 | |
| SCGB2A1* | Unmethylated | Methylated | Methylated in lung cancer | NM_002407.1 | 18 | |
| VHL | Unmethylated | Methylated | Mutated in sporadic cell renal carcinoma and von Hippel-Lindau syndrome | NT_022517.17 | 19 | |
| PPP1R3A* | Methylated | Unmethylated | Deregulated in colorectal cancer | NM_002711.2 | 20 | |
| WISP3 | Methylated | Unmethylated | Loss of expression in inflammatory breast cancer | NM_003880.2 | 21 | |
| Genes enhancing proliferation | ||||||
| ADORA3 | Methylated | Unmethylated | Activation of the NFκB pathway | NM_020683.5 | 22 | |
| AIRE | Methylated | Unmethylated | Regulated by the PKC pathway | NM_000383.1 | 23 | |
| CARD15 | Methylated | Unmethylated | Activation of the NFκB pathway | NM_022162.1 | 24 | |
| FABP7 | Methylated | Unmethylated | Regulated by the MAPK/ERK pathway | NM_001446.3 | 25 | |
| LOC340061 | Methylated | Unmethylated | Activation of the NFκB pathway | NM_198282.1 | 26 | |
| PRF1 | Methylated | Unmethylated | Enhancing the MAPK pathway | NM_005041.3 | 27 | |
| UNC5CL | Methylated | Unmethylated | Activation of the NFκB pathway | NM_173561.1 | 28 | |
| Genes facilitating tumorigenesis | ||||||
| ANGPT2 | Methylated | Unmethylated | Preferentially expressed in unmutated CLL | NM_001147.1 | 29 | |
| IFNB1 | Methylated | Unmethylated | Shown to enhance B-cell proliferation | NM_002176.2 | 30 | |
| URP2 | Methylated | Unmethylated | Implicated in facilitating CLL tumorigenesis | NM_031471.4 | 31 | |
| Antiapoptotic genes | ||||||
| BCL2 | Methylated | Unmethylated | Known antiapoptotic gene, up-regulated in many cancers | NT_025028.13 | 32 | |
| PLD1 | Unmethylated | Methylated | Stimulates cell-cycle progression by repressing p53 regulation of p21 | NM_002662.2 | 33 | |
| Proapoptotic genes | ||||||
| BCL10 | Unmethylated | Methylated | Proapoptotic genes, involved in p53 mediated cell death | NM_003921.2 | 34 | |
| IL19 | Methylated | Unmethylated | Regulating apoptosis by activation of the STAT3 pathway | NM_013371.2 | 35 | |
| Genes involved in prognosis | ||||||
| NGFR | Unmethylated | Methylated | Highly expressed in favorable prognostic ALL patients | NM_002507.1 | 36 | |
| IGHV unmutated and IGHV3-21 CLL | ||||||
| Tumor-suppressor genes | ||||||
| ABI3* | Methylated | Unmethylated | Lost in cancer cell lines, implicated in lung metastasis | NM_016428.3 | 17 | |
| GPX3 | Methylated | Unmethylated | Deleted or methylated in prostate cancer | NM_002084.2 | 37 | |
| IGSF4* | Methylated | Unmethylated | Down-regulated by methylation in neuroblastoma | NM_014333.2 | 38 | |
| SERPIND5* | Methylated | Unmethylated | Tumor-suppressive properties in breast cancer | NM_002639.2 | 39 | |
| SLC22A18 | Unmethylated | Methylated | Deregulated in several cancer forms | NM_183233.1 | 40 | |
| Genes repressing proliferation | ||||||
| LDOC1 | Unmethylated | Methylated | Implicated in repressing the NFκB pathway | NM_012317.2 | 41 | |
| ZNF540 | Methylated | Unmethylated | Involved in down-regulating the MAPK pathway | NM_152606.2 | 42 | |
| Genes facilitating tumorigenesis | ||||||
| IL17RC | Unmethylated | Methylated | Shown to facilitate tumorigenesis in prostate cancer | NM_032732.2 | 43 | |
| ANGPT2 | Unmethylated | Methylated | Expressed in IGHV unmutated CLL | NM_001147.1 | 29 | |
