Abstract
Loss of function of tumor suppressor genes, such as PTEN, CEBPΑ, and CTNNA1 (encoding the α-catenin protein), has been found to play an essential role in leukemogenesis. However, whether these genes genetically interact remains largely unknown. Here, we show that PTEN-mammalian target of rapamycin signaling acts upstream to dictate the ratio of wild-type p42 C/EBPα to its dominant-negative p30 isoform, which critically determines whether p30 C/EBPα (lower p42/p30 ratio) or p42 C/EBPα (higher p42/p30 ratio) binds to the proximal promoter of the retained CTNNA1 allele. Binding of p30 C/EBPα recruits the polycomb repressive complex 2 to suppress CTNNA1 transcription through repressive H3K27me3 modification, whereas binding of p42 C/EBPα relieves this repression and promotes CTNNA1 expression through activating H3K4me3 modification. Loss of Pten function in mice and zebrafish induces myelodysplasia with abnormal invasiveness of myeloid progenitors accompanied by significant reductions in both wild-type C/EBPα and α-catenin protein. Importantly, frame-shift mutations in either PTEN or CEBPA were detected exclusively in the primary LICs with low CTNNA1 expression. This study uncovers a novel molecular pathway, PTEN-C/EBPα-CTNNA1, which is evolutionarily conserved and might be therapeutically targeted to eradicate LICs with low CTNNA1 expression.
Introduction
The α-catenin protein (encoded by the CTNNA1 gene) functions as a connector linking the E-cadherin/β-catenin complex to the filamentous actin cytoskeleton at adherens junctions.1 In mice, conditional ablation of α-catenin in skin and neural progenitor cells causes epithelial and cortical hyperplasia because of abnormal activation of the Ras-MAPK cascade and hedgehog signaling, respectively,2,3 whereas retroviral insertions that disrupt the 5′ end of the CTNNA1 gene induce murine myeloid leukemia.4 Loss-of-function mutations (including exon skipping or homologous genomic micro-deletion) or decreased α-catenin protein levels have been reported in human cancer cell lines derived from various solid tumors and have been shown to correlate with disease progression and invasiveness.1 Restoration of α-catenin expression in these α-catenin-deficient cells attenuates their ability to form tumors in vivo.5 More recently, several groups showed that CTNNA1 is expressed in normal hematopoietic stem cells (HSCs); however, its expression is significantly lower in human leukemia-initiating cells (LICs) in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML).6-11 These results suggest that CTNNA1 is an essential tumor suppressor gene involved in leukemia stem cell transformation, and they imply that any mechanism that could be exploited to restore CTNNA1 expression represents a potential therapeutic strategy to eliminate LICs expressing low levels of CTNNA1 without adversely affecting normal stem cells.
Phosphatase and tensin homolog (PTEN), a negative regulator of phosphoinositide-3 kinase/Akt/mammalian target of rapamycin (mTOR) signaling, and CCAAT enhancer binding protein alpha (CEBPA, encoding C/EBPα protein) are frequently mutated in hematologic malignancies.12,13 Although elegant studies have shown that conditional ablation of Pten in murine HSCs promotes the formation of LICs and that the development of AML can be rescued by rapamycin, a selective inhibitor of mTOR activity,14-16 the downstream targets mediating these pathologic effects are largely unknown. Heterozygous mutations within the amino terminus of the CEBPA gene eliminate the expression of the 42-kDa wild-type C/EBPα protein (p42 C/EBPα), but they do not affect the 30-kDa isoform initiated further downstream (p30 C/EBPα). It has been shown that the truncated p30 C/EBPα protein not only compromises the DNA-binding ability of p42 C/EBPα, inhibiting trans-activation of key granulocytic target genes in a dominant-negative manner,13 but it also binds to the promoters of a unique set of target genes to suppress their transcription.17 Murine hematopoietic progenitor cells expressing only p30 C/EBPα induce a myeloid malignancy that is sustained by LICs with a committed myeloid molecular signature.14-16 These results suggest that p30 C/EBPα is a functionally independent transcription factor that does not act solely as a dominant-negative regulator of p42 C/EBPα. Although progress has been made on the role of these regulators, whether and how these myeloid suppressors (PTEN, C/EBPα, and CTNNA1) genetically interact and synergize to promote myeloid development and transformation remain largely unknown.
We recently showed that the CTNNA1 gene, located in the proximal 5q31.1 critically deleted region, is expressed in normal HSCs (CD34+CD38−CD90+Lin−) but is significantly lower (10%-30% of normal levels) in LICs (CD34+CD38−CD123+Lin−) and in the myeloid leukemia cell line HL-60 with del(5q) because of DNA promoter methylation of the retained CTNNA1 allele.7,10 The HL-60 cell line is an excellent cellular model to investigate the mechanisms underlying the transcriptional silencing of CTNNA1 and the pathogenesis of MDS and AML in that it also harbors a deletion of the 5q31 region and has very low CTNNA1 expression (< 10% of normal levels) from the retained CTNNA1 allele because of DNA promoter methylation and histone deacetylation.7 Retrovirally mediated restoration of CTNNA1 expression in HL-60 cells results in reduced proliferation and increased apoptotic cell death. These data are consistent with a model in which genomic loss of one allele accompanied by epigenetic inactivation of the retained CTNNA1 allele contributes to the malignant transformation of normal HSCs.7
Because inhibitors of DNA methyltransferases and histone deacetylases have shown only limited response rates in clinical trials,18,19 we began using the HL-60 cell line as a model to explore additional mechanisms or upstream signaling pathways responsible for the transcriptional silencing of CTNNA1 in HL-60 cells and primary LICs. Our results reveal a novel, evolutionarily conserved PTEN-C/EBPα-CTNNA1 axis playing an important role in the development and transformation of HSCs and myeloid progenitors in humans, mice and zebrafish. This protein axis might be targeted to eliminate LICs expressing low levels of CTNNA1.
Methods
Plasmids and antibodies
All plasmids encoding small hairpin RNAs (shRNAs) of the polycomb repressive complex 2 (PRC2) have been described.20 The suppressor of zeste 12 (SUZ12)-specific sequence20 was cloned into the pRetroSuper plasmid.21 The p42, p30 C/EBPα, and NF-Yam29 were cloned into the retroviral MigR1 vector. Wild-type PTEN were cloned into lentiviral PRRL vector. Antibodies against NF-Ya (Rockland Immunochemicals), AML-1 (Active Motif), C/EBPα (C1-14AA), C/EBPα (C2-C18) (Santa Cruz Biotechnology), C/EBPα (N), enhancer of zeste 2 (EZH2), phospho-eIF4E (Ser209), eIF4E, PTEN (138G6; Cell Signaling Technology), β-actin (Sigma-Aldrich), and SUZ12 (Abcam) are all commercially available. All antibodies directed against histone modifications were purchased from Abcam and Upstate Biotechnology. A rabbit polyclonal antiserum against zebrafish C/EBPα protein was generated using a C-terminal peptide of zebrafish C/EBPα as an antigenic source.
Cell lines, retroviral infection, and reagents
HL-60, NB4, KG-1, HEK293FT, and PT67 packaging cells were cultured as described previously.7,20 HEK293FT cells were transfected using calcium phosphate transfection methods. The pRetroSuper-based retrovirus was produced with PT67 packaging cells (Clontech) and subsequently used to infect NB4 or HL-60 cells as described previously.20 MSCV-based MigR1 retrovirus and PRRL-based lentivirus were produced by transfecting MigR1 or PRRL, vsvg, and gag/pol or δ8.9 plasmids into HEK293FT cells. Trichostatin A (TSA), 5-aza-2′-deoxycytidine (DAC), and 2-aminopurine were purchased from Sigma-Aldrich. Rapamycin was purchased from Calbiochem.
