Abstract
Platelets play a key role in hemostasis and various diseases including arterial thrombosis. Glycoprotein VI (GPVI) mediates adhesion to collagen structures exposed at sites of vascular injury and subsequent platelet activation. We determined the effects of specific activation of GPVI on the human platelet proteome. Isolated human platelets were stimulated with an activating monoclonal antibody specific for GPVI. Platelet proteins were analyzed by 2-dimensional difference gel electrophoresis (2D-DIGE) and mass spectrometry. We identified 8 differentially abundant proteins associated with cell signaling, metabolism, organization and rearrangement of the cytoskeleton, and membrane trafficking. Differentially abundant proteins included aldose reductase (AR), beta-centractin, charged multivesicular body protein 3, Src substrate cortactin, ERp57, and pleckstrin. Importantly, GPVI-modulated protein abundance was functionally relevant. Correspondingly, AR enzyme activity significantly increased upon GPVI activation and inhibition of AR resulted in reduced platelet aggregation. Furthermore, ERp57 was released upon ligation of platelet GPVI and increased the activity of tissue factor, a major initiator of blood coagulation. In summary, GPVI activation results in differential changes in abundance of platelet proteins, including AR and ERp57, which support platelet aggregation and platelet-dependent coagulation. These results provide further insight into the mechanisms that underlie platelet activation through the GPVI receptor and may help to identify novel pharmacologic targets.
Introduction
Platelets play a fundamental role in bleeding control and wound healing, but also in the pathophysiology of various diseases such as atherosclerosis and arterial thrombosis. After vascular injury, platelets accumulate rapidly at the site of endothelial disrupture binding to exposed subendothelial matrix proteins. As collagen represents the main constituent of the vascular wall, platelet-collagen interactions are critical events in the pathophysiology of cardiovascular diseases.1
Platelet collagen receptors are divided into those directly interacting with collagen, including glycoprotein VI (GPVI), the integrin α2β1, and CD36, and those that interact indirectly through collagen-bound von Willebrand factor, including GPIbα and the integrin αIIbβ3 (reviewed in Clemetson and Clemetson2 ). Platelet GPVI has been identified as the major direct platelet collagen receptor.3 GPVI is a 60- to 65-kDa type I transmembrane glycoprotein that belongs to the immunoglobulin superfamily.4 In humans, GPVI forms a complex with the Fc receptor γ-chain on the cell surface.5,6 Ligand binding to GPVI triggers tyrosine phosphorylation of the immunoreceptor tyrosine-based activation motif of the Fc receptor γ-chain, initiating downstream signaling via Syk kinases, LAT, SLP-76, and phospholipase C. Furthermore, stimulation of GPVI induces platelet activation leading to secretion and inside-out signaling to the integrins α2β1 and αIIbβ3 that stabilize platelet interaction with the arterial wall and mediate platelet aggregation.7,8 Platelets deficient in GPVI show loss of collagen-induced adhesion and aggregation in vitro.3,9 Likewise, function blocking anti-GPVI monoclonal antibodies attenuate ex vivo platelet aggregation in response to collagen and collagen-related peptide, which mimics the collagen triple helix.10,11 Recently, we demonstrated in mice in vivo that GPVI is a major determinant of arterial thrombus formation.12 Thus, platelet collagen receptors, and especially GPVI, have emerged as a highly interesting target for novel antiplatelet drugs.13
Recent advances in mRNA and protein analysis are expected to significantly contribute to a better understanding of the fundamentals of platelet biology. Because platelets do not have a nucleus, proteomics technology is the method of choice to provide data on protein expression in these cells. In the present study, we applied 2-dimensional difference gel electrophoresis (2D-DIGE) to investigate differential changes in the human platelet proteome after stimulation of the GPVI receptor. This elaborate technique combines considerable advantages compared with classic 2D-gel electrophoresis: (1) direct comparison of stimulated sample and control in one gel, (2) inclusion of an internal pooled protein standard that facilitates the normalization of different gels, and (3) the reliable quantitation of proteins within a high dynamic range. Thus, 2D-DIGE has emerged as a powerful tool to comprehensively assess changes in the cytoplasmic platelet proteome while minimizing interassay variability.14,15 Using 2D-DIGE and mass spectrometry, we here determined that specific activation of platelet GPVI results in a significant change in abundance of several proteins. Consecutively, we also analyzed functional changes of 2 identified proteins and their relevance for platelet aggregation and blood coagulation.
Methods
For more information, see the supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Antibody generation
The monoclonal antibody directed against human GPVI was generated as previously described.16 In brief, Lou/C rats were immunized with an adenovirally expressed human GPVI-Fc fusion protein. The latter represents a soluble form of GPVI with the extracellular domain of human GPVI fused to the human Fc domain. Antibodies were screened and purified as described.16 HGP4C9 monoclonal antibody (immunoglobulin G1 [IgG1] subtype) specifically bound to GPVI-Fc but not control Fc (not shown). RmC7H8, raised in rats against an irrelevant human antigen, served as control monoclonal antibody (mAb).
Platelet preparation
Whole blood (100 mL) was drawn without stasis from 7 healthy blood donors between 22 and 36 years of age (2 male, 5 female) directly into 20% ACD buffer (85mM sodium citrate, 64.9mM citric acid, 111mM D-glucose). Approval was obtained from the Technical Institute of Munich institutional review board for these studies, and informed consent was obtained in accordance with the Declaration of Helsinki. Blood was centrifuged for 20 minutes at 206g without a break. The supernatant platelet-rich plasma was subjected to gel filtration using several sepharose 2B columns in parallel with a gel bed of 50 mL each, 10-cm column length, and a diameter of 2.5 cm. A maximum of 10 mL of platelet-rich plasma was loaded onto each column and gravity flow was applied for gel filtration. Gel filtration buffer (137mM sodium chloride, 2.7mM potassium chloride, 1mM magnesium chloride, 5.5mM D-glucose, 3mM sodium dihydrogen orthophosphate, 3.5mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 0.35% [wt/vol] bovine serum albumin, pH 7.35) was used for equilibration of sepharose 2B columns and elution of platelets in fractions of 2 mL. Amount and concentration of platelets in the resulting suspension were analyzed using a Sysmex 3 part differential blood cell analyzer (KX-21; Sysmex Europe GmbH). To avoid contamination with von Willebrand factor, the fractions were collected until the concentration maximum of platelets was reached.17 Fractions containing platelets were mixed, acidified with 2.5 times volume washing buffer (90mM sodium chloride, 5mM potassium chloride, 36mM sodium citrate, 5mM D-glucose, 10mM ethylenediaminetetraacetic acid, pH 6.5 to prevent clotting) and spun down (12 minutes, 530g, 20°C). The pellet was resuspended in 1 mL of washing buffer and the platelet count was analyzed again. One-half of each sample was designated for activation, and the other half was intended as control; thus each pair of sample and control consisted of platelets of a single donor and underwent the same process. Total platelet recovery per milliliter ranged between 6.2 × 108 and 14.8 × 108, with a purity of more than 99%. Bradford assay measurements of protein concentration yielded between 1 and 3 mg of protein per original blood sample.