| FHL2 | Methylated | Unmethylated | NM_201556.1 | |||
| S100A14 | Unmethylated | Methylated | NM_020672.1 | |||
| Antiapoptotic genes | ||||||
| BCL2 | Unmethylated | Methylated | Known antiapoptotic gene, up-regulated in many cancers | NT_025028.13 | 32 | |
| PLD1 | Methylated | Unmethylated | Stimulates cell-cycle progression by repressing p53 regulation of p21 | NM_002662.2 | 33 | |
| IGHV mutated and IGHV3-21 CLL | ||||||
| Genes enhancing proliferation | ||||||
| LOC340061 | Methylated | Unmethylated | Activation of the NFκB pathway | NM_198282.1 | 26 | |
| PRF1 | Methylated | Unmethylated | Enhancing the MAPK pathway | NM_005041.3 | 27 | |
| RASGRP3 | Methylated | Unmethylated | Regulated B-cell proliferation by facilitating B-cell receptor–RAS signaling | NM_170672.1 | 44 | |
| Genes facilitating tumorigenesis | ||||||
| LMO2 | Methylated | Unmethylated | Known proto-oncogene, expressed in human B-cell lymphomas | NM_005574.2 | 45 | |
| URP2 | Methylated | Unmethylated | Implicated in facilitating CLL tumorgenesis | NM_031471.4 | 31 | |
| NCRMS | Methylated | Unmethylated | XR_000219.2 | |||
| Proapoptotic genes | ||||||
| TP53I3 | Methylated | Unmethylated | Shown to induce p53-mediated cell death | NM_004881.2 | 46 | |
| BCL10 | Unmethylated | Methylated | Proapoptotic genes, involved in p53-mediated cell death | NM_003921.2 | 34 | |
| Genes involved in prognosis | ||||||
| DNTT | Methylated | Unmethylated | Associated with poor prognosis in acute myeloid leukemia | NM_004088.3 | 47 |
These genes represent candidate tumor-suppressor genes.
Confirmation of array data using MSP-PCR and RQ-PCR
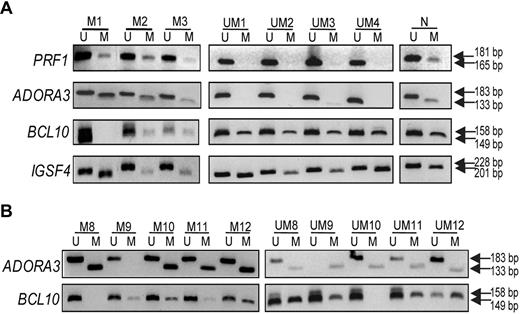
To confirm the array data, we selected 4 genes for MSP-PCR: that is, PRF1 and ADORA3 (methylated in IGHV mutated CLL samples) and BCL10 and IGSF4 (methylated in IGHV unmutated CLL samples). PRF1 and ADORA3 are mainly involved in cell proliferation by constitutive activation of the mitogen-activated protein kinase (MAPK) and NFκB pathways, respectively,22,27 whereas BCL10 is an antiapoptotic gene34 and IGSF4 is a candidate tumor suppressor gene.38 Figure 3A illustrates the results for 3 IGHV mutated and 4 unmutated CLL samples, verifying that PRF1 and ADORA3 were mostly methylated in mutated samples, whereas mostly BCL10 and IGSF4 were methylated in unmutated samples. In addition, we selected 10 additional IGHV mutated and unmutated CLL samples (not included in the array study), and analyzed the methylation status for ADORA3 and BCL10, which further confirmed the divergent methylation status between the mutated/unmutated CLL subgroups (Figure 3B).
Validation experiments using MSP-PCR. MSP-PCR was performed for the PRF1, ADORA3, BCL10, and IGSF4 genes, using bisulfite-treated DNA from IGHV mutated samples (M1-3 to M8-12) and IGHV unmutated samples (UM1-4 and UM8-12). (A) Data for samples analyzed in the methylation array; (B) results from additional CLL samples are included. Analysis of each gene in panels A and B was performed during the same experiment and on the same gel; vertical line(s) have been inserted to indicate a repositioned gel lane. N indicates normal PBMC sample; M, methylation-specific primers; and U, unmethylation-specific primers.