Immunoprecipitation and ChIP
For coimmunoprecipitation assays, HL-60 cells were washed in phosphate-buffered saline and lysed with coimmunoprecipitation buffer (10% glycerol, 2mM ethylenediaminetetraacetic acid, 50mM Tris-HCl, pH 7.5, 150mM NaCl, 0.5% NP-40, 1mM NaF). Specific antibodies were added to the lysates for 8 to 16 hours with 20 μL of slurry protein A or G Sepharose beads saturated with bovine serum albumin (Upstate Biotechnology). The immunoprecipitates were then washed 3 or 4 times with coimmunoprecipitation buffer and loaded for sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Details for chromatin immunoprecipitation (ChIP) are available in supplemental Data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Bioinformatics
The sequences of the CTNNA1 promoter were retrieved from the genome database (http://genome.ucsc.edu/). Bioinformatics analyses of the promoter were performed with VISTA servers (http://genome.lbl.gov/vista/index.shtml). Syntenic analysis was performed as described previously.22
RNA preparation, real-time quantitative RT-PCR, and mutation analysis
Cells were washed twice with phosphate-buffered saline and transferred to an RNase-free tube containing Trizol (Invitrogen). Total RNAs were extracted according to the manufacturer's manual and diluted in diethyl pyrocarbonate-treated H2O. Total RNAs (1 μg) were used as templates and reverse transcriptions were performed in a volume of 20 μL. The cDNAs were diluted to 500 μL with ddH2O, and 3 μL was used for each reverse-transcribed polymerase chain reaction (RT-PCR) reaction. Relative gene expression levels were calculated as follows: % of GAPDH expression = 100/[2CT(gene) − CT(GAPDH)]. Primer and probe sequences are listed in supplemental Table 1. Real-time quantitative PCR reactions consisted of 200nM of each primer, 3 μL of cDNA, 5 μL of 2Í qPCR Mastermix (Toyobo) in a total volume of 10 μL. All primers and probes were purchased from Biosune. PCR conditions were as follows: 2 minutes at 50°C, 3 minutes at 94°C, followed by 40 cycles of 15 seconds at 94°C, and 1 minute at 60°C. The C/EBPα gene was sequence with primers as reported.13
Western blotting
Cells were homogenized in lysis buffer (20mM Tris-HCl pH 7.4, 150mM NaCl, 5mM ethylenediaminetetraacetic acid, 10% glycerol, and 0.1% Triton X-100) containing protease inhibitor cocktail and phosphatase inhibitor (Roche Diagnostics). Proteins were separated on a 10% to 12% sodium dodecyl sulfate–polyacrylamide gel and transferred electrophoretically to a nitrocellulose membrane (Schleicher & Schuell). The membrane was blocked and then incubated with the indicated antibodies followed by incubation with an appropriate horseradish peroxidase–conjugated secondary antibody (1:10 000).
Fish care, morpholinos, microinjection, and whole-mount mRNA in situ hybridization
Zebrafish maintenance, breeding, and staging were performed as described previously.23 The Ptena and Ptenb morpholino antisense oligonucleotides used in this study have been described previously24 and were purchased from Gene Tools. Morpholinos were diluted to different concentrations with Danieu buffer. Capped mRNAs were synthesized with Massage machine kit according to the manufacturer's instructions (Ambion). Microinjections into zebrafish embryos were performed with a Harvard Apparatus microinjector at the 1-cell stage. Whole-mount in situ hybridization with Dig-labeled antisense probe was performed as described previously.23
Human and mouse samples
Bone marrow samples were collected from patients in accordance with the Declaration of Helsinki after having obtained their informed consent and with Institutional Review Board approval from the Institute of Health Sciences and the Shanghai Institutes for Biological Sciences. Human LICs and normal HSCs were obtained as previously described7 with informed consent. Fluorescence-activated cell sorter (FACS)–sorted murine Lin−c-Kit+ myeloid progenitors and proliferative mononuclear cells (MNCs) were obtained from the bone marrow of Pten−/− mice.15
Results
Increased H3K27 trimethylation of the proximal promoter of the retained CTNNA1 allele in del(5q) cells
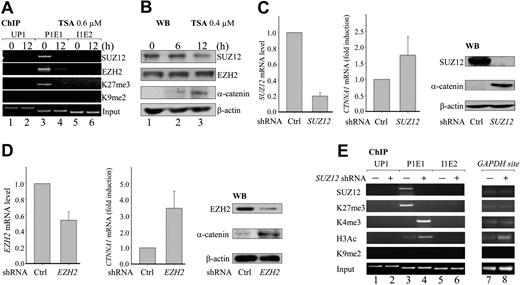
To examine whether histone methylation events are involved in inactivating the promoter of the retained CTNNA1 allele in del(5q) HL-60 cells, we first performed ChIP analysis with a panel of antibodies against methylated histones (H3K4, H3K9, H3K27, H3K79, and H4K20) and acetylated histones H3/H4 (H3ac/H4ac). Independent primer pairs for 5 distinct amplicons (UP1, P1, P1E1, P1E1-2, and I1E2) spanning 35 kb of the retained CTNNA1 genomic region were synthesized to detect potential histone modifications (Figure 1A). The results show an abundant enrichment of repressive histone trimethylation at lysine 27 (H3K27me3) at the contiguous P1 and P1E1 sites of the proximal promoter in HL-60 cells. This modification was only weakly detected in non-del(5q) NB4 cells and KG-1 cells that harbor a deletion of 5q31-33 with normal CTNNA1 levels7 (Figure 1B arrows; supplemental Figure 1). Concomitantly, reduced levels of activating H3K4 methylation and H3 acetylation at the P1E1 region were also detected in HL-60 cells compared with NB4 cells (Figure 1B asterisks). These results suggest that H3K27me3 might be involved in the transcriptional silencing of the retained CTNNA1 allele.
Elevated H3K27 trimethylation levels in the proximal promoter of the retained CTNNA1 allele. (A) A diagram of the human CTNNA1 genomic locus and the regions analyzed by ChIP. TSS indicates transcriptional start site. (B) ChIP analyses of histone modifications with the indicated antibodies in HL-60 and NB4 leukemia cell lines. (C-D) Semiquantitative RT-PCR and ChIP analyses of CTNNA1 expression and histone modifications in HL-60 cells treated with TSA for the indicated time. (E) Semiquantitative RT-PCR and ChIP analyses of CTNNA1 expression and H3K27me3 in HL-60 cells treated with DAC for the indicated times. (F) ChIP analyses of CTNNA1 promoter for H3K27me3 enrichment in FACS-sorted LICs (CD34+CD38−CD123+lineage−) from patient samples with or without del(5q). Enrichment of H3K27me3 was calculated by: [ChIP-P1E1(LICs)/INPUT-P1E1(LICs)]/[ChIP-P1E1(HL-60)/INPUT-P1E1(HL-60)]. ● represents case V with del(5q), which expresses a normal amount of CTNNA1.7
Elevated H3K27 trimethylation levels in the proximal promoter of the retained CTNNA1 allele. (A) A diagram of the human CTNNA1 genomic locus and the regions analyzed by ChIP. TSS indicates transcriptional start site. (B) ChIP analyses of histone modifications with the indicated antibodies in HL-60 and NB4 leukemia cell lines. (C-D) Semiquantitative RT-PCR and ChIP analyses of CTNNA1 expression and histone modifications in HL-60 cells treated with TSA for the indicated time. (E) Semiquantitative RT-PCR and ChIP analyses of CTNNA1 expression and H3K27me3 in HL-60 cells treated with DAC for the indicated times. (F) ChIP analyses of CTNNA1 promoter for H3K27me3 enrichment in FACS-sorted LICs (CD34+CD38−CD123+lineage−) from patient samples with or without del(5q). Enrichment of H3K27me3 was calculated by: [ChIP-P1E1(LICs)/INPUT-P1E1(LICs)]/[ChIP-P1E1(HL-60)/INPUT-P1E1(HL-60)]. ● represents case V with del(5q), which expresses a normal amount of CTNNA1.7
We previously demonstrated that treatment of HL-60 cells with TSA), a histone deacetylase inhibitor) or 5-aza-2′-deoxycytidine (DAC, a DNA methyltransferase inhibitor) significantly up-regulated CTNNA1 transcripts and α-catenin protein in HL-60 cells (Figure 1C).7 We therefore investigated whether these inhibitors would modulate H3K27me3 and H3K4me3 levels in the P1E1 region, and whether H3K27me3 contributes to this transcriptional silencing. In contrast to NB4 and KG-1 cells (supplemental Figure 1), treatment of HL-60 cells with 0.6μM TSA significantly decreased H3K27me3 levels at the P1E1 site, with a coincident increase in the levels of H3K4me3 and H4 acetylation in a time-dependent fashion (Figure 1D). Similarly, treatment of HL-60 cells with 1.0μM DAC for 48 hours also induced a significant reduction of H3K27me3 at the P1 site (Figure 1E). No changes were observed at the GAPDH locus (supplemental Figure 2). In addition, analysis of FACS-sorted LICs (CD34+CD38−CD123+) from previously reported clinical samples with and without del(5q), for which sufficient material was available,7 showed that H3K27me3 was higher in samples with reduced CTNNA1 expression (VII and VIII)7 than in samples with normal CTNNA1 levels (cases V, XIII, XIV, XV, XVI, XIX, and XX7 ; Figure 1F). These results indicate that the repressive histone mark H3K27me3 might contribute to the epigenetic silencing of the retained CTNNA1 allele.