Platelet activation
For activation, 2 × 108 platelets per milliliter in Tyrode buffer (137mM sodium chloride, 12.1mM sodium hydrogen carbonate, 2.61mM potassium chloride, 0.1% [wt/vol] bovine serum albumin, 0.1% [wt/vol] d-glucose, 10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4) were mixed with magnesium chloride and calcium chloride to a final concentration of 1mM each. For flow cytometric analysis, platelet activation was performed with different concentrations of HGP4C9 monoclonal antibody for 10 minutes. Subsequently, 1 μg/mL HGP4C9 was used for proteome analysis, and also for further functional studies on platelets. To preclude unspecific binding of the constant region of the antibody, control samples were treated with an isotype control IgG1 antibody (RmC7H8, 1 μg/mL) for 10 minutes. After activation, the suspensions were pelleted (12 minutes, 530g, 20°C), and platelet pellets were shock frozen in liquid nitrogen and stored at −80°C. The known platelet agonist adenosine diphosphate (ADP) was additionally used for activation and subsequent fluorescence-activated cell sorting (FACS) analysis; nonstimulated controls were treated with phosphate-buffered saline.
2D-DIGE
Proteins were labeled with spectrally resolvable fluorescent cyanine dyes before further separation according to their charge and size, respectively.18 For each of the 7 biologic replicates, one analytic 2D-DIGE gel was prepared for pH 4.5 to 6.8, and one for pH 6.25 to 8.6, each containing a total of 150 μg of protein (50 μg each of cyanine 5 [Cy5]-labeled proteins of control platelets, Cy3-labeled proteins of GPVI-activated platelets, and Cy2-labeled internal pooled standard proteins). The process of detection of differentially abundant spots was carried out in several consecutive steps including minimal labeling of the samples, 2D-gel electrophoresis consisting of isoelectric focusing (IEF) and sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE), acquisition of the gel images, and DeCyder (2-D Differential Analysis software v6.5; GE Healthcare) analysis. The whole strategy consisted of 14 protein gels and thus a total of 42 readout images for the 3 different labeling dyes. Changes in protein abundance were considered biologically relevant, if (1) the spot was present in all 7 gels, (2) the average ratio exceeded plus or minus 2.0 for pH 4 to 7 gels and plus or minus 1.5 for pH 6 to 9 gels, and (3) the P value was less than .05.
Isoelectric focusing (IEF) was performed using the Multiphor II IEF system (GE Healthcare Biosciences). Samples were applied onto 18-cm immobilized pH gradient strips pH 4 to 7 and pH 6 to 9. DryStrips (GE Healthcare) were rehydrated overnight in buffer (7 M urea, 2 M thiourea, 4% [wt/vol] 3-[(3-cholamidopropyl) dimethylammonio]-1-propane sulphonate (CHAPS), 1% [vol/vol] Pharmalyte (GE Healthcare), pH 3-10, 13mM 1,4-dithioerythritol (DTE), 0.8% [vol/vol] bromphenol blue; for pH gradient pH 6-9, DryStrips DTE was replaced by 0.012% [vol/vol] DeStreak [GE Healthcare] reagent). Anodic sample cup loading was used for all analytic and pH 6 to 9 preparative samples, and in-gel rehydration was used for pH 4 to 7 preparative samples. Analytic and preparative samples on pH 4 to 7 gradients were focused for a total of 44 kVh, analytic samples on pH 6 to 9 gradients were focused for a total of 34.5 kVh, and preparative samples on pH 6 to 9 gradients were focused for a total of 55 kVh. SDS-PAGE was performed in a Protean II xi device (Bio-Rad) using a 2-cm 4% polyacrylamide stacking gel and a 12% polyacrylamide separation gel. The immobilized pH gradient strips were loaded onto the SDS–polyacrylamide gels and fixed with 0.5% (wt/vol) agarose in running buffer (25mM tris(hydroxymethyl)aminomethane, 192mM glycine, 3.4mM SDS). Gels were run at 9°C, 30 mA/gel for 30 minutes and 40 mA/gel for 3 hours in running buffer. Please refer to the supplemental Methods for details.
For preparative gels, 500 μg of unlabeled platelet protein was applied to each gel. The gels were poststained with Roti-Blue (Carl Roth) colloidal Coomassie stain for evaluation of total protein content. All spots of interest were manually picked from the gels (supplemental Figure 1). From the location of the spots in the gel, the approximate protein size and the isoelectric point (pI) were estimated for each excised spot. All spots were digested with trypsin and subjected to mass spectrometry.
Mass spectrometry and database search
We applied a 4800 matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometer (Applied Biosystems) for analysis of mass spectrometry (MS) and tandem MS (MS/MS) spectra, and a Thermo LTQ mass spectrometer (Thermo Electron) for liquid chromatography (LC) MS/MS analysis. We used the Mascot V2.1 (Matrix Science) algorithm for database search. Please refer to the supplemental Methods for details.
Western blot
Washed human platelets were activated as described in “Platelet activation,” and both platelets and platelet releasates were collected. Probes were lysed in prechilled radioimmunoprecipitation assay buffer.19 After 30 minutes of incubation on ice, cell debris was removed by centrifugation at 16 000g for 15 minutes. From each sample, 40 μg of total protein (or 30 μg of protein from platelet supernatants) was separated under reducing conditions on a 4% to 20% precast Novex tris(hydroxymethyl)aminomethane-glycine gradient gel (Invitrogen) and transferred to a Hybond nitrocellulose membrane (Amersham Pharmacia Biotech). The blots were probed with antibodies directed against aldose reductase (Santa Cruz Biotechnology), charged multivesicular body protein 3 (CHMP3; Santa Cruz Biotechnology), cortactin (Cell Signaling Technology), ERp57 (Abnova), and pleckstrin (Acris), and visualized by enhanced chemiluminescence using a commercially available kit (Pierce). Equal loading of each lane was assessed using an antibody recognizing total beta-tubulin (Cell Signaling Technology).