Validation experiments using MSP-PCR. MSP-PCR was performed for the PRF1, ADORA3, BCL10, and IGSF4 genes, using bisulfite-treated DNA from IGHV mutated samples (M1-3 to M8-12) and IGHV unmutated samples (UM1-4 and UM8-12). (A) Data for samples analyzed in the methylation array; (B) results from additional CLL samples are included. Analysis of each gene in panels A and B was performed during the same experiment and on the same gel; vertical line(s) have been inserted to indicate a repositioned gel lane. N indicates normal PBMC sample; M, methylation-specific primers; and U, unmethylation-specific primers.
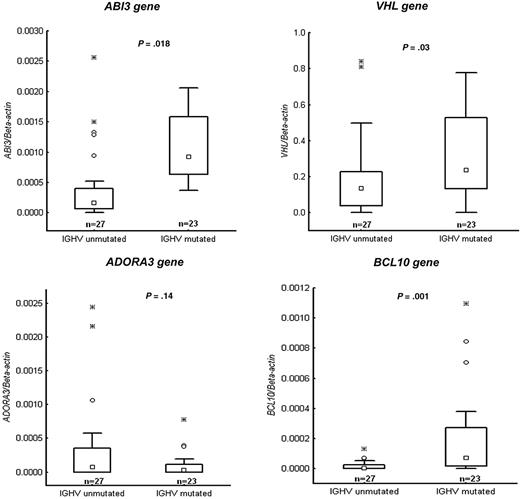
After conformation of the methylation status of the selected genes, we correlated these data with mRNA expression levels of the respective genes as well as 3 additional genes (ie, VHL, ABI3, and NGFR) using RQ-PCR. Figure 4 demonstrates RQ-PCR results for 4 of the genes (VHL, ABI3, ADORA3, and BCL-10) in a CLL cohort of 50 samples, confirming that the mRNA expression levels correlated strongly with the methylation status. Of the 4 genes analyzed, 3 genes (VHL, ABI3, and BCL-10) showed a significant difference between the IGHV mutated and unmutated subgroups. Furthermore, RT-PCR results for the PRF1 and NGFR genes (supplemental Figure 1) were analyzed on the same 4 IGHV mutated and unmutated samples included in the MSP-PCR analyses, further verifying the MSP-PCR data.
Validation experiments using quantitative RT-PCR. Relative expression data presented as box plots for the following genes: VHL, ABI3, ADORA3, and BCL10. Twenty-seven IGHV unmutated and 23 IGHV mutated CLL cases were investigated that had been not included in the methylation array analysis. Values of P are indicated above each box plot. Boxes indicate the interquartile range (25%-75%) with the smaller inner square indicating the median. The whiskers show the minimum and maximum values, except for outliers (○).
Validation experiments using quantitative RT-PCR. Relative expression data presented as box plots for the following genes: VHL, ABI3, ADORA3, and BCL10. Twenty-seven IGHV unmutated and 23 IGHV mutated CLL cases were investigated that had been not included in the methylation array analysis. Values of P are indicated above each box plot. Boxes indicate the interquartile range (25%-75%) with the smaller inner square indicating the median. The whiskers show the minimum and maximum values, except for outliers (○).
Bisulfite sequencing of methylated tumor suppressor genes
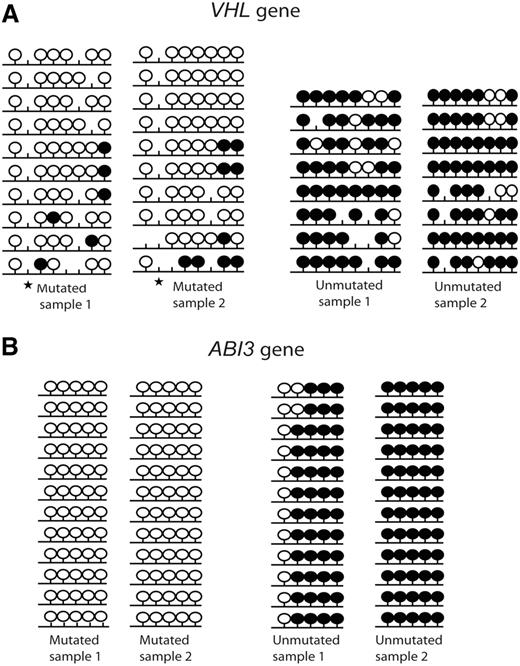
Two candidate tumor suppressor genes (VHL and ABI3) that displayed significant differences in gene expression levels between IGHV mutated and unmutated CLL samples were selected for bisulfite sequencing and subcloning to confirm the degree of methylation using 2 IGHV mutated and 2 IGHV unmutated samples. The bisulfite sequencing region covered the CpG sites which were included in the array and showed a significant difference in the MI values between IGHV mutated and IGHV unmutated samples. Both genes exhibited a high degree of hypermethylation in the IGHV unmutated samples compared with the IGHV mutated samples (Figure 5).
