Abstract
Induced pluripotent stem cells (iPSCs) can be generated from various differentiated cell types by the expression of a set of defined transcription factors. So far, iPSCs have been generated from primary cells, but it is unclear whether human cancer cell lines can be reprogrammed. Here we describe the generation and characterization of iPSCs derived from human chronic myeloid leukemia cells. We show that, despite the presence of oncogenic mutations, these cells acquired pluripotency by the expression of 4 transcription factors and underwent differentiation into cell types derived of all 3 germ layers during teratoma formation. Interestingly, although the parental cell line was strictly dependent on continuous signaling of the BCR-ABL oncogene, also termed oncogene addiction, reprogrammed cells lost this dependency and became resistant to the BCR-ABL inhibitor imatinib. This finding indicates that the therapeutic agent imatinib targets cells in a specific epigenetic differentiated cell state, and this may contribute to its inability to fully eradicate disease in chronic myeloid leukemia patients.
Introduction
Tumors develop in the context of a particular developmental state by acquiring mutations in oncogenes and tumor suppressor genes. Oncogenic mutations are often observed in a tissue- or cell type-specific manner.1 Similarly, in hereditary cancer syndromes, patients are predisposed to tumor development in specific tissues despite the presence of mutated cancer genes in all somatic cells. This suggests that the effects of cancer-relevant mutations are highly influenced by the state of a particular cell, including cell environment and differentiation. One way of studying the interaction of oncogenic mutations with different tissue types is to reprogram an established cancer cell line to pluripotency. Although this has been achieved for cell lines of experimentally induced mouse tumors through nuclear transfer,2 this process was not applied to human cancer because of technical constraints. The recent advance to reprogram differentiated cell types through expression of a combination of transcription factors3,4 has allowed induced pluripotent stem cells (iPSCs) to be generated from various cell types, including cells of the hematopoietic lineage.5,6 However, until now, this has not been achieved for human tumor cells.
Methods
Detailed methods are provided in the supplemental Methods data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All animal experiments were approved by the committee of animal care from Whitehead Institute.
Results and discussion
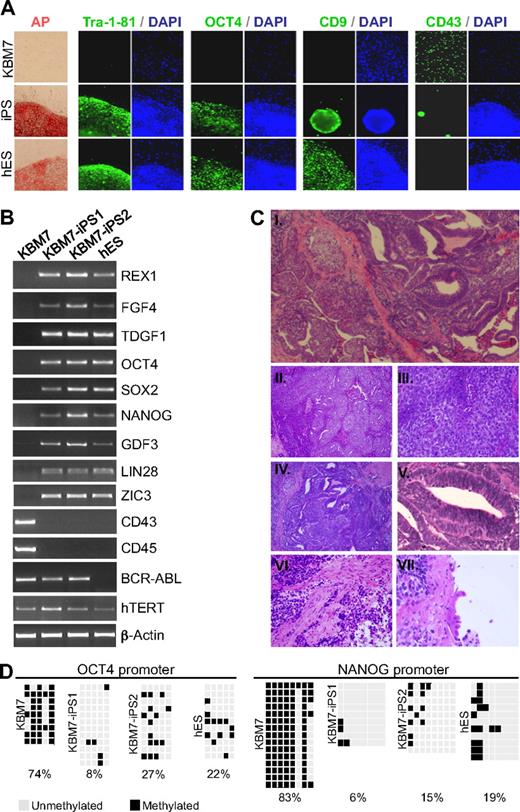
To address whether human cancer cells can be reprogrammed into iPSCs, we used KBM7 cells derived from blast crisis stage chronic myeloid leukemia (CML).7 A near-diploid subclone of KBM7 cells was used carrying a Philadelphia translocation. We infected KBM7 cells with retroviruses expressing 4 reprogramming factors: OCT4, SOX2, c-MYC, and KLF4. After 21 days, rare colonies with characteristic embryonic stem (ES) cell morphologies were expanded. Two cancer-derived iPS cell lines KBM7-iPS1 and KBM7-iPS2 that grew as adherent, flat colonies (Figure 1A) were characterized further. KBM7-iPS1 and KBM7-iPS2 expressed the ES cell markers alkaline phosphatase, CD9, Tra-1-81, and OCT4 and had lost expression of the hematopoietic marker CD43 (Figure 1A). Reverse-transcription polymerase chain reaction revealed that the reprogrammed clones expressed REX1, FGF4, TDGF1, NANOG, GDF3, LIN28, and ZIC3 as well as endogenous transcripts of OCT4 and SOX2 at levels similar to human ES cells (Figure 1B). Both pan-hematopoietic markers CD43 and CD45 were no longer expressed (Figure 1B; supplemental Figure 1). These cancer-derived iPSCs are distinct from pluripotent cells generated from normal cells, as is evident from expression of the BCR-ABL oncogene (Figure 1B; supplemental Figure 2) and their abnormal karyotype indicative of chromosomal instability (supplemental Figure 2). Until now, attempts to reprogram other CML cell lines have not been successful (data not shown).
Generation of iPSCs derived from human KBM7 leukemic cancer cells. (A) Introduction of c-MYC, OCT4, SOX2, and KLF4 into KBM7 cells led to the formation of rare adherent colonies that displayed morphologic similarity to human embryonic stem (ES) cells as well as high alkaline phosphatase activity. Reprogrammed KBM7 cells homogeneously stained for pluripotency markers Tra-1-81 and OCT4 and CD9 but did not stain for hematopoietic CD43 (green immunofluorescence; nuclear counterstain in blue). (B) Total RNA from KBM7 cells, reprogrammed KBM7 cells, and human ES cells was isolated and analyzed by reverse-transcribed polymerase chain reaction for the expression of human ES cell characteristic transcripts REX1, FGF4, and TDGF1, endogenous OCT4 and SOX2, NANOG, GDF3, LIN28, and ZIC3, the hematopoietic specific transcripts CD43 and CD45, and the CML specific BCR-ABL fusion transcript. (C) Subcutaneous injection of cancer induced pluripotent stem cells (iPSCs; KBM7-iPS2) into non-obese diabetic severe combined immunodeficiency (NOD-SCID) mice led to formation of teratoma. Hematoxylin and eosin-stained sections of the tumor (i, original magnification ×40) revealed extensive areas of embryonal carcinoma (ii, original magnification ×100; iii, original magnification ×400) and ectoderm-derived primitive neuroectodermal tissue (iv, original magnification ×100; v, original magnification ×400). Differentiation into muscle (mesodermal; vi, original magnification ×400), and ciliated respiratory epithelium (endodermal; vii, original magnification ×600) was also present. (D) Methylation analysis of the OCT4 and NANOG promoter region in KBM7 cells, the generated iPSCs, and hES cells. Light gray squares represent unmethylated; black squares, methylated cytosine-phosphate-guanosine. Numbers indicate the percentage methylated cytosine-phosphate-guanosine.
Generation of iPSCs derived from human KBM7 leukemic cancer cells. (A) Introduction of c-MYC, OCT4, SOX2, and KLF4 into KBM7 cells led to the formation of rare adherent colonies that displayed morphologic similarity to human embryonic stem (ES) cells as well as high alkaline phosphatase activity. Reprogrammed KBM7 cells homogeneously stained for pluripotency markers Tra-1-81 and OCT4 and CD9 but did not stain for hematopoietic CD43 (green immunofluorescence; nuclear counterstain in blue). (B) Total RNA from KBM7 cells, reprogrammed KBM7 cells, and human ES cells was isolated and analyzed by reverse-transcribed polymerase chain reaction for the expression of human ES cell characteristic transcripts REX1, FGF4, and TDGF1, endogenous OCT4 and SOX2, NANOG, GDF3, LIN28, and ZIC3, the hematopoietic specific transcripts CD43 and CD45, and the CML specific BCR-ABL fusion transcript. (C) Subcutaneous injection of cancer induced pluripotent stem cells (iPSCs; KBM7-iPS2) into non-obese diabetic severe combined immunodeficiency (NOD-SCID) mice led to formation of teratoma. Hematoxylin and eosin-stained sections of the tumor (i, original magnification ×40) revealed extensive areas of embryonal carcinoma (ii, original magnification ×100; iii, original magnification ×400) and ectoderm-derived primitive neuroectodermal tissue (iv, original magnification ×100; v, original magnification ×400). Differentiation into muscle (mesodermal; vi, original magnification ×400), and ciliated respiratory epithelium (endodermal; vii, original magnification ×600) was also present. (D) Methylation analysis of the OCT4 and NANOG promoter region in KBM7 cells, the generated iPSCs, and hES cells. Light gray squares represent unmethylated; black squares, methylated cytosine-phosphate-guanosine. Numbers indicate the percentage methylated cytosine-phosphate-guanosine.
Not all 4 reprogramming factors are strictly required for the generation of iPSCs from somatic cells.8–10 We asked whether all 4 factors were required for reprogramming of cancer cells. Unexpectedly, removal of c-MYC from the reprogramming mixture resulted in cell death. Removal of OCT4, SOX2, or KLF4 had a less dramatic phenotype and adherent colonies formed. However, these colonies maintained CD43 marker expression and did not exhibit ES cell morphology (supplemental Figure 3), suggesting that none of the 4 factors is dispensable.
In the chronic phase of CML, the BCR-ABL mutation drives cell expansion but cells retain the capacity to undergo differentiation.11 Subsequently, secondary lesions lead to blast crisis, which is characterized by the inability to undergo differentiation.12 We addressed whether iPSCs derived from KBM7 cells, which resemble the blast phase of CML, were able to undergo differentiation. The most stringent test to validate that reprogrammed cells have gained pluripotency is teratoma formation.13 Cancer-derived iPSCs (KBM7-iPS2) were subcutaneously injected into nonobese diabetic severe combined immunodeficiency (NOD-SCID) mice. This resulted in teratoma formation (Figure 1C; supplemental Figure 4). Histologic analysis demonstrated the presence of tissues representing all 3 germ layers. Large tumor areas contained primitive neuroectodermal tissue (ectoderm-derived) and structures resembling embryonal carcinoma. Focal areas were observed containing structures closely resembling ciliated respiratory epithelium (endodermal) and muscle (mesodermal). In vitro differentiation assays demonstrated that these cells generated embryoid bodies and differentiate into Nestin-expressing neuroepithelial rosette-like structures and βIII-tubulin-positive cells of neuronal morphology (supplemental Figure 5).
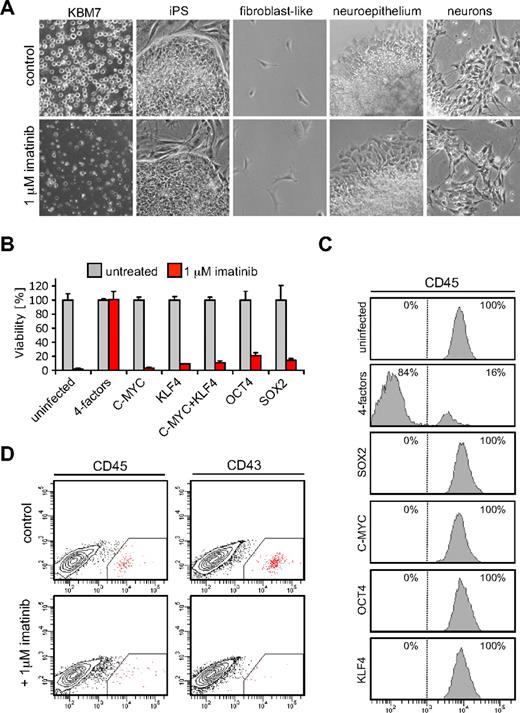
Exquisite dependency of CML cells on BCR-ABL signaling forms the basis for successful clinical suppression of disease by imatinib (STI-571; Gleevec).14,15 The mechanism of this dependency is poorly understood. It has been suggested that the accumulation of genetic alterations in tumor cells leads to a new genetic program in which the acquired oncogenic lesion becomes an indispensable component.16 We used cancer-derived iPSCs to study oncogene dependence. In contrast to parental KBM7 cells, KBM7-iPS1 and KBM7-iPS2 were completely resistant to imatinib (Figure 2A). Loss of oncogene addiction was observed in iPSCs but also in neuronal cells or fibroblast-like cells derived from iPS2 cells (Figure 2A). Imatinib resistance was not observed when each transcription factor was individually introduced into KBM7 cells (Figure 2B) and was observed 7 days after infection with the 4 transcription factors (Figure 2B). The majority of cells lost expression of the pan-hematopoietic marker CD45 (Figure 2C), suggesting a certain level of dedifferentiation that was not observed when cells expressed either transcription factor alone (Figure 2C). After 1 week, expression of the 4 factors induced partial demethylation of the OCT4 and NANOG promoters and NANOG was detected (supplemental Figure 6A). Although these cells showed signs of dedifferentiation, they did not yet fully resemble pluripotent ES cells as apparent from absence of LIN28 and E-cadherin expression (supplemental Figure 6B). We addressed whether differentiation toward the hematopoietic lineage would restore oncogene addiction. Embryoid bodies derived from KBM7-iPSCs were cultured in serum-free medium supplemented with hematopoietic cytokines. Although further experiments are needed to address to what extent these cells are able to differentiate into all lineages, cells positive for CD34, CD43, and CD45 could be observed indicative of hematopoietic differentiation. Addition of imatinib to the culture medium for a period of 3 days strongly reduced the number of cells that were positive for the hematopoietic markers, whereas the viability of the nonhematopoietic cells was unaffected (Figure 2D; supplemental Figure 7). These experiments suggest that, despite the presence of oncogenic lesions and aberrant wiring of regulatory circuits in KBM7 cells, the process of reprogramming readily abolishes BCR-ABL dependency, which is restored by differentiation into the hematopoietic lineage.
Reprogramming of KBM7 cells results in escape from oncogene addiction. (A) KBM7-iPSCs acquired insensitivity to treatment with 1μM of the BCR-ABL kinase inhibitor imatinib (Gleevec), whereas parental KBM7 cells were highly sensitive. KBM7 cells, reprogrammed KBM7-iPS2 cells, and nonhematopoietic differentiated derivatives were cultured without (top row) or with the addition of 1μM imatinib for 3 days (bottom row). All images were acquired with a standard Nikon microscope with a 10× objective. (B) Viability of KBM7 cells infected with retroviral constructs encoding indicated cDNAs on imatinib treatment 7 days after infection. Only when the 4 factors were expressed in combination, KBM7 cells acquired insensitivity to imatinib. Cell viability was measured using an XTT (2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) conversion assay. Values given are percentages of uninfected control cultures. Data are the mean plus or minus SD values from a typical experiment performed in triplicate. (C) Flow cytometry histograms of KBM7 cells 7 days after infection with retroviruses expressing indicated cDNAs showed that a combination of the 4 reprogramming factors led to loss of expression of CD45, a pan-hematopoietic marker. (D) Flow cytometric analysis of KBM7-iPSCs differentiated into the hematopoietic lineage showed distinct subpopulation stained by CD45 and CD43 antibodies. Imatinib treatment resulted in a reduction of these subpopulations.
Reprogramming of KBM7 cells results in escape from oncogene addiction. (A) KBM7-iPSCs acquired insensitivity to treatment with 1μM of the BCR-ABL kinase inhibitor imatinib (Gleevec), whereas parental KBM7 cells were highly sensitive. KBM7 cells, reprogrammed KBM7-iPS2 cells, and nonhematopoietic differentiated derivatives were cultured without (top row) or with the addition of 1μM imatinib for 3 days (bottom row). All images were acquired with a standard Nikon microscope with a 10× objective. (B) Viability of KBM7 cells infected with retroviral constructs encoding indicated cDNAs on imatinib treatment 7 days after infection. Only when the 4 factors were expressed in combination, KBM7 cells acquired insensitivity to imatinib. Cell viability was measured using an XTT (2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) conversion assay. Values given are percentages of uninfected control cultures. Data are the mean plus or minus SD values from a typical experiment performed in triplicate. (C) Flow cytometry histograms of KBM7 cells 7 days after infection with retroviruses expressing indicated cDNAs showed that a combination of the 4 reprogramming factors led to loss of expression of CD45, a pan-hematopoietic marker. (D) Flow cytometric analysis of KBM7-iPSCs differentiated into the hematopoietic lineage showed distinct subpopulation stained by CD45 and CD43 antibodies. Imatinib treatment resulted in a reduction of these subpopulations.
We show here that a human cancer cell line can be reprogrammed into a state of restricted pluripotency. It remains to be seen whether other human cancer cells can be reprogrammed into iPSCs. Interestingly, the oncogenic mutations present in the reprogrammed KBM7-iPSCs do not fully prevent cellular differentiation occurring during teratoma formation, although we cannot exclude that the degree of maturation is less extensive than observed in teratomas derived from noncancerous cells. Finally, our results could suggest that a specific differentiated epigenetic cell state is needed to maintain BCR-ABL dependency, providing a rationale for heterogeneity in response to therapeutics targeting oncogene addiction. This may contribute to the inability of BCR-ABL inhibitors to fully cure disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Maisam Mitalipova, Frank Soldner, Menno Creyghton, and Wenjun Guo for reagents and helpful discussion.
T.R.B. was supported by the Kimmel Foundation and the Whitehead Institute Fellows program. R.J. was supported by the National Institutes of Health (grants RO1-HD045022 and R37-CA084198).
National Institutes of Health
Authorship
Contribution: J.E.C., J.P., and T.R.B. designed and performed research, analyzed data, and wrote the paper; M.V., V.A.B., S.G., and F.D.C. designed and conducted research and analyzed data; and M.W. and R.J. analyzed research and wrote the paper.
Conflict-of-interest disclosure: R.J. is an advisor to Stemgent and a cofounder of Fate Therapeutics. The remaining authors declare no competing financial interests.
The current affiliation for S.G. is University Pathologists, Roger Williams Medical Center/Boston University School of Medicine, Providence, RI. The current affiliation for F.D.C. is Harvard University, Department of Stem Cell and Regenerative Biology, Children's Hospital Boston, Boston, MA. The current affiliation for M.W. is Institute for Stem Cell Biology and Regenerative Medicine, Stanford University School of Medicine, Palo Alto, CA.
Correspondence: Thijn R. Brummelkamp, Whitehead Institute for Biomedical Research, Nine Cambridge Center, Cambridge, MA 02142; e-mail: brummelkamp@wi.mit.edu.
References
Author notes
J.E.C. and J.P. contributed equally to this study.