Flow cytometric analysis
Evaluation of surface expression of platelet P-selectin (CD62P), CD40 ligand (CD40L), and integrin αIIbβ3 (glycoprotein [GP]IIbIIIa) with specific monoclonal antibodies (mAbs) was conducted on a FACSCalibur (BD Biosciences) as described in detail previously.7,19 The following mAbs were used: anti-CD62P (clone CLB Thromb/6; Biozol), anti-CD40L (clone 24-31; Calbiochem), and PAC-1 (340507; BD Biosciences). PAC-1 is a murine monoclonal antibody directed against the platelet fibrinogen receptor, integrin αIIbβ3, that binds to activated but not unstimulated platelets.20
Platelet aggregation
Optical aggregometry was performed as described21 using HGP4C9 (1 μg/mL). For inhibition of aldose reductase (AR), platelets were preincubated with epalrestat (3 or 10μM; Sequoia Research Products) or 3-O-methyl-quercetin (1 or 10μM; Sequoia Research Products) for 10 minutes before stimulation with HGP4C9.
Aldose reductase activity assay
Aldose reductase activity was measured as described previously22 with slight modifications. Briefly, reduction of nicotinamide adenine dinucleotide phosphate (NADPH) was assessed photometrically by measuring the decrease of absorption at 340 nm in a reaction mixture consisting of 140 μL of sodium phosphate buffer (final concentration, 10mM; pH 6.2), 20 μL of NADPH (final concentration, 100μM), and 20 μL of platelet lysate (final concentration, 1 mg/mL). The reaction was started by addition of 20 μL of DL-glyceraldehyde (final concentration, 100mM; all reagents from Sigma-Aldrich). Absorption was measured at room temperature for 3 minutes. All values were corrected for blanks containing all components of the reaction except for the substrate. AR activity was normalized to protein content as measured by bicinchoninic acid protein (BCA) assay (Sigma-Aldrich) and expressed in milliunits per microgram (nanomoles NADPH oxidized per minute per microgram of protein). For inhibition of aldose reductase, platelets were preincubated with 10μM epalrestat (Sequoia Research Products) for 10 minutes before stimulation with HGP4C9.
Procoagulant activity
Functional relevance of GPVI-dependent regulation of ERp57 was determined using a previously described coagulation assay that quantifies factor Xa formation.23,24 The method uses a chromogenic substrate specific for factor Xa (S2222; Chromogenix), the product of hydrolysis being directly proportional to the amount of factor Xa present.23 Human platelets were incubated with 1 μg/mL HGP4C9 or control IgG (RmC7H8) for 10 minutes. After activation, platelets were centrifuged for 10 minutes at 600g and platelet supernatants (P-SNs) were retrieved. P-SNs were incubated with human recombinant tissue factor (Innovin, dilution 1:500; Dade Behring) in the presence of anti-ERp57 (1:50 dilution; Abcam) or isotype control antibody. Each value was determined in triplicate measurements.
Statistics
Data are presented as mean (± SEM) and analyzed using Student t test or 1-way analysis of variance with a Tukey posthoc test where appropriate. P values less than .05 were considered statistically significant.
Results
Stimulation of GPVI induces platelet activation
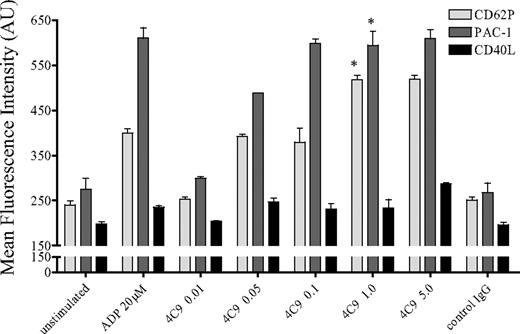
Interaction of collagen with platelets induces aggregation and secretion, which results in thrombus formation in vivo.12 To evaluate the role of GPVI for platelet activation, we stimulated the GPVI receptor with the specific mAb HGP4C9 or an irrelevant nonbinding control IgG1. Using FACS analysis, we determined that ligation of GPVI through mAb HGP4C9 resulted in a substantial, dose-dependent release of the α-granule protein P-selectin (CD62P) and, to a lesser extent, also of the cytosolic cytokine CD40L. In addition, GPVI signaling induced activation of the integrin αIIbβ3 as indicated by PAC-1 binding (Figure 1). The effects of GPVI ligation on P-selectin and PAC-1 binding were comparable with platelet activation induced by 20μM ADP. As higher concentrations did not translate into stronger platelet activation, platelets were activated with 1 μg/mL HGP4C9 throughout all experiments. Importantly, preincubation of HGP4C9 with a soluble GPVI-Fc fusion protein, consisting of the extracellular domain of GPVI and human C-terminal Fc tag,16 completely abolished platelet activation in a concentration-dependent manner (supplemental Figure 2A). This supports the notion that HGP4C9 specifically binds to GPVI. The Fab fragment of HGP4C9 did not cause platelet activation and secretion. This indicates that platelet activation by HGP4C9 depends on cross-linking of GPVI via the full IgG (supplemental Figure 2B) and probably involves Fcγ receptor signaling.
Stimulation of GPVI receptor with the mAb HGP4C9 induces platelet activation. P-selectin (CD62P), activated integrin αIIbβ3 (PAC-1), and CD40L surface expression was determined on isolated human platelets after stimulation with ADP (20μM), different concentrations of mAb HGP4C9 (0.01-5 μg/mL), or control IgG1 (5 μg/mL) for 10 minutes (n = 4-6, mean ± SEM, *P < .001 for HGP4C9 1 μg/mL vs control). Results are given as mean fluorescence intensity (arbitrary units, AU).
Stimulation of GPVI receptor with the mAb HGP4C9 induces platelet activation. P-selectin (CD62P), activated integrin αIIbβ3 (PAC-1), and CD40L surface expression was determined on isolated human platelets after stimulation with ADP (20μM), different concentrations of mAb HGP4C9 (0.01-5 μg/mL), or control IgG1 (5 μg/mL) for 10 minutes (n = 4-6, mean ± SEM, *P < .001 for HGP4C9 1 μg/mL vs control). Results are given as mean fluorescence intensity (arbitrary units, AU).
2D-DIGE
In all HGP4C9-treated samples of isolated and purified platelets, we identified the GPVI antibody, indicating that the antibody HGP4C9 had actually bound to the platelets. In contrast, no IgG could be detected by 2D-DIGE when we exposed platelets to an isotype-matched control antibody (IgG1), thus indicating the specificity of the GPVI-platelet interaction.