Bisulfite sequencing of 2 tumor suppressor genes. (A) VHL and (B) ABI3. Results from 2 IGHV mutated and 2 IGHV unmutated CLL samples are shown. Black lollipops indicate methylated CpG sites; and white lollipops, unmethylated CpG sites. ★ indicates a polymorphic site.
Bisulfite sequencing of 2 tumor suppressor genes. (A) VHL and (B) ABI3. Results from 2 IGHV mutated and 2 IGHV unmutated CLL samples are shown. Black lollipops indicate methylated CpG sites; and white lollipops, unmethylated CpG sites. ★ indicates a polymorphic site.
Reexpression of methylated genes using a methyl inhibitor
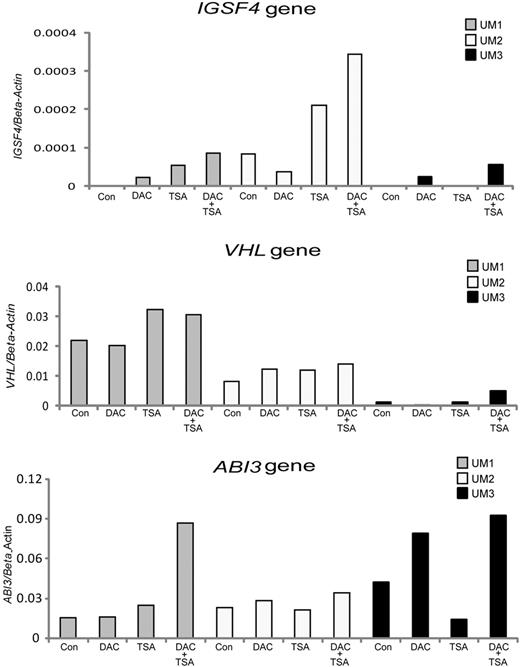
To investigate the role of DNA methylation in transcriptional regulation of gene promoters, we examined the effects of DNA methyl transferase inhibitor DAC and/or histone deacetylase inhibitor TSA treatment on gene expression in 3 IGHV unmutated and 3 IGHV mutated CLL samples. Four genes found to be mainly hypermethylated in unmutated samples (IGSF4, VHL, ABI3, and NGFR) were selected for reexpression analysis. An increase in the activation of the IGSF4, ABI3, and VHL genes was observed in response to DAC or TSA treatment, especially when applied in combination, in several of the IGHV unmutated CLL samples compared with the corresponding untreated samples (Figure 6). Reactivation of the NGFR gene is shown in supplemental Figure 2. Similar data were obtained for all the above-mentioned genes using the CLL EBV transformed cell line HG3 (data not shown).
RT-PCR analysis showing the reexpression of methylated tumor suppressor genes in unmutated CLL. Three different candidate tumor suppressor genes—(A) IGSF4, (B) ABI3, and (C) VHL—were investigated in 3 IGHV unmutated CLL samples (UM1 to UM3, shown in different colors) with and without treatment using the methyl inhibitor DAC, the HDAC inhibitor TSA, or in combination. Con indicates control.
RT-PCR analysis showing the reexpression of methylated tumor suppressor genes in unmutated CLL. Three different candidate tumor suppressor genes—(A) IGSF4, (B) ABI3, and (C) VHL—were investigated in 3 IGHV unmutated CLL samples (UM1 to UM3, shown in different colors) with and without treatment using the methyl inhibitor DAC, the HDAC inhibitor TSA, or in combination. Con indicates control.
Discussion
In hematologic malignancies, genes involved in important cellular pathways have been described to be affected by CpG island methylation in association with transcriptional silencing.48 The role of aberrant DNA methylation and individual DNA promoter methylation has been investigated in CLL (eg, ZAP-70, TWIST 2),6,7 although there have been few comprehensive studies dedicated to uncover the methylation status on a global level. Using a genome-wide methylation array, we here for the first time identified distinct methylation profiles in prognostic subsets of CLL.