PRC2 is essential for H3K27me3-mediated transcriptional suppression of the retained CTNNA1 allele
PRC2 contains SUZ12, EZH2, and embryonic ectoderm development (EED) and mediates H3K27 methylation through the catalytic SET domain of EZH2 in various cellular contexts,25 We therefore asked whether components of PRC2 colocalize with H3K27me3 and whether such colocalization might be affected by TSA treatment. ChIP analysis was performed with antibodies specifically directed against SUZ12, EZH2, H3K27me3, and H3K9me2. The results show that both SUZ12 and EZH2 specifically occupy the same region as H3K27me3 at the P1E1 site (Figure 2A lane 3), but not at the UP1 and I1E2 sites (Figure 2A lanes 1 and 5). Treatment of HL-60 cells with 0.6μM TSA for 12 hours caused a massive reduction of the enrichment of SUZ12, EZH2, and H3K27me3 (Figure 2A lane 4). TSA treatment did not change SUZ12 or EZH2 protein levels (Figure 2B lanes 1-3), suggesting that TSA more probably relieved transcriptional silencing by inhibiting the recruitment of SUZ12 and EZH2 than by degrading these proteins.
PRC2 suppresses CTNNA1 expression through H3K27 trimethylation. (A) HL-60 cells were treated with 0.6μM TSA for 0 and 12 hours. ChIP analyses were performed with the indicated antibodies. (B) Western blot analyses of EZH2, SUZ12, and α-catenin proteins in HL-60 cells treated with 0.4μM TSA for the indicated times. (C-D) HL-60 cells were infected with retroviral constructs generating SUZ12- and EZH2-specific shRNA. Knockdown of SUZ12 (C) and EZH2 (D), as well as levels of CTNNA1 expression, were determined by both real-time quantitative RT-PCR and Western blot analysis. (E) HL-60 cells were infected with retroviral SUZ12 shRNA, and ChIP analyses were performed with the indicated antibodies at the UP1, P1E1, and I1E2 sites, or at the promoter of the GAPDH gene. Knockdown of SUZ12 resulted in a massive reduction of the repressive H3K27me3, with concomitant increase of the activating H3K4me3 marked specifically at the P1E1 site.
PRC2 suppresses CTNNA1 expression through H3K27 trimethylation. (A) HL-60 cells were treated with 0.6μM TSA for 0 and 12 hours. ChIP analyses were performed with the indicated antibodies. (B) Western blot analyses of EZH2, SUZ12, and α-catenin proteins in HL-60 cells treated with 0.4μM TSA for the indicated times. (C-D) HL-60 cells were infected with retroviral constructs generating SUZ12- and EZH2-specific shRNA. Knockdown of SUZ12 (C) and EZH2 (D), as well as levels of CTNNA1 expression, were determined by both real-time quantitative RT-PCR and Western blot analysis. (E) HL-60 cells were infected with retroviral SUZ12 shRNA, and ChIP analyses were performed with the indicated antibodies at the UP1, P1E1, and I1E2 sites, or at the promoter of the GAPDH gene. Knockdown of SUZ12 resulted in a massive reduction of the repressive H3K27me3, with concomitant increase of the activating H3K4me3 marked specifically at the P1E1 site.
We next infected HL-60 cells with retroviruses expressing previously reported shRNAs against EZH2 and SUZ12.20 Significant reductions of SUZ12 and EZH2 transcripts (80% and 50%, respectively) and proteins were achieved, compared with control shRNA-infected cells under the same conditions (Figure 2C-D). As a result, robust increases in both CTNNA1 transcripts and α-catenin protein were detected (Figure 2C-D). Furthermore, knockdown of SUZ12 also decreased the levels of repressive H3K27me3 and was associated with a simultaneous increase in activating H3K4me3 and H3 acetylation marks at the P1E1 site (Figure 2E lanes 3 and 4). No obvious changes in H3K27 or H3K4 were observed at other sites (UP1, I1E2, and GAPDH loci; Figure 2E lanes 1 and 2, 5 and 6, and 7 and 8, respectively). Taken together, these results indicate that the PRC2 complex is recruited to the proximal promoter of the retained CTNNA1 allele to silence its transcription through H3K27 trimethylation.
The ratio of p42 versus p30 C/EBPα is critical for PRC2 recruitment and H3K27 trimethylation
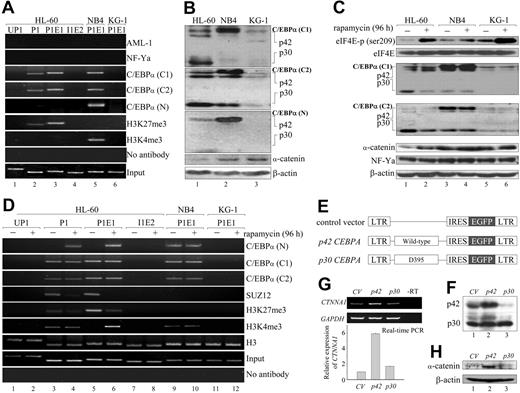
In Drosophila, PcG proteins recognize and exert their activity through specific DNA sequences known as polycomb response elements.26 In mammalian cells, such elements have not been identified. Instead, it has been shown that transcription factors recognize specific DNA-binding sequences and are required to direct proper positioning of PRC2 proteins on chromatin; or other cofactors, such as histone deacetylase (HDAC), recruit PRC2 complexes by interacting with EED.27,28 In an attempt to identify potential factors that could regulate PRC2-mediated suppression, we analyzed the CTNNA1 promoter 5.0 kb upstream of the transcriptional start site and found potential binding sites for 4 transcription factors (AML-1, MZF-1, NF-Y, and C/EBPα) that have previously been shown to contribute to myeloid development and self-renewal and transformation of HSCs29-31 (Figure 1A). Neither knockdown of AML-1 and MZF-1 using retrovirally mediated shRNA32 nor suppression of heterotrimeric NF-Y function through retrovirally mediated overexpression of NF-Yam29 (a dominant-negative form containing a mutation in the DNA-binding domain)31 had any effect on the level of CTNNA1 expression (supplemental Figure 3A-C; and data not shown). ChIP experiments with specific antibodies directed against AML-1 and NF-Ya31,33 further revealed that the proximal promoter of the CTNNA1 allele was not occupied by these transcription factors (Figure 3A top; supplemental Figure 3D-E). These results suggest that AML-1, NF-Ya, and MZF-1 are unlikely to contribute to the transcriptional suppression of CTNNA1 expression.