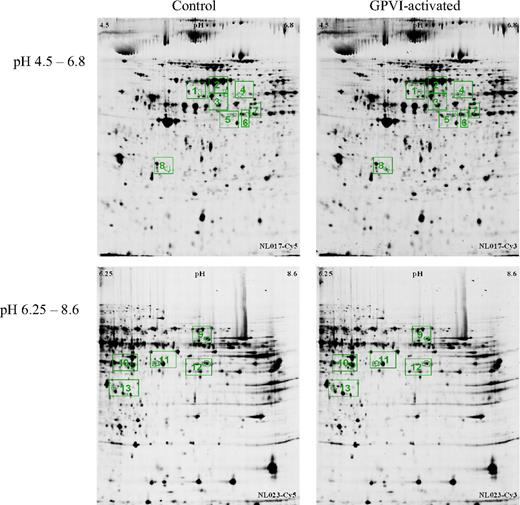
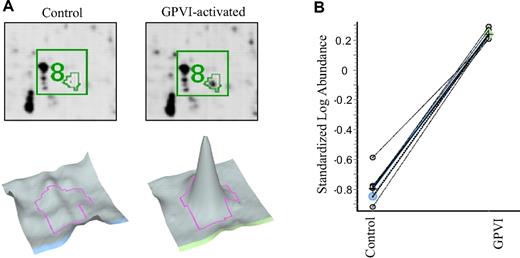
We investigated the differential platelet proteome in 7 healthy human donors. Final evaluation of differences between control and GPVI-activated states led to a list of 13 differentially abundant protein spots (Table 1). Eight of these protein spots were found in the pH range 4.5 to 6.8, all of which were increased in the GPVI-activated samples with a minimum factor of 2. The remaining 5 protein spots were found in the pH range 6.25 to 8.6, of which 1 was increased and 4 were decreased in the GPVI-activated sample, with a minimum factor of plus or minus 1.5 (Figures 2–3 and Table 1). Figure 3 illustrates the high reproducibility of the observed abundance alterations, showing, as an example, the quantification of spot 8 (identified as “charged multivesicular body protein 3”) in 7 biologic replicates. As illustrated in supplemental Figure 3, all differentially abundant proteins were examined for variability within the 7 biologic replicates.
List of differentially abundant protein spots with corresponding average ratio and DeCyder P values
| Spot no. . | Protein . | Average ratio . | DeCyder P . | SC, % . | IS . | PS . | Accession no. . | No. of identified peptides . | Mass database, Da . | Mass gel, Da . | pI database . | pI gel . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ig gamma-1 chain C region (rat) | 5.3 | 1.3e−05 | 37 | 728 | 23 | 36 493 | 55 000 | 6.43 | 5.6 | ||
| 2 | Ig gamma-1 chain C region (rat) | 4.8 | 4.2e−11 | 26 | 380 | 10 | 35 923 | 55 000 | 6.43 | 5.9 | ||
| 3 | Ig gamma-1 chain C region (rat) | 3.0 | 3.8e−7 | 26 | 626 | P30101 | 22 | 36 493 | 55 000 | 6.43 | 5.9 | |
| ERp57 | 10 | 110 | P30101 | 3 | 57 146 | 5.98 | ||||||
| 4 | Src substrate cortactin | 2.4 | 1.2e−08 | 37 | 97 | 209 | Q14247 | 1 | 61 770 | 50 000 | 5.24 | 6.1 |
| 5 | Pleckstrin | 6.6 | 7.4e−03 | 24 | 189 | 223 | P08567 | 3 | 40 456 | 45 000 | 8.32 | 6.0 |
| 6 | Pleckstrin | 5.4 | 9.8e−03 | 43 | 371 | 459 | P08567 | 4 | 40 456 | 45 000 | 8.50 | 6.1 |
| 7 | Pleckstrin | 5.0 | 1.2e−02 | 22 | 302 | 328 | P08567 | 4 | 40 456 | 45 000 | 8.50 | 6.2 |
| beta-centractin | 35 | 64 | 143 | P42025 | 1 | 42 381 | 5.98 | |||||
| 8 | Charged multivesicular body protein 3 | 10.6 | 8.5e−12 | 16 | 122 | Q9CQ10 | 3 | 24 983 | 29 000 | 5.10 | 5.3 | |
| 9 | Pyruvate kinase isozymes M1/M2 | −1.6 | 3.2e−04 | 50 | 355 | 559 | P14618 | 7 | 58 339 | 55 000 | 7.95 | 7.5 |
| N(2) N(2)-dimethylguanosine tRNA methyltransferase | 16 | 11 | 59 | Q9NXH9 | 0 | 73 386 | 7.77 | |||||
| 10 | Pleckstrin | −1.5 | 2.7e−02 | 37 | 426 | 499 | P08567 | 6 | 40 471 | 45 000 | 8.50 | 6.6 |
| 11 | Pleckstrin | −1.6 | 3.8e−02 | 30 | 231 | 275 | P08567 | 4 | 40 471 | 45 000 | 8.50 | 6.9 |
| 12 | Pleckstrin | −1.5 | 1.1e−02 | 30 | 177 | 226 | P08567 | 3 | 40 471 | 45 000 | 8.50 | 7.5 |
| 13 | Aldose reductase | 1.5 | 4.6e−02 | 14 | 448 | P45376 | 32 | 36 099 | 36 000 | 6.56 | 6.4 |
| Spot no. . | Protein . | Average ratio . | DeCyder P . | SC, % . | IS . | PS . | Accession no. . | No. of identified peptides . | Mass database, Da . | Mass gel, Da . | pI database . | pI gel . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ig gamma-1 chain C region (rat) | 5.3 | 1.3e−05 | 37 | 728 | 23 | 36 493 | 55 000 | 6.43 | 5.6 | ||
| 2 | Ig gamma-1 chain C region (rat) | 4.8 | 4.2e−11 | 26 | 380 | 10 | 35 923 | 55 000 | 6.43 | 5.9 | ||
| 3 | Ig gamma-1 chain C region (rat) | 3.0 | 3.8e−7 | 26 | 626 | P30101 | 22 | 36 493 | 55 000 | 6.43 | 5.9 | |
| ERp57 | 10 | 110 | P30101 | 3 | 57 146 | 5.98 | ||||||
| 4 | Src substrate cortactin | 2.4 | 1.2e−08 | 37 | 97 | 209 | Q14247 | 1 | 61 770 | 50 000 | 5.24 | 6.1 |
| 5 | Pleckstrin | 6.6 | 7.4e−03 | 24 | 189 | 223 | P08567 | 3 | 40 456 | 45 000 | 8.32 | 6.0 |
| 6 | Pleckstrin | 5.4 | 9.8e−03 | 43 | 371 | 459 | P08567 | 4 | 40 456 | 45 000 | 8.50 | 6.1 |
| 7 | Pleckstrin | 5.0 | 1.2e−02 | 22 | 302 | 328 | P08567 | 4 | 40 456 | 45 000 | 8.50 | 6.2 |
| beta-centractin | 35 | 64 | 143 | P42025 | 1 | 42 381 | 5.98 | |||||
| 8 | Charged multivesicular body protein 3 | 10.6 | 8.5e−12 | 16 | 122 | Q9CQ10 | 3 | 24 983 | 29 000 | 5.10 | 5.3 | |
| 9 | Pyruvate kinase isozymes M1/M2 | −1.6 | 3.2e−04 | 50 | 355 | 559 | P14618 | 7 | 58 339 | 55 000 | 7.95 | 7.5 |
| N(2) N(2)-dimethylguanosine tRNA methyltransferase | 16 | 11 | 59 | Q9NXH9 | 0 | 73 386 | 7.77 | |||||
| 10 | Pleckstrin | −1.5 | 2.7e−02 | 37 | 426 | 499 | P08567 | 6 | 40 471 | 45 000 | 8.50 | 6.6 |
| 11 | Pleckstrin | −1.6 | 3.8e−02 | 30 | 231 | 275 | P08567 | 4 | 40 471 | 45 000 | 8.50 | 6.9 |
| 12 | Pleckstrin | −1.5 | 1.1e−02 | 30 | 177 | 226 | P08567 | 3 | 40 471 | 45 000 | 8.50 | 7.5 |
| 13 | Aldose reductase | 1.5 | 4.6e−02 | 14 | 448 | P45376 | 32 | 36 099 | 36 000 | 6.56 | 6.4 |
A positive algebraic sign indicates an increase of intensity in the GPVI-activated samples; a negative algebraic sign indicates a decrease. Data were generated using DeCyder software.