In tumor cells, a disrupted methylation pattern is often observed featuring global hypomethylation and region-specific hypermethylation of certain gene promoters, compared with their normal cellular counterparts. In the present study, the overall degree of methylation, indicated as MI values, of all CLL primary samples demonstrated lower methylation levels compared with normal healthy controls (Figure 1), which is in line with previous findings.49 We also noticed that the EBV-transformed CLL cell lines displayed very different methylation patterns compared with CLL primary samples with lower MI values. This is perhaps not unexpected considering their higher proliferative capacity.
To identify differentially methylated genes between CLL subgroups, we applied a stringent bioinformatic approach which revealed significantly different methylation profiles between IGHV unmutated and mutated CLL as well as between IGHV mutated/unmutated and IGHV3-21 CLL (Figure 2). Many of the identified genes were found to be involved in important cellular pathways, where dysregulation of these genes could significantly contribute to leukemogenesis. For instance, we identified known or candidate tumor suppressor genes that were preferentially methylated in the poor-prognostic IGHV unmutated (7 genes) and IGHV3-21 (1 gene) subgroups (Table 2). Although many of these genes have been implicated in several other cancers (Table 2), their role in CLL has not been investigated. Some of the known or candidate tumor suppressor genes that were specifically methylated in IGHV unmutated CLL were: ABI3, implicated in lung metastasis,17 VHL, mutated in sporadic renal cell carcinoma as well as in the hereditary syndrome von Hippel-Lindau19 and IGSF4, methylated in neuroblastoma.38 Furthermore, we performed validation experiments to verify the methylation data from the arrays. For instance, the high degree of methylation and the corresponding decreased gene expression of 2 genes, VHL and ABI3, were confirmed by both bisulfite sequencing and RQ-PCR in IGHV unmutated CLL (Figure 4 and 5). Taken together, several tumor suppressor genes were found methylated particularly in poor-prognostic, unmutated CLL, while they remained unmethylated and hence expressed in IGHV mutated CLL.
Previous studies implicate that certain cellular pathways, such as NFκB, phosphatidylinositol 3-kinase (PI3K)/Akt and MAPK/ERK, are dysregulated in CLL B cells leading to activation of antiapoptotic pathways. Our data strengthens this idea and further indicates that certain genes involved in such proliferative pathways may be controlled by DNA methylation in CLL. In particular, we identified expression of genes that enhance constant cell proliferation and invasion in the IGHV unmutated CLL subgroup (Table 2). Some of the genes included were ADORA3 and PRF1 genes, that activate the NFκB and MAPK pathways, respectively, and which were expressed in the poor-prognostic CLL subsets compared with IGHV mutated CLL.26,27 In addition, genes involved in repressing the NFκB pathway (LCOD1 gene)29 and MAPK (ZNF540 gene)36 pathways were generally methylated in both IGHV unmutated and IGHV3-21 samples in contrast to IGHV mutated CLL. The methylation status and gene expression were confirmed for 2 such genes—for example, ADORA3 and PRF1—using both MSP-PCR and RQ-PCR in IGHV unmutated versus mutated CLL (Figure 3 and 4). Moreover, genes previously reported to be involved in facilitating tumorigenesis within other cancer types were noted to be preferentially unmethylated in the IGHV unmutated (7 genes) and IGHV3-21 (4 genes) CLL subgroups in comparison to IGHV mutated CLL (Table 2). For example, IFNB1, shown to enhance B-cell proliferation,30 and IL17RC, involved in facilitating tumorigenesis in prostate cancer,43 were unmethylated in IGHV unmutated samples. Thus, the data indicates that poor-prognostic CLL subsets favor expression of genes aiding tumor proliferation.
Both IGHV unmutated CLL and IGHV mutated CLL also showed deregulation of anti- and proapoptotic genes. For example, BCL2 and PLD1, which are known antiapoptotic gene,32,33 remained unmethylated in IGHV unmutated and IGHV mutated CLL samples, respectively. On the other hand, proapoptotic genes such as BCL1034 and TP5313, shown to induce p53-mediated cell death,46 were methylated in IGHV unmutated and IGHV mutated CLL samples, respectively. As shown in Figure 3, we could confirm both the differential methylation status and expression level of BCL10 in mutated and unmutated CLL.