Involvement of C/EBPα isoforms in PRC2-mediated H3K27me3 and CTNNA1 expression from the retained allele. (A) ChIP analyses were performed with the indicated antibodies in the HL-60, NB4, and KG-1 leukemia cell lines. Note that the C/EBPα (N) antibody recognizes only the p42 wild-type full-length C/EBPα protein, whereas C/EBPα (C1) and C/EBPα (C2) recognize both p42 and p30 C/EBPα. (B) Western blot analysis of p42, p30 C/EBPα, and α-catenin proteins in HL-60, NB4, and KG-1 leukemia cells. Note that the ratio of p42/p30 is significantly reduced only in HL-60 cells. (C) HL-60, NB4, and KG-1 cells were treated with 10nM rapamycin for 96 hours followed by Western blot analyses with the indicated antibodies. (D) ChIP analyses were performed with the indicated antibodies in HL-60, NB4, and KG-1 cells treated with 10nM rapamycin for 96 hours. (E-H) Effects of p42 and p30 C/EBPα overexpression on CTNNA1 levels. Retrovirally mediated overexpression of p42 and p30 C/EBPα (E) in HL-60 cells as determined by Western blot analysis (F). The levels of CTNNA1 transcripts and α-catenin protein were determined by real-time quantitative RT-PCR (G) and Western blot analysis (H), respectively.
Involvement of C/EBPα isoforms in PRC2-mediated H3K27me3 and CTNNA1 expression from the retained allele. (A) ChIP analyses were performed with the indicated antibodies in the HL-60, NB4, and KG-1 leukemia cell lines. Note that the C/EBPα (N) antibody recognizes only the p42 wild-type full-length C/EBPα protein, whereas C/EBPα (C1) and C/EBPα (C2) recognize both p42 and p30 C/EBPα. (B) Western blot analysis of p42, p30 C/EBPα, and α-catenin proteins in HL-60, NB4, and KG-1 leukemia cells. Note that the ratio of p42/p30 is significantly reduced only in HL-60 cells. (C) HL-60, NB4, and KG-1 cells were treated with 10nM rapamycin for 96 hours followed by Western blot analyses with the indicated antibodies. (D) ChIP analyses were performed with the indicated antibodies in HL-60, NB4, and KG-1 cells treated with 10nM rapamycin for 96 hours. (E-H) Effects of p42 and p30 C/EBPα overexpression on CTNNA1 levels. Retrovirally mediated overexpression of p42 and p30 C/EBPα (E) in HL-60 cells as determined by Western blot analysis (F). The levels of CTNNA1 transcripts and α-catenin protein were determined by real-time quantitative RT-PCR (G) and Western blot analysis (H), respectively.
ChIP assays using specific antibodies directed against the C-terminus (C1 and C2) or the N-terminus of C/EBPα (N) revealed that only p30 C/EBPα, but not p42 C/EBPα, was specifically associated with the P1 and P1E1 regions of the CTNNA1 promoter solely in HL-60 cells (Figure 3A lanes 2 and 3). This unanticipated finding suggests a lack of functional p42 C/EBPα protein in HL-60 cells. As expected, Western blot analyses using C/EBPα antibodies (C1, C2, and N) consistently revealed substantially lower p42 C/EBPα levels and a concomitant increase in p30 C/EBPα only in HL-60 cells, resulting in a significantly reduced ratio of p42 to p30 C/EBPα (∼ 0.1:1; Figure 3B lanes 1-3). Because N-terminal frame-shift mutations resulting in increased translation of the p30 C/EBPα isoform have been observed in approximately 10% of AML patients,13 we sequenced the full-length CEBPA cDNA in HL-60 cells, and no mutations were found (data not shown).
It has been shown in 3T3-L1 adipocytes that expression of the p42 versus the p30 C/EBPα isoform can be regulated via an evolutionarily conserved cis-regulatory upstream open reading frame (u-ORF) in the 5′-untranslated region that senses the activities of eIF4E and eIF2, which are downstream effectors of the mTOR.34,35 mTOR is a protein serine/threonine kinase that is positively regulated by the proto-oncoprotein AKT and antagonized by the PTEN tumor suppressor.36,37 Interestingly, enhanced eIF4E and eIF2 activities accompanied by concomitant up-regulation of p30 C/EBPα have been found in mammary- and intestinal-epithelial cancer cells.34
To assess whether increased levels of the p30 isoform result from the activation of mTOR signaling, we treated HL-60 cells with rapamycin, an mTOR-specific inhibitor, and observed a dose- and time-dependent reduction in p30 C/EBPα (supplemental Figure 4). Treatment of HL-60 cells with 10nM rapamycin for 96 hours not only significantly increased the levels of phosphorylated eIF4E (p-eIF4E) but also consistently reduced p30 C/EBPα levels to 25% of control levels (Figure 3C lanes 1 and 2). A concomitant increase in α-catenin protein was also observed that did not affect p42 C/EBPα or NF-Ya protein levels (Figure 3C). No such changes were detected in NB4 and KG-1 cells (Figure 3C lanes 3-6).
To explore the mechanism by which rapamycin restored CTNNA1 expression, we treated HL-60 cells with 10nM rapamycin and performed ChIP assays. We found that p42 C/EBPα (N[b]) was strongly associated with the proximal promoter and a concomitant decrease in SUZ12 recruitment and H3K27me3 levels, coincident with higher H3K4me3 levels at the P1E1 region (Figure 3D lanes 5 and 6). These results suggest that inhibition of mTOR signaling results in departure of the PRC2 complex, allowing p42 C/EBPα to bind to the proximal promoter and resulting in the recruitment of an as-yet-unknown H3K4 methyltransferase to mediate H3K4me3.
To further confirm these observations, we tested whether directly increasing the p42/p30 C/EBPα ratio would restore CTNNA1 expression. Retrovirally mediated overexpression of p42 C/EBPα in HL-60 cells increased the p42/p30 ratio from 0.2:1 to 1:1 (Figure 3E-F lanes 1 and 2), whereas overexpression of p30 C/EBPα further lowered the ratio (< 0.1:1; Figure 3F lane 3). This resulted in a 6-fold increase in CTNNA1 transcripts and robust induction of α-catenin protein in the p42-, but not the p30-overexpressing, HL-60 cells (Figure 3G-H lanes 1-3). In contrast, overexpression of either p42 or p30 in NB4 cells did not have any effect on α-catenin levels (supplemental Figure 5A-C). Taken together, these results indicate that an abnormally low p42/p30 ratio is critical for the PRC2-mediated epigenetic suppression of the retained CTNNA1 allele.
PTEN acts upstream to determine p42/p30 C/EBPα ratio and CTNNA1 expression
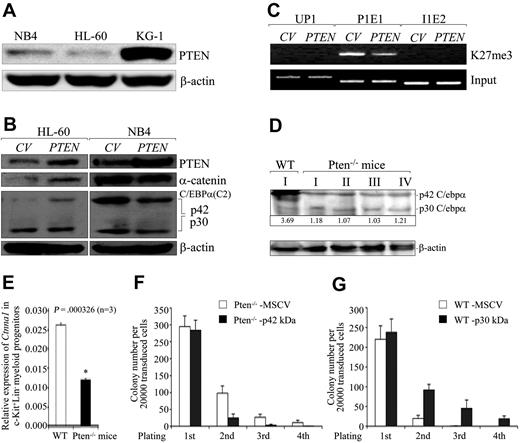
As we studied the molecular mechanisms responsible for mTOR activation and the reduced p42/p30 C/EBPα ratio, we found a very low level of PTEN protein in HL-60 cells compared with NB4 and KG-1 cells (Figure 4A). This result, which is consistent with previous observations that HL-60 cells have few PTEN transcripts, cannot be a consequence of gene silencing by hypermethylation.38 Lentivirally mediated restoration of PTEN expression in HL-60 cells induced significant up-regulation of α-catenin protein, coincident with an elevated p42/p30 C/EBPα ratio (Figure 4B left) and reduced H3K27me3 recruitment specifically at the P1E1 site (Figure 4C). No such changes were observed in NB4 cells overexpressing PTEN(Figure 4B right). Notably, the mechanism responsible for increasing the p42/p30 C/EBPα ratio in PTEN-transduced versus rapamycin-treated HL-60 cells appeared to be distinct: PTEN up-regulated p42 C/EBPα, whereas rapamycin down-regulated, p30 C/EBPα (Figures 3C, 4B).
PTEN acts upstream to determine the p42/p30 C/EBPα ratio and CTNNA1 expression. (A) Western blot analysis of PTEN protein levels in HL-60, NB4, and KG-1 cells. (B) Effects of lentivirally mediated overexpression of PTEN on α-catenin protein levels in HL-60 and NB4 cells as determined by Western blot analysis. (C) Effects of lentivirus-mediated overexpression of PTEN on H3K27me3 modification at the P1E1 site. (D-E) Western blot and quantitative real-time RT-PCR analyses of murine C/ebpα protein and Ctnna1 transcripts in MNCs and FACS-sorted Lin−c-Kit+ myeloid progenitors from Pten−/− knockout mice,15 respectively. The Arabic numerals at the bottom of panel D denote the p42/p30 ratio. (F) FACS-sorted c-Kit+ myeloid progenitors from Pten−/− knockout mice were infected with MSCV-IRES-GFP or MSCV-p42 C/ebpα-IRES-GFP. FACS-GFP positive cells were serially plated on methylcellulose and colonies were counted (n = 3). (G) FACS-sorted c-Kit+ myeloid progenitors from wild-type mice were infected with MSCV-IRES-GFP or MSCV-p30 C/ebpα-IRES-GFP. FACS-sorted GFP+ cells were serially plated on methylcellulose and colonies were counted (n = 3).
PTEN acts upstream to determine the p42/p30 C/EBPα ratio and CTNNA1 expression. (A) Western blot analysis of PTEN protein levels in HL-60, NB4, and KG-1 cells. (B) Effects of lentivirally mediated overexpression of PTEN on α-catenin protein levels in HL-60 and NB4 cells as determined by Western blot analysis. (C) Effects of lentivirus-mediated overexpression of PTEN on H3K27me3 modification at the P1E1 site. (D-E) Western blot and quantitative real-time RT-PCR analyses of murine C/ebpα protein and Ctnna1 transcripts in MNCs and FACS-sorted Lin−c-Kit+ myeloid progenitors from Pten−/− knockout mice,15 respectively. The Arabic numerals at the bottom of panel D denote the p42/p30 ratio. (F) FACS-sorted c-Kit+ myeloid progenitors from Pten−/− knockout mice were infected with MSCV-IRES-GFP or MSCV-p42 C/ebpα-IRES-GFP. FACS-GFP positive cells were serially plated on methylcellulose and colonies were counted (n = 3). (G) FACS-sorted c-Kit+ myeloid progenitors from wild-type mice were infected with MSCV-IRES-GFP or MSCV-p30 C/ebpα-IRES-GFP. FACS-sorted GFP+ cells were serially plated on methylcellulose and colonies were counted (n = 3).
To further examine whether the epistatic interactions among PTEN, p42 C/EBPα, and CTNNA1 transcription were conserved in mice, we determined the levels of p42 C/ebpα protein and Ctnna1 transcription in abnormally proliferating MNCs derived from the bone marrow of Pten−/− mice.15 Of 10 Pten−/− mice analyzed, all demonstrated significantly reduced p42 C/ebpα proteins and an increased expression of p30 C/ebpα (therefore, a dramatically lower p42/p30 ratio; Figure 4D; supplemental Figure 5D), and this was accompanied by an approximately 50% decline in Ctnna1 transcripts in FACS-sorted c-Kit+Lin− hematopoietic progenitors from Pten−/− mice (Figure 4E). Restoration of p42 C/ebpα levels reversed the elevated serial replating capacity of Pten−/− hematopoietic progenitor cells (Figure 4F), whereas overexpression of p30 C/ebpα significantly enhanced the serial replating capacity of wild-type c-Kit+Lin− cells (Figure 4G), suggesting that p30 C/ebpα promoted proliferation or blocked differentiation of hematopoietic progenitor cells.
Knockdown of Ptenb reduces wild-type C/ebpα and α-catenin protein levels and induces myelodysplasia during zebrafish embryogenesis
The zebrafish genome contains 2 Pten genes encoding proteins (Ptena and Ptenb) that exhibit 88% and 86% identity to the human PTEN protein, respectively.24 Interestingly, genomic synteny is conserved between zebrafish Ptenb and the human PTEN locus (supplemental Figure 6A), suggesting that Ptenb is more probably the ortholog of human PTEN.
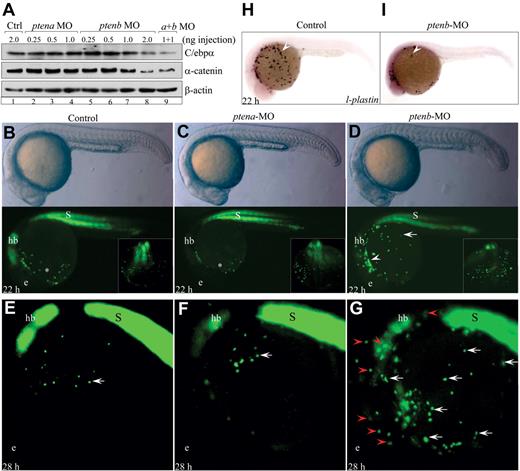
It has been shown that antisense morpholino oligonucleotide-mediated knockdown of either Ptena or Ptenb significantly increases the levels of phosphorylated Akt to 1.2- and 2.3-fold, respectively, and induced several developmental defects during zebrafish embryogenesis.24 We first determined the levels of α-catenin protein and wild-type C/ebpα in Ptena- and Ptenb-deficient embryos by microinjecting the same morpholino oligonucleotides into one-cell stage embryos. As shown in Figure 5A, a dose-dependent decrease in the levels of α-catenin and wild-type C/ebpα protein was observed only in the 22 hpf embryos injected with 1 to 2 ng of the Ptenb-specific morpholino (lanes 7 and 8). In contrast, injection of 1 to 2 ng of either Ptena or control morpholino had no effect on α-catenin or wild-type C/ebpα levels (lanes 1-4). Combined injection of Ptena and Ptenb morpholinos (1 ng of each) did not further reduce α-catenin or wild-type C/ebpα levels (lane 9), suggesting that Ptena was not involved in α-catenin or C/ebpα regulation. These results strongly suggest an evolutionarily conserved function for the PTEN-C/EBPα network in the regulation of CTNNA1 transcription. Consistent with this notion, embryonic ChIP analysis showed that an 8-fold enrichment of H3K27me3 at the F5 site of the zebrafish Ctnna1 promoter (∼ 4.0 kb) was observed only in the Ptenb-deficient embryos injected with 2 ng of Ptenb morpholino (supplemental Figure 6B bottom). Interestingly, the region represented by the F5 site appears to be conserved between the zebrafish and human CTNNA1 promoters (supplemental Figure 6B top).
An evolutionarily conserved function of Ptenb regulates C/ebpα and α-catenin levels and prevents myelodysplasia in zebrafish embryos. (A) Western blot analysis of C/ebpα and α-catenin proteins in embryos injected with the indicated morpholino oligonucleotides at 22 hpf. Note that the polyclonal antibody used detected only zebrafish wild-type p38-kDa C/ebpα protein. (B-D) Morphology of embryos and appearance of EGFP+ myeloid progenitors in Tg(zpu.1:EGFP) transgenic embryos injected with the indicated morpholinos at 22 hpf. hb indicates hindbrain; and s, somite. (E-G) EGFP+ myeloid progenitors in Tg(zpu.1:EGFP) transgenic embryos injected with the indicated morpholinos at 28 hpf. hb indicates hindbrain; and s, somite. All embryos are shown in lateral view with the head to the left. (H-I) Whole-mount mRNA in situ hybridization analysis of l-plastin expression in control and ptenb-deficient embryos at 22 hpf.
An evolutionarily conserved function of Ptenb regulates C/ebpα and α-catenin levels and prevents myelodysplasia in zebrafish embryos. (A) Western blot analysis of C/ebpα and α-catenin proteins in embryos injected with the indicated morpholino oligonucleotides at 22 hpf. Note that the polyclonal antibody used detected only zebrafish wild-type p38-kDa C/ebpα protein. (B-D) Morphology of embryos and appearance of EGFP+ myeloid progenitors in Tg(zpu.1:EGFP) transgenic embryos injected with the indicated morpholinos at 22 hpf. hb indicates hindbrain; and s, somite. (E-G) EGFP+ myeloid progenitors in Tg(zpu.1:EGFP) transgenic embryos injected with the indicated morpholinos at 28 hpf. hb indicates hindbrain; and s, somite. All embryos are shown in lateral view with the head to the left. (H-I) Whole-mount mRNA in situ hybridization analysis of l-plastin expression in control and ptenb-deficient embryos at 22 hpf.
We next microinjected Ptena and Ptenb morpholino oligonucleotides into one-cell stage embryos derived from the established Tg(zpu.1:EGFP) transgenic line39,40 to evaluate myeloid progenitor development. The relative stability of the enhanced green fluorescent protein (EGFP) protein allowed EGFP+ progenitors to be observed at a later time when pu.1 mRNA was no longer detected because of either the physiologic wane of primitive myelopoiesis or the degradation of pu.1 mRNA in pten-deficient myeloid progenitors. In control and Ptena-injected 22 hpf embryos, approximately 30 EGFP+ myeloid progenitors had migrated out from the head mesenchyme into the lower part of the yolk sac (Figure 5B-C asterisks). In contrast, a significantly greater number of EGFP+ (80-120 cells) myeloid progenitors were detected over the entire yolk sac surface with many cells still maintaining within the head mesenchyme (Figure 5D arrow and arrowhead). As the development advanced, only 10 to 15 EGFP+ myeloid progenitors were observed over the yolk sac of control embryos at 28 hpf (Figure 5E-F arrows), whereas the outgrowth and abnormal distribution of EGFP+ myeloid progenitors became apparent (Figure 5G white arrows; n = 91 of 121). Some EGFP+ cells were abnormally localized within distinct territories of the developing brain, suggesting an ectopic invasiveness (Figure 5G red arrowheads).
Whole-mount mRNA in situ hybridization was performed to evaluate myeloid differentiation with several myeloid-specific markers of functionally mature macrophages (l-plastin),41 myelomonocytes (lysozyme C),42 and neutrophils (mpo).43 Compared with control and Ptena-injected embryos, a significant reduction in the numbers of l-plastin+, mpo+, and lysozyme C+ mature myeloid cells was detected in Ptenb-deficient embryos at 22 hpf (Figure 5H-I arrows; supplemental Figure 7A arrow; data not shown). To dynamically trace the differentiation and maturation of Ptenb-deficient myelomonocytes, a stable transgenic zebrafish, Tg(zlyz:EGFP), expressing EGFP under the control of the lysozyme C promoter, was analyzed.42 A robust decrease in EGFP+ myelomonocytes was observed as early as 22 hpf in embryos injected with either Ptenb or Ptena/Ptenb morpholinos compared with those injected with control or Ptena morpholino (supplemental Figure 7B arrows). At 36 hpf, an approximately 10-fold decrease in the number of EGFP+ myelomonocytes and a slightly curled tail24 were observed in Ptenb- and Ptena/Ptenb-deficient embryos (supplemental Figure 7C arrow and asterisks). Moreover, treatment of Ptenb-deficient Tg(zlyz:EGFP) transgenic embryos at 22 hpf with 300 nm rapamycin for 14 hpf resulted in a significant recovery of EGFP+ myelomonocytes (supplemental Figure 7D). No detectable changes in scl (a marker of primitive hematopoietic stem cells) or gata-1 (a marker of erythroid progenitors) expression were observed (supplemental Figure 8). These results strongly suggest that loss of Ptenb function during zebrafish embryogenesis caused abnormal myeloid development characterized by outgrowth, differentiation blockade, and invasive migration of myeloid progenitors.
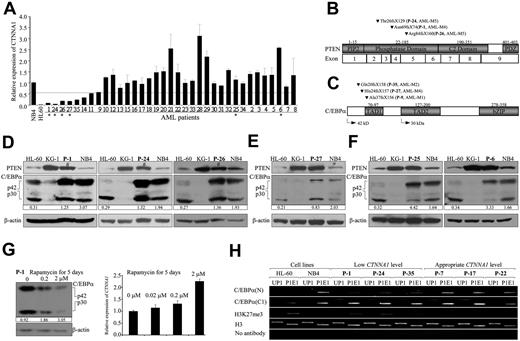
Disruption of the PTEN-C/EBPα-CTNNA1 axis in human AML
To address the pathologic significance of the PTEN-C/EBPα-CTNNA1 axis in human MDS/AML patients, we first determined the CTNNA1 levels by real-time RT-PCR in FACS-sorted LICs (CD34+CD38−CD123+Lin−; purity > 90%) from 35 MDS/AML patients. The percentage of morphologically identifiable leukemic blast cells in the bone marrow or peripheral blood of these patients ranged from 1.5% to 92%, and most of the patients did not have detectable cytogenetic abnormalities (supplemental Table 2). Of 35 patients, 8 (P-1, P-24, P-26, P-27, P-35, P-14, P-11, and P-9) have significantly reduced CTNNA1 transcript levels in their LICs compared with normal HSCs and control NB4 cells (Figure 6A dashed line). We then sequenced the entire PTEN and CEBPA coding regions. Frame-shift mutations (3 in PTEN and 3 in CEBPA) were identified in 6 of 8 patients expressing low levels of CTNNA1 transcripts (Figure 6B-C). No such mutations were detected in the patients P-14 and P-11 and in the other 27 patients expressing appropriate levels of CTNNA1 (Figure 6A). The 3 frame-shift mutations in PTEN resulting from insertions or deletions (supplemental Table 2) were predicted to cause truncation of the PTEN protein, whereas the other 3 frame-shift mutations in CEBPA resulting from deletions were predicted to eliminate the expression of the p42 C/EBPα without affecting the p30 C/EBPα isoform, whose transcription is initiated further downstream.13
Frame-shift mutations are exclusively detected in primary LICs with low CTNNA1 transcripts. (A) Quantitative real-time RT-PCR analysis of CTNNA1 transcripts in FACS-sorted LICs (CD34+CD38−CD123+Lin−) from MDS or AML patients. The down-regulation of CTNNA1 transcripts was arbitrarily defined as 60% reduction (dashed line) compared with normal HSCs and the control NB4 cell line. (B-C) Summary of frame-shift mutations detected in the PTEN (B) and CEBPA (C) genes. Note that these mutations were only identified in 6 of 8 patient's LICs expressing low levels of CTNNA1 transcripts. (D) Western blot analysis with protein lysates extracted from the MNCs of patients expressing low CTNNA1 and carrying frame-shift mutations in the PTEN gene (P-1, P-24, and P-26). Note the reduction of the p42/p30 ratio in patients P-1, P-24, and P-26 who had frame-shift mutations in the PTEN gene. #Wild-type PTEN protein that is probably expressed from the normal blood cells or blasts within the heterogeneous MNC mixture. (E) Western blot analysis with protein lysates extracted from MNCs of patient expressing low CTNNA1 and carrying N-terminal frame-shift mutations in the CEBPA gene (P-27). (F) Western blot analysis with protein lysates extracted from MNCs of patients expressing appropriate levels of CTNNA1 and carrying no mutations in either PTEN or CEBPA (P-25 and P-6). (G) The MNCs derived from patient P-1 were treated with rapamycin at the indicated concentrations for 5 days. Western blot analysis showed a dose-dependent increase in the p40/p30 C/EBPα ratio (left) and a coincident up-regulation of CTNNA1 transcripts as determined by quantitative real-time RT-PCR analysis (right). The Arabic numbers at the bottom of panels D through G denote the p42/p30 ratio. (H) ChIP analyses with the indicated antibodies in primary MNCs of patients expressing low (P-1, P-24, and P-35) and appropriate CTNNA1 levels (P-7, P-17, and P-22).
Frame-shift mutations are exclusively detected in primary LICs with low CTNNA1 transcripts. (A) Quantitative real-time RT-PCR analysis of CTNNA1 transcripts in FACS-sorted LICs (CD34+CD38−CD123+Lin−) from MDS or AML patients. The down-regulation of CTNNA1 transcripts was arbitrarily defined as 60% reduction (dashed line) compared with normal HSCs and the control NB4 cell line. (B-C) Summary of frame-shift mutations detected in the PTEN (B) and CEBPA (C) genes. Note that these mutations were only identified in 6 of 8 patient's LICs expressing low levels of CTNNA1 transcripts. (D) Western blot analysis with protein lysates extracted from the MNCs of patients expressing low CTNNA1 and carrying frame-shift mutations in the PTEN gene (P-1, P-24, and P-26). Note the reduction of the p42/p30 ratio in patients P-1, P-24, and P-26 who had frame-shift mutations in the PTEN gene. #Wild-type PTEN protein that is probably expressed from the normal blood cells or blasts within the heterogeneous MNC mixture. (E) Western blot analysis with protein lysates extracted from MNCs of patient expressing low CTNNA1 and carrying N-terminal frame-shift mutations in the CEBPA gene (P-27). (F) Western blot analysis with protein lysates extracted from MNCs of patients expressing appropriate levels of CTNNA1 and carrying no mutations in either PTEN or CEBPA (P-25 and P-6). (G) The MNCs derived from patient P-1 were treated with rapamycin at the indicated concentrations for 5 days. Western blot analysis showed a dose-dependent increase in the p40/p30 C/EBPα ratio (left) and a coincident up-regulation of CTNNA1 transcripts as determined by quantitative real-time RT-PCR analysis (right). The Arabic numbers at the bottom of panels D through G denote the p42/p30 ratio. (H) ChIP analyses with the indicated antibodies in primary MNCs of patients expressing low (P-1, P-24, and P-35) and appropriate CTNNA1 levels (P-7, P-17, and P-22).
If PTEN acts upstream to dictate the p42/p30 C/EBPα ratio, one would expect that the p42/p30 ratio would decrease in patients P-1, P-24, and P-26 who had frame-shift mutations in the PTEN gene. Indeed, Western blot analyses with protein lysates extracted from MNCs showed 60%, 32%, and 30% reductions in the p42/p30 ratio in patients P-1, P-24, and P-26, respectively, compared with the p42/p30 ratio of NB4 cells (Figure 6D). As expected, patient P-27, who had an N-terminal frame-shift mutation in the CEBPA gene but did not have a PTEN mutation, showed a 59% reduction in the p42/p30 ratio (Figure 6E), whereas patients P-25 and P-6, who expressed appropriate levels of CTNNA1 transcripts and did not have detectable mutations in either PTEN or CEBPA, had an appropriate p42/p30 C/EBPα ratio (Figure 6F).
Because patient P-1 has a predicted loss-of-function mutation in the PTEN gene and a significantly reduced p42/p30 ratio (Figure 6B-D), we asked whether treatment of MNCs derived from this patient with rapamycin would reverse the p42/p30 ratio. As shown in Figure 6G, a dose-dependent increase in the p42/p30 ratio was detected: treatment with 0.2 and 2μM rapamycin resulted in 2.0- and 3.3-fold up-regulation of the p42/p30 ratio, respectively, compared with MNC cells treated with vehicle (Figure 6G left panel). Consequently, a 2-fold up-regulation in the levels of CTNNA1 transcripts was observed in MNCs treated with 2μM rapamycin (Figure 6G right panel). Furthermore, using ChIP assay, we evaluated the enrichment of histone H3K27me3 and the C/EBPα binding in the proximal promoter of CTNNA1 gene in the primary leukemic blasts of patients P-1, P-24, and P-35 with low CTNNA1 level, and patients P-7, P-17, and P-22 with appropriate level of CTNNA1. The results showed that the enrichment of H3K27me3 in the patients P-1, P-24, and P-35 is more abundant than that in patients P-7, P-17, and P-22 (Figure 6H). On the other hand, the binding activity of p42 C/EBPα to the P1E1 site is significantly weaker in the patients P-1, P-24, and P-35, compared with patients P-7, P-17, and P-22 (Figure 6H). Thus, there is a similar chromatin pattern of H3K27me3 modification and interaction with C/EBPα between primary samples and HL60 cell line.
Discussion
In this study, we report that PTEN deficiency and activation of mTOR signaling decrease the p42/p30 C/EBPα ratio, leading to excessive p30 C/EBPα binding to the proximal promoter of CTNNA1 and subsequently recruiting the PRC2 complex to transcriptionally silence the retained CTNNA1 allele through histone 3 trimethylation at lysine 27 (H3K27me3). Importantly, reductions in CTNNA1 transcript and protein levels were also observed in Pten-deficient murine myeloid progenitors (Lin−c-Kit+) and zebrafish embryos, respectively. Translational control of C/EBPα isoforms (p42 vs p30) by PTEN-mTOR signaling appeared to be essential for PTEN-mediated CTNNA1 expression. Restoration of PTEN expression or inhibition of mTOR signaling by rapamycin treatment in HL-60 cells consistently increased the p42/p30 C/EBPα ratio, resulting in the replacement of p30 C/EBPα with p42 C/EBPα and an increase in histone 3 trimethylation at lysine 4; this relieved the transcriptional silencing and restored the α-catenin protein. The lack or significant reduction of p42 C/EBPα protein in Pten-deficient murine bone marrow MNCs and zebrafish embryos strongly supports the suggestion that C/EBPα is an immediate downstream target of PTEN-mTOR signaling and that CTNNA1 is a novel, evolutionarily conserved downstream target of the PTEN-C/EBPα axis.
The other 2 important findings are that CTNNA1 is a downstream target of the C/EBPα transcription factor and that PTEN-C/EBPα signaling regulates PcG protein recruitment. Previous studies have documented a 10-fold reduction of CTNNA1 transcripts in AML cases with either CEBPA mutations or CEBPA promoter methylation, suggesting that low levels of CTNNA1 in C/EBPα-deficient AML may be a critical step in leukemic transformation.9,11,44 However, the underlying molecular mechanism remains unclear. Our finding that the ratio of p42 to p30 C/EBPα is critically involved in the transcription regulation of CTNNA1 provides a reasonable explanation: reduction of the p42/p30 ratio results in enhanced p30 binding to the CTNNA1 promoter, which suppresses its transcription by recruiting the repressive PRC2 complex. An increase in the p42/p30 ratio enables p42 to replace p30 and bind to the CTNNA1 promoter and recruit the activating H3K4me3 enzymatic complex. Unfortunately, the precise mechanism by which an elevated p42/p30 ratio converts a repressive PRC2-mediated H3K27me3 to an activating H3K4me3 remains to be elucidated. We were unable to detect a direct interaction of p30 C/EBPα with SUZ12 or EZH2 but able to confirm the reported interaction of EZH2 with SUZ12 by coimmunoprecipitation analysis (supplemental Figure 9), suggesting that p30 C/EBPα recruits the PRC2 component through an unknown factor. p30 C/EBPα has been shown to bind to the promoters of MPP11, p84N5, and SMYD2 to suppress their transcription,17 and p30 C/EBPα-mediated sumoylation of p42 C/EBPα has been shown to recruit HDAC and EED to induce H3K27me3.27,45 Based on these findings, we propose a model that PTEN deficiency-mediated mTOR activation decreases the p42/p30 C/EBPα ratio, allowing excessive p30 C/EBPα to bind to the CTNNA1 promoter and directly or indirectly associate with the transcriptional-silencing complex containing PRC2, HDAC, and DNA methyltransferase (supplemental Figure 10A). Lowering the p30 C/EBPα concentration through inhibiting mTOR activation releases the “sequestered” p42 C/EBPα from the p42:p30 heterodimer,46 allowing it to bind to the promoter and recruit an unknown complex that is able to catalyze H3K4me3 (supplemental Figure 10B).
Our studies also reveal that the PTEN-C/EBPα-CTNNA1 axis is evolutionarily conserved and that functional deficiency of this axis promotes the formation of LICs and malignant myeloid progenitors, resulting in the development of a myelodysplasia-like syndrome and leukemia in zebrafish and mice. More importantly, the integrity of PTEN-C/EBPα signaling was exclusively disrupted in 6 of 8 AML patients expressing reduced levels of CTNNA1 transcripts. No such disruptions were observed in the other 27 MDS/AML patients expressing appropriate levels of CTNNA1 transcripts. In addition, of 35 patients, 2 bZIP mutations of C/EBPα were detected: patient P-27 carried a bZIP in-frame insertion mutation (940-941insAGA), and the other patient P-15 had a single base alternation (866G→A; Arg→His) (data not shown). Previous studies have shown that the frequency of mutations in both N-terminus and bZIP domain in AML patients is more than 50%.9,47-49 Although patient P-27 had a combinational mutation in both N-terminus and bZIP domain, the proposed model on the PTEN/C/EBPα/CTNNA1 axis predicts that the bZIP in-frame insertion (940-941insAGA) in patient P-27 appears to be unlikely to completely abrogate the p30 C/EBPα functions.
We predict that this conserved axis will not only substantiate current AML classification, but it will also provide strong rationale for targeted therapy to eradicate LICs expressing CTNNA1 transcripts at low levels.6,7 It has recently been shown that treatment with rapamycin not only decreases the in vitro clonogenic capability of CD34+ cells from high-risk MDS patients, but it also eliminates LICs from Pten−/− knockout mice without damaging normal hematopoietic progenitors.16,50,51 In addition, depleting cellular PRC2 components with 3-deazaneplanocin A or abolishing H3K27 methylation by UTX and JMJD3 demethylases might benefit MDS/AML patients with low CTNNA1 levels.52,53
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Tenen at Harvard Institutes of Medicine, Di Croce at Center de Regulacio Genomica, Barcelona, and Thomas Look and John Kanki at Dana-Farber Cancer Institute for providing C/EBPα, pRetroSuper, NF-Yam29 plasmids, and Tg(zpu.1:EGFP) transgenic fish, respectively. We thank anonymous reviewer No. 2 for the insightful comments and suggestions and all the members of our laboratory for helpful discussions.
This work was supported in part by the National Basic Research Program of China (2007CB947003), the National Natural Science Foundation of China (30525019, 30830047 and 30771185), Science and Technology Commission of Shanghai Municipality (09XD1404700), the Innovation Program of the Chinese Academy of Sciences (KSCX2-YW-R-03, KSCX1-YW-22-03) and PhD Programs Foundation of Ministry of Education of China (20060248085).
Authorship
Contribution: C.-T.F., K.-Y.Z., Y.-F.L., S.T.M., Y.-F.F., C.-G.R., Z.-W.D., and J.-W.Z. performed experiments and analyzed data; J.-Q.M., Y.-J.L., M. Dong, Y.J., Y.C., M. Deng, W.Z., and B.C. assisted with experiments; C.-T.F., K.-Y.Z., and T.X.L. designed research plan and wrote the paper; and P.B., S.-J.C., Z.C., M.W.B., J.Z., and J.-W.Z. critically read the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ting Xi Liu, Laboratory of Development and Diseases, Institute of Health Sciences, Rm 408, Bldg 1, 225 South Chong Qing Rd, Shanghai 200025, People's Republic of China; e-mail: txliu@sibs.ac.cn.
References
Author notes
C.-T.F. and K.-Y.Z. contributed equally to this study.

![Figure 1. Elevated H3K27 trimethylation levels in the proximal promoter of the retained CTNNA1 allele. (A) A diagram of the human CTNNA1 genomic locus and the regions analyzed by ChIP. TSS indicates transcriptional start site. (B) ChIP analyses of histone modifications with the indicated antibodies in HL-60 and NB4 leukemia cell lines. (C-D) Semiquantitative RT-PCR and ChIP analyses of CTNNA1 expression and histone modifications in HL-60 cells treated with TSA for the indicated time. (E) Semiquantitative RT-PCR and ChIP analyses of CTNNA1 expression and H3K27me3 in HL-60 cells treated with DAC for the indicated times. (F) ChIP analyses of CTNNA1 promoter for H3K27me3 enrichment in FACS-sorted LICs (CD34+CD38−CD123+lineage−) from patient samples with or without del(5q). Enrichment of H3K27me3 was calculated by: [ChIP-P1E1(LICs)/INPUT-P1E1(LICs)]/[ChIP-P1E1(HL-60)/INPUT-P1E1(HL-60)]. ● represents case V with del(5q), which expresses a normal amount of CTNNA1.7](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/23/10.1182_blood-2009-11-255778/4/m_zh89991054030001.jpeg?Expires=1765889848&Signature=i1H3kQeXrMLV-8019BnpkuJq0wiaQjeDvoJ1S~eAvGOcVSOcFGl0ovXsHTcCgSorJq4qfGstVtyoggM6BGWHndk2OYWyCiTEIfvtaCkkeHhbRDXnSIKrSl3R3ZJYMO2rRXYoZf9vfYDIk9IqTpgh-M4vgx03uOKAfF~Qizoq082xK~u4mhEeLsGAfL2SqTrzl1Cdt4aCEhE-xRcPLOWZqYnvbzaSbGlRDwk9Tr1A37XHfQUo8pjETyFMcfjQxq6sjbUQi5oUjckRllX4f8k2IBBXY2CkupdfPOgfvYv9zZJtQhV4GvOB1fc4D1oRqRyN8ifJT3C4s5SkL3dDTm5fLQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)