2D-DIGE analysis of human platelets incubated with control IgG1 (left column), or after activation of GPVI receptor (HGP4C9 mAb 1 μg/mL, right column) at different pH gradients: 4.5-6.8 (top row) and 6.25-8.6 (bottom row). Images show representative gels of proteins labeled with the fluorescent cyanine dyes Cy5 (control) and Cy3 (GPVI-activated sample). Differentially abundant proteins are framed in green: protein spots 1 to 8 (pH 4.5-6.8, top row), and protein spots 9-13 (pH 6.25-8.6, bottom row).
2D-DIGE analysis of human platelets incubated with control IgG1 (left column), or after activation of GPVI receptor (HGP4C9 mAb 1 μg/mL, right column) at different pH gradients: 4.5-6.8 (top row) and 6.25-8.6 (bottom row). Images show representative gels of proteins labeled with the fluorescent cyanine dyes Cy5 (control) and Cy3 (GPVI-activated sample). Differentially abundant proteins are framed in green: protein spots 1 to 8 (pH 4.5-6.8, top row), and protein spots 9-13 (pH 6.25-8.6, bottom row).
Example of a protein analysis of spot 8. (A) Enlarged image of 2D-DIGE showing protein spot 8. Platelets were incubated with control IgG1 (left) or HGP4C9 for GPVI activation (right) in the pH range 4.5 to 6.8. Below, corresponding 3D images of protein spot 8 are shown. (B) Graph view of spot pair 8 in 7 biologic replicates. The log standardized abundance is derived from the normalized spot volume standardized against the in-gel standard of each gel.
Example of a protein analysis of spot 8. (A) Enlarged image of 2D-DIGE showing protein spot 8. Platelets were incubated with control IgG1 (left) or HGP4C9 for GPVI activation (right) in the pH range 4.5 to 6.8. Below, corresponding 3D images of protein spot 8 are shown. (B) Graph view of spot pair 8 in 7 biologic replicates. The log standardized abundance is derived from the normalized spot volume standardized against the in-gel standard of each gel.
All 13 spots were proportionally increased or decreased in the single gels, and consistency among the biologic replicates could be observed as indicated by P values (Table 1) and 3D representation of all spots (supplemental Figure 3). By reviewing all 3D images, the correct matching and change in abundance could be confirmed.
Identification of proteins of interest
All 13 spots of interest were manually picked from the pH 4.5 to 6.8 and pH 6.8 to 8.25 gels (supplemental Figure 1A-B) and subjected to mass spectrometry. Using matrix-assisted laser desorption/ionization tandem time-of-flight analysis, spots 4 to 7 and 9 to 12 could be identified (Table 1). Spots 1, 2, 3, 8, and 13 were subjected to nano–LC–electrospray ionization (ESI) MS/MS. Importantly, by combining both mass spectrometry methods, all 13 differentially abundant protein spots could be determined (Table 1). Identified proteins included pleckstrin, beta-centractin, Src substrate cortactin, charged multivesicular body protein 3, aldose reductase, and ERp57. These proteins are involved in diverse cell functions such as signaling, metabolism, organization and rearrangement of the cytoskeleton, and membrane trafficking.
Changes in abundance of some proteins, for example, aldose reductase (Figure 4A) and cortactin (supplemental Figure 4B), could be verified using classical SDS-PAGE and Western blot (WB) analysis. However, the WB results obtained for CHMP3 did not reproduce the expression pattern seen using the proteomic approach (Table 1, supplemental Figure 4A). According to Table 1, an increase in CHMP3 abundance is expected in the 4C9-stimulated platelets compared with IgG control. These changes were not seen using WB. Further, WB analysis of ERp57 in platelet lysates showed no differences between 4C9 and IgG treatment. However, we found a significant increase in ERp57 in the platelet releasate (Figure 5A). Finally, we detected minor changes in the total pleckstrin amount by WB (supplemental Figure 4C). However, the protein was identified in 6 different spots using 2D-DIGE (Table 1), and the calculated average ratio suggested both an increase (spots 5-7) and a decrease (spots 10-12) between the 4C9- and IgG-treated samples.
GPVI-ligation triggers aldose reductase activation. (A) Detection of aldose reductase (AR) in platelets by Western blot analysis. Cellular lysates from platelets treated with control IgG (RmC7H8) or GPVI antibody HGP4C9 (1 μg/mL) for 10 minutes were obtained, and AR expression (36-kDa protein) was determined by Western blot analysis. (B) Aldose reductase activity was measured as described previously by photometric quantification of NADPH reduction.22 AR activity was normalized to protein content as measured by BCA assay and expressed in milliunits per microgram (nanomoles NADPH oxidized per minute per microgram of protein). For inhibition of aldose reductase (AR) platelets were preincubated with 10μM epalrestat for 10 minutes before stimulation with HGP4C9 (n = 6, mean ± SEM, *P < .05 for HGP4C9 vs IgG [solvent] and **P < .01 for HGP4C9 with solvent vs epalrestat). (C) Individual data of the human blood donors indicate the effects of platelet stimulation by HGP4C9 compared with control IgG. (1) Individual effects and (2) mean ± SEM (n = 6, *P < .05). (D) Activation of GPVI induces platelet aggregation involving aldose reductase. Aggregation of washed human platelets was induced by the GPVI antibody HGP4C9 (1 μg/mL), and light transmission was assessed in the presence of different aldose reductase inhibitors (epalrestat or quercetin) or solvent. Results are shown as aggregation in percentage of light transmission (n = 4-6, mean ± SEM, *P < .01 for solvent vs 10μM epalrestat and vs 10μM quercetin).
GPVI-ligation triggers aldose reductase activation. (A) Detection of aldose reductase (AR) in platelets by Western blot analysis. Cellular lysates from platelets treated with control IgG (RmC7H8) or GPVI antibody HGP4C9 (1 μg/mL) for 10 minutes were obtained, and AR expression (36-kDa protein) was determined by Western blot analysis. (B) Aldose reductase activity was measured as described previously by photometric quantification of NADPH reduction.22 AR activity was normalized to protein content as measured by BCA assay and expressed in milliunits per microgram (nanomoles NADPH oxidized per minute per microgram of protein). For inhibition of aldose reductase (AR) platelets were preincubated with 10μM epalrestat for 10 minutes before stimulation with HGP4C9 (n = 6, mean ± SEM, *P < .05 for HGP4C9 vs IgG [solvent] and **P < .01 for HGP4C9 with solvent vs epalrestat). (C) Individual data of the human blood donors indicate the effects of platelet stimulation by HGP4C9 compared with control IgG. (1) Individual effects and (2) mean ± SEM (n = 6, *P < .05). (D) Activation of GPVI induces platelet aggregation involving aldose reductase. Aggregation of washed human platelets was induced by the GPVI antibody HGP4C9 (1 μg/mL), and light transmission was assessed in the presence of different aldose reductase inhibitors (epalrestat or quercetin) or solvent. Results are shown as aggregation in percentage of light transmission (n = 4-6, mean ± SEM, *P < .01 for solvent vs 10μM epalrestat and vs 10μM quercetin).
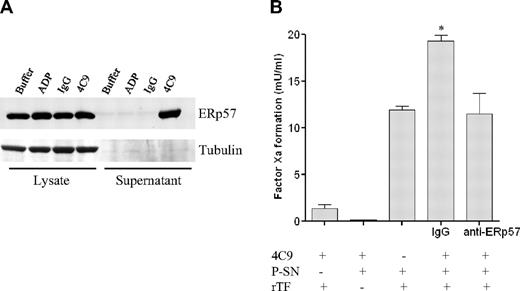
GPVI-activated platelets release ERp57. (A) Detection of ERp57 in platelets and platelet releasates by Western blot analysis. Cellular lysates and supernatants were obtained from platelets treated with either phosphate-buffered saline buffer, control IgG (RmC7H8), GPVI antibody HGP4C9 (1 μg/mL), or ADP (20μM) for 10 minutes, and ERp57 expression (57-kDa protein) was determined by Western blot analysis. (B) GPVI-activated platelets release functionally active ERp57, which stimulates blood coagulation in the presence of recombinant tissue factor (rTF). Supernatants (P-SNs) recovered from HGP4C9 (1 μg/mL)–activated platelets (2 × 107) were incubated with recombinant human TF (Innovin) plus anti-ERp57 or control antibody (IgG) for 15 minutes. TF-induced factor Xa formation was quantified by hydrolysis of the specific substrate S2222 (n = 3, mean ± SEM, *P < .05 IgG vs anti-PDI antibody).
GPVI-activated platelets release ERp57. (A) Detection of ERp57 in platelets and platelet releasates by Western blot analysis. Cellular lysates and supernatants were obtained from platelets treated with either phosphate-buffered saline buffer, control IgG (RmC7H8), GPVI antibody HGP4C9 (1 μg/mL), or ADP (20μM) for 10 minutes, and ERp57 expression (57-kDa protein) was determined by Western blot analysis. (B) GPVI-activated platelets release functionally active ERp57, which stimulates blood coagulation in the presence of recombinant tissue factor (rTF). Supernatants (P-SNs) recovered from HGP4C9 (1 μg/mL)–activated platelets (2 × 107) were incubated with recombinant human TF (Innovin) plus anti-ERp57 or control antibody (IgG) for 15 minutes. TF-induced factor Xa formation was quantified by hydrolysis of the specific substrate S2222 (n = 3, mean ± SEM, *P < .05 IgG vs anti-PDI antibody).
Functional relevance of GPVI-mediated changes in platelet proteome
As determined by 2D-DIGE, activation of GPVI results in differential changes in protein abundance in platelets. To address the biologic relevance of these abundance alterations, we determined the functional role of 2 of the newly identified candidate proteins for GPVI-induced platelet responses. We show here that the abundance of aldose reductase (AR) is increased in platelets after GPVI ligation. Results were verified by WB (Figure 4A). Interestingly, inhibitors of AR have recently been described to influence platelet function in rats.26,27 Therefore, we first focused on the role of AR for GPVI-induced platelet activation. We measured AR activity in platelets in response to GPVI ligation using a recently described assay.22 Importantly, AR activity significantly increased after activation of GPVI by HGP4C9 antibody (Figure 4B-C). In contrast, enzyme activity was virtually abrogated in the presence of the aldose reductase inhibitor epalrestat (Figure 4B). Interestingly, epalrestat also reduced AR activity in platelets incubated with IgG, indicating a constitutive AR activity in platelets. Next, we addressed the role of AR for GPVI-induced platelet aggregation. Although incubation with HGP4C9 resulted in robust aggregation of isolated platelets (Figure 4D), preincubation with inhibitors of AR dose-dependently reduced platelet aggregation. Thus, we have identified a novel pathway, where ligation of GPVI leads to increased abundance of AR protein, which in turn fosters platelet aggregation.
In addition to AR, we show here by 2D-DIGE that also the abundance of ERp57 is increased upon GPVI ligation. ERp57 represents a prominent member of the disulfide isomerase family, and is a close homologue of the enzyme protein disulfide isomerase (PDI).28 This is interesting, because we found only recently that PDI triggers conversion of tissue factor (TF) from the functionally inactive to the active form.23 This process, also referred to as TF decryption, represents an initial step in coagulation activation. Platelets release PDI upon activation and thereby support TF-dependent blood coagulation at sites of vessel damage in mice.23 Hence, we hypothesized here that ligation of GPVI may contribute to platelet-dependent coagulation via ERp57. Using WB analysis, we show that ERp57 is released upon GPVI activation (Figure 5A). To address this further, we used an in vitro coagulation assay that quantifies tissue factor–induced factor Xa formation. Indeed, platelet-dependent TF activation was significantly increased after GPVI activation (Figure 5B). In addition, selective suppression of ERp57 by a specific antibody blunted TF activation induced by GPVI-activated platelets. Hence, we show here, for the first time, that GPVI triggers platelet-dependent coagulation in a process that involves ERp57.
Discussion
Platelet activation through GPVI, the major platelet collagen receptor, is a central event during platelet adhesion and thrombus formation after vascular injury.12 However, the underlying signaling events, including changes in protein expression, remain elusive. Previous studies have shown that platelet activation with the thrombin receptor-activating peptide or the GPVI agonist collagen-related peptide induces diverse changes of the platelet proteome including phosphorylation events.29,30 Thereby, novel signaling proteins (eg, adapter protein Dok-2 and type I transmembrane protein G6f) could be identified.29,30 Some changes described were specific for GPVI, for example, tyrosine phosphorylation of G6f was found to occur in response to collagen but not in response to the G protein–coupled receptor agonists thrombin and ADP.30 However, previous publications used classic 2-dimensional gel electrophoresis or sample prefractionation followed by 1-dimensional (1D) PAGE. Although these methods have proven their feasibility to identify protein changes, there are still limitations regarding their reproducibility and sensitivity.31 Therefore, we used the 2-dimensional fluorescence difference gel electrophoresis (2D-DIGE) technique as the currently most sophisticated approach for quantitative 2D gel approaches.
Platelets are rapidly activated after their incubation with various agonists. This immediately results in functional changes, such as firm adhesion and aggregation. Likewise, 10 minutes of stimulation with our GPVI antibody results in robust platelet activation and aggregation. In the present paper, we therefore addressed changes in protein abundance that appear in parallel to platelet activation and may therefore contribute to alterations in platelet function. We directly compared human platelets activated through GPVI receptor with platelets from the same donor and sample preparation incubated with control IgG. Incubation with the anti-GPVI mAb (1 μg/mL) induced significant platelet activation, which was accompanied by differential changes in abundance of 8 proteins.
Alterations in abundance of some proteins (aldose reductase [AR], cortactin) were clearly verified by SDS-PAGE and WB analysis. However, results from other proteins (CHMP3) were not reproduced using the conventional immunoblotting approach. This discrepancy could be because of the limited dynamic range of WB quantification methods compared with the superior fluorescence-based quantification used in the 2D-DIGE approach. Moreover, in contrast to our 2D DIGE approach, 1D Western blots cannot detect abundance differences within 1 protein species caused by, for example posttranslational modifications such as phosphorylation. We have detected 6 individual spots for pleckstrin and observed a shift of pleckstrin protein spots to a lower isoelectric point (Table 1), suggesting an increase of phosphorylated pleckstrin isoforms and thus a decrease of unphosphorylated isoforms, although the total amount of pleckstrin remains largely stable. This aspect should be clarified by future studies addressing the platelet phosphoproteome. However, because of the constant amount of total pleckstrin, these changes may not be identified by classical SDS-PAGE and Western blot analysis with a pleckstrin antibody.
In general, detection of proteins also depends on their solubility, and proteins altered in abundance by GPVI binding represent very soluble proteins. It is unlikely, however, that the observed changes in protein abundance are caused by differences in solubility, or accessibility, between the GPVI-stimulated and IgG-treated platelets. Instead, the highly reproducible quantitative protein profiles within each group (GPVI and control antibody treated), as indicated by the P values of proteins with altered abundance in the DIGE analysis, demonstrate the high reproducibility of the complete analysis procedure including the initial solubilization of thrombocyte proteins.
Among those proteins that showed significant alterations in abundance, we identified the enzyme AR, which is critically involved in the cell carbohydrate metabolism. Glucose that is not consumed by glycolysis, for example, in patients with high blood glucose, may alternatively enter the polyol pathway where AR reduces it to sorbitol (reviewed in Kador et al32 ).
In our study, GPVI-induced platelet activation resulted in a significant increase of AR (Table 1). This was accompanied by enhanced AR enzyme activity (Figure 4B-C). Conversely, inhibition of AR activity resulted in reduced platelet aggregation (Figure 4D). Together, these data indicate for the first time that AR enzyme activity is enhanced after ligation of GPVI and fosters platelet aggregation. From a clinical point of view, the role of AR for GPVI-dependent platelet activation is striking. AR inhibitors have been reported to prevent secondary complications of diabetes mellitus.33 In diabetic patients, hexokinase becomes saturated as a result of elevated glucose levels and the fraction of glucose metabolized by AR increases.34 It is generally assumed that this enhanced utilization of the AR-dependent polyol pathway is associated with an increased oxidative stress, contributing to the microvascular and macrovascular diabetic complications.22,35 Correspondingly, chronic pharmacologic inhibition of AR has been shown to reduce the frequency of polyneuropathy and also retinopathy in diabetic patients.33,36 Notably, hyperaggregability of platelets is another hallmark of diabetes mellitus. In fact, alterations of platelet function are considered to play a prominent role in the pathogenesis of diabetic microangiopathies and macroangiopathies.37–40 Interestingly, we have recently shown that patients with type 2 diabetes exhibit an enhanced platelet surface expression of the collagen receptor GPVI compared with persons without diabetes, and that stimulation of GPVI results in increased platelet activation and secretion in diabetic subjects.7 Based on our present finding that GPVI triggers enhanced AR enzyme activity in platelets, it is tempting to speculate that AR inhibitors—in addition to their established beneficial effects on microvascular complications of diabetes—may provide a novel therapeutic strategy for the prevention of thrombotic vascular complications in diabetic patients. This will have to be addressed in detail in future.
Among the proteins identified in the present study, we also found an increase in abundance of ERp57. ERp57 belongs to the superfamily of thioredoxins and is found mainly in the endoplasmic reticulum.28 ERp57 exhibits not only enzymatic functions catalyzing disulfide bond formation and isomerization, but also chaperone functions that inhibit protein aggregation.41 ERp57 associates with the ER membrane protein calreticulin that plays a role in a variety of cellular functions including calcium storage and signaling.42 ERp57 and protein disulfide isomerase (PDI) are close homologues and very similar in their domain architecture.41 Although the 2 enzymes recognize common substrate proteins, they can also have different physiologic functions in vitro and in vivo.43
Interestingly, blocking PDI inhibits several platelet activation pathways, including platelet aggregation, secretion, and fibrinogen binding.44 In addition, PDI secreted from platelets stimulates activity of TF in isolated cells and also microparticles. We have shown only recently that PDI is released from adherent platelets at the site of vascular injury in vivo, which triggers TF-dependent blood coagulation in mice.23 However, platelet activation may result in the release of different thiol isomerase enzymes.45 In the present paper, we demonstrate that activation of platelet GPVI increases the release of ERp57, which augments TF activity (Figure 5). Therefore, stimulation of platelet GPVI may represent a novel mechanism that triggers platelet activation and, consecutively, also blood coagulation.
Another protein identified here was pleckstrin, a major substrate of protein kinase C in platelets, which is known to play an important role in cell signaling and cytoskeletal changes and reorganization. Although pleckstrin homology (PH) domains occur in a wide range of proteins involved in intracellular signaling or as constituents of the cytoskeleton, pleckstrin is one of the rare proteins to contain 2 PH domains (reviewed in Toker and Cantley46 ). PH domains form a structurally conserved family that is associated with many regulatory pathways within the cell. In detail, pleckstrin is closely linked to phosphoinositide signaling as it binds specifically to phosphoinositides, for example, to PtdIns(4,5)P2 and Ins(1,4,5)P3.46
In addition, we determined the differential expression of beta-centractin (CTRN2), also referred to as arp-l47 or actin-RPV in vertebrates,48 which is an actin-related protein localized to microtubule-associated structures.49 As a major component of the dynactin complex, CTRN2 is involved in the regulation of cytoplasmic dynein-mediated functions.50
Cortactin is a multidomain protein that is phosphorylated downstream of Src tyrosine kinases and has potential roles in linking signaling molecules with activators of actin assembly. Actin cytoskeletal reorganization downstream of cellular Src plays an important role in growth factor and integrin signaling.51 Only recently it was found that cortactin phosphorylation by Src enhances arp2/3-mediated actin polymerization.51
Proteins that make up the endosomal sorting complex required for transport are involved in the sorting and trafficking of membrane proteins into multivesicular bodies. Charged multivesicular body protein 3 (CHMP3), a subunit of endosomal sorting complex required for transport, has been proposed to play an important role in cytokinesis such as the fusion of multivesicular endosomes with lysosomes.52
It is possible that the observed effects of GPVI ligation on platelet activation and proteomic changes are not completely specific for GPVI, but instead may be part of common pathways after platelet activation via different stimuli. However, immunoblotting of platelet supernatants indicated differences in the release of ERp57 after stimulation with GPVI antibody compared with ADP (Figure 5A), although both agonists induced similar platelet activation as determined by flow cytometry (Figure 1). These findings indicate that stimulation of GPVI leads to specific changes within platelets, and that the observed effects are not an epiphenomenon of platelet activation per se (as induced by ADP). However, this question should be further addressed in future proteomic studies by juxtaposing platelet samples stimulated with different platelet agonists.
In conclusion, we used an elaborate proteomic approach to identify proteins that change in abundance upon activation of the GPVI receptor. The proteins identified are involved in cell signaling (pleckstrin, ERp57), cytoskeletal rearrangement (pleckstrin, cortactin, beta-centractin), membrane trafficking (CHMP3), stress response (ERp57), and metabolic activity (aldose reductase). Importantly, we also provided evidence for a functional relevance of the GPVI-induced changes in the platelet proteome. We believe that the presented data contribute to the understanding of the mechanisms that underlie platelet activation and thrombus formation after vascular injury. This may ultimately lead to the identification of novel targets for future therapeutic interventions in thrombotic disorders.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We acknowledge the excellent technical assistance of Sandra Kerstan.
This study was supported by grants of the German Foundation of Heart Research and the Dr Helmut Legerlotz-Stiftung (C.S.), and grants from the Deutsche Forschungsgemeinschaft (DFG; Ma2186-3/1 and Ma2186-4/1; S.M.). S.M. is a Heisenberg fellow of the DFG.
Authorship
Contribution: C.S. designed and performed research, analyzed data, and wrote the paper; N.V.L. performed research, analyzed data, and wrote the paper; T.F. performed research, contributed vital analytic tools, and analyzed data; M.L., S.P., and C.A.G. performed research and analyzed data; E.K. contributed vital new reagents; M.K. and A.G.K. performed research; B.E. and K.L. contributed vital analytic tools and analyzed data; S.M. designed the research, analyzed data, and wrote the paper; and G.J.A. designed the research, contributed vital analytic tools, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christian Schulz, Deutsches Herzzentrum, Technische Universität München, Lazarettstrasse 36, D-80636 München, Germany; e-mail: schulz@dhm.mhn.de.
References
Author notes
C.S. and N.V.L. contributed equally to this work.

![Figure 4. GPVI-ligation triggers aldose reductase activation. (A) Detection of aldose reductase (AR) in platelets by Western blot analysis. Cellular lysates from platelets treated with control IgG (RmC7H8) or GPVI antibody HGP4C9 (1 μg/mL) for 10 minutes were obtained, and AR expression (36-kDa protein) was determined by Western blot analysis. (B) Aldose reductase activity was measured as described previously by photometric quantification of NADPH reduction.22 AR activity was normalized to protein content as measured by BCA assay and expressed in milliunits per microgram (nanomoles NADPH oxidized per minute per microgram of protein). For inhibition of aldose reductase (AR) platelets were preincubated with 10μM epalrestat for 10 minutes before stimulation with HGP4C9 (n = 6, mean ± SEM, *P < .05 for HGP4C9 vs IgG [solvent] and **P < .01 for HGP4C9 with solvent vs epalrestat). (C) Individual data of the human blood donors indicate the effects of platelet stimulation by HGP4C9 compared with control IgG. (1) Individual effects and (2) mean ± SEM (n = 6, *P < .05). (D) Activation of GPVI induces platelet aggregation involving aldose reductase. Aggregation of washed human platelets was induced by the GPVI antibody HGP4C9 (1 μg/mL), and light transmission was assessed in the presence of different aldose reductase inhibitors (epalrestat or quercetin) or solvent. Results are shown as aggregation in percentage of light transmission (n = 4-6, mean ± SEM, *P < .01 for solvent vs 10μM epalrestat and vs 10μM quercetin).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/20/10.1182_blood-2009-07-230268/5/m_zh89991050470004.jpeg?Expires=1770149666&Signature=v2GmwPknljuDgOMAlxOZF-JIzwd2V6F6LR7qgshwpyXHqCbPwPVUB6zyepH4RLbKg-qyoSlZeJsrZbXVebSuiWd6WFZ7n1TmFuZlnARUTCnMQV48Fx9YzuEnt776XV5nj475G1fOFw4wJ3okfKvDwZZ55jHy~TKSjeZz7FyaPH8qlzH2PyWFwTwMQmpxsP7z0NziAk6b17rWhrZ0~yOYvWR2YIEdfQVD7i2v1E4agLt4KvKVVeg2jeMPpbTqL6yKs5-bKmeT7znomkCe2U7Wzg78FXNewqrtAaBIFJW93BbygoUM0LZiUlvOHQSZGyG0pB~koesXFDQ-kivZgHBy6g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)