Interestingly, we also identified certain genes that have been related to prognosis, for instance, the ANGPT2 gene which has been shown to be expressed preferentially in IGHV unmutated CLL samples.29 Our array data indeed demonstrated that this gene was methylated in mutated CLL while being unmethylated in unmutated CLL. In addition, expression of the NGFR gene, which has been associated with favorable prognosis in ALL,36 was unmethylated in IGHV mutated CLL and showed increased expression on the RNA level within this subset (supplemental Figure 1). However, further analysis of the ANGPT2 and NGFR genes in a larger CLL cohort is needed to assess their prognostic impact.
Apart from the “traditional” genes, the HumanMethylation27 BeadChip array also contains 110 miRNA promoters. However, none of these miRNAs met the criteria set to define a significant difference in methylation between the studied subsets. Evidently, this does not exclude that methylation changes may exist between CLL subsets considering that only a fraction of miRNA promoters were covered by the array.
Unlike genetic changes, epigenetic changes are reversible using demethylating agents such as DAC. Consequently, the role of these agents as treatment options is currently under investigation in hematologic malignancies including CLL.50,51 In this study, we demonstrated that inhibition of DNA methylation using methyl- and histone deacetyl (HDAC) inhibitors could induce expression of methylated tumor suppressor genes (eg, IGSF4, ABI3, and VHL) in unmutated CLL primary samples. We generally did not observe high induction of gene expression with methyl inhibitor treatment, probably because CLL primary cells do not proliferate at a high rate under in vitro conditions, although the effect of the drugs was pronounced when applied in combination. These data indeed strengthen the role of DNA methylation in regulating promoter expression and open up the possibility of targeted therapy, although this has to be studied further.
Gene expression microarray studies have previously revealed different gene expression profiles between IGHV mutated and IGHV unmutated/IGHV3-21 CLL patients.52-54 However, the majority of the identified genes in the present study could not be found in these gene expression studies.52-54 There could be several possible explanations for this: for example, incomplete overlap with genes included on the expression arrays and different criteria applied for identifying genes differing between subsets. Gene expression can also be silenced by other mechanisms than methylation—for instance, mutations or deletions—which will not be identified on methylation arrays. In addition, we cannot exclude the possibility that differentially methylated genes may not always correlate with a significant difference in gene expression. Because this is one of the first studies using the whole-genome Illumina array, we decided to apply stringent criteria to reduce false-positive genes. On the other hand, we may indeed have lost some important genes, which would have come up using less stringent criteria. That notwithstanding, we believe that our confirmation experiments using different techniques such as MSP-PCR, RQ-PCR, and bisulfite sequencing strengthen the validity of our data.
In conclusion, the observation that the distinct methylation profiles encompassed candidate genes involved in cellular pathways regulating proliferation and apoptosis in IGHV unmutated and mutated CLL underlines a critical role for epigenetic changes during leukemogenesis. Most identified tumor suppressor genes were specifically silenced in IGHV unmutated CLL, implicating the role of these genes in the pathogenesis of this subgroup. Specific inhibition of expression of unmethylated genes involved in facilitating tumorigenesis and reexpression of methylated candidate tumor suppressor genes within the poor-prognostic subgroups could represent a possible route for drug therapy. Hopefully, our results may help to further unravel the pathogenesis of CLL and identify possible novel biomarkers and targets for drug therapy in CLL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Swedish Cancer Society, the Swedish Research Council, the Medical Faculty of Uppsala University, Uppsala University Hospital, and Lion's Cancer Research Foundation in Uppsala, Sweden. ABBEST, Ireland, provided a stipend for N.C.
Authorship
Contribution: M.K. and N.C. performed research, analyzed data, and wrote the paper; H.G. and A.I. performed bioinformatic analyses and analyzed data; C.E. performed microarray analysis and analyzed data; F.R. analyzed data; and R.R. supervised the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard Rosenquist, MD, PhD, Department of Genetics and Pathology, Rudbeck Laboratory, Uppsala University, SE-751-85 Uppsala, Sweden; e-mail: richard.rosenquist@genpat.uu.se.
References
Author notes
M.K. and N.C. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal