Abstract
Among the different phenotypes of von Willebrand disease (VWD) type 2A, we identified a particular subgroup with a high frequency of 29%, characterized by a relative decrease of large von Willebrand factor (VWF) multimers and decreased A Disintegrin And Metalloproteinase with ThromboSpondin type 1 motifs, member 13 (ADAMTS13)–mediated proteolysis previously described in a single family as VWD type IIE (VWD2A/IIE). Phenotype and genotype of 57 patients from 38 unrelated families displaying a particular multimer pattern resembling the original VWD2A/IIE were studied. Pathogenicity of candidate mutations was confirmed by expression studies and phenotypic characterization of recombinant mutants. Specific mutations were identified in all patients. Twenty-two different mutations, most of them affecting cysteine residues, 17 of them being novel, are clustering mainly in the VWF D3 domain and correlate with the VWD2A/IIE phenotype. An intracellular retention of most mutants and/or a defect of multimerization seem to be the main pathogenic molecular mechanisms. ADAMTS13 proteolysis of mutant VWF was not different from wild-type VWF in a static assay, suggesting that reduced in vivo proteolysis is not an intrinsic property of mutant VWF. Our study identified a distinct VWD subtype with a common molecular background which contributes significantly to the heterogeneous spectrum of VWD.
Introduction
Von Willebrand disease (VWD), an inherited hemorrhagic disorder, is caused by either quantitative (types 1 and 3) or qualitative (type 2) defects of von Willebrand factor (VWF), a large multimeric protein playing a key role in primary hemostasis. The first comprehensive classification of VWD1 included numerous different types, 2 that were distinguished mainly by analyzing the multimer structure of VWF by sodium dodecyl sulfate–agarose gel electrophoresis of plasma VWF.2 One particular type was more common (IIA); others were described only in single families. Among them was the type IIE, characterized by a relative reduction of the VWF high-molecular-weight multimers (HMWMs), an abnormal structure of individual VWF oligomers and dominant inheritance.3
Individual oligomers from normal plasma appear as a triplet, consisting of a main band, flanked by 2 satellite bands being the products of proteolysis.4 By electrophoresis at higher resolution this triplet resolves into a quintuplet of 5 bands, the central band, 2 flanking inner subbands and 2 flanking outer subbands, respectively.5 Whereas in patients with type IIA the outer satellite bands appear pronounced because of enhanced susceptibility of VWF to proteolysis by the VWF specific metalloprotease ADAMTS13 (A Disintegrin And Metalloproteinase with ThromboSpondin type 1 motif, member 13),3,5 in patients with type IIE, proteolysis of VWF is reduced.3 This correlates with abnormal oligomers, consisting of a pronounced central band, flanked by only the inner subbands. By electrophoresis at low resolution, the abnormal oligomers appear as only one single broad band.
In the current classification of VWD,6 several different phenotypes with a reduction or lack of HMWM and corresponding decreased platelet-dependent function are designated VWD type 2A, which, however, comprises several different phenotypes with particular molecular pathomechanisms, ranging from dimerization7 and multimerization8 defects, impaired intracellular transport and secretion9-11 to enhanced proteolysis of mutant VWF (mVWF) multimers in the circulation.12,13 Defects of multimerization can be caused by an impaired intermolecular disulfide bonding of VWF dimers at their amino-terminals of the mature VWF subunit in VWD type 2A phenotype IIC. The respective mutations of the latter phenotype are located within the D2 domain of the VWF propeptide.8,14,15
It has been suggested that because of the presence of specific consensus sequences (CGLC) in the propeptide D1 and D2 domains, common to disulfide isomerases, these particular domains could play a role in catalyzing the intermolecular disulfide bonding at the amino-terminal of the mature VWF subunit.16 The hypothetical “substrate” for this disulfide isomerase activity is located in the D3 domain of VWF where probably several cysteine residues participate in mature VWF intermolecular amino-terminal disulfide bonding.17,18 Cysteines C1099, C1142, and at least 1 of 3 subsequent cysteines C1222, C1225, and C1227, respectively, probably participate directly in this intermolecular binding.18,19 Thus, mutations in this domain, particularly those affecting cysteine residues, could potentially have an effect on multimer formation.
Among patients with VWD type 2A, there is a distinct phenotype with relative reduction of HMWMs, and an aberrant triplet structure with lack of the typical triplet subbands. This phenotype, with a relatively high frequency of 29% among patients with VWD type 2A diagnosed in our laboratory (U.B.),20 shows a multimer and triplet structure similar to the previously published phenotype IIE.3 Such a high prevalence of this phenotype has not been reported to date. Up to now only one mutation in VWD2A phenotype IIE (VWD2A/IIE) has been identified. It is a frameshift mutation in exon 52 and causes a defect of dimerization at the carboxy-terminal VWF monomer.21 However, analyzing the VWF 3′ region in additional patients was unsuccessful. Therefore, we extended our mutation screening and analyzed the complete coding sequence of the VWF gene in patients with VWD2A/IIE.
Methods
Patients
Approximately 8000 samples per year are referred to our reference laboratory (U.B.) to confirm and/or to refine the diagnosis of VWD. In approximately 15% of them VWD is diagnosed by measuring standard parameters and classified by VWF multimer analysis. In cases with an unusual aberrant multimer pattern, a molecular genetic analysis is suggested as additional test. Fifty-seven patients from 38 families displaying a particular multimer pattern in accordance with the original VWD2A/IIE3 were available for molecular studies. Patients' informed consent was obtained by the referring doctors. The study was in accordance with the amended version of the Declaration of Helsinki and was approved under the respective institutional guidelines of the referring and participating institutions.
Diagnosis of VWD2A/IIE
Conventional diagnostic tests were carried out with citrated plasma (0.011M trisodium citrate) of patients. Leukocytes were obtained from whole citrated blood or EDTA (ethylenediaminetetraacetic acid) blood for subsequent DNA extraction. VWF:antigen (VWF:Ag22 ) and VWF:collagen binding (VWF:CB23 ) with the use of equine collagen (Horm; Nycomed; a mixture of 95% collagen type I and 5% type III) as binding substrate in comparison to a normal plasma pool were obtained as standard parameters. FVIII:C and Ristocetin-Cofactor (VWF:RCo) data are incomplete. These parameters had to be analyzed by the referring institutions (because of their sensitivity to prolonged transport) and were either not done or not communicated in many of the investigated cases. Multimer analysis was performed in all cases by sodium dodecyl sulfate–agarose electrophoresis gels2 with luminescent visualization.24,25 The luminescent blot was stored on electronic media by photo imaging (FluorChem 8000; Alpha Innotech Corp).
Mutation screening
Nucleotide numbering, amino acid numbering, and nomenclature of mutations were according to recent recommendations.26 High-molecular-weight genomic DNA was prepared from leukocyte buffy coats by standard techniques and was used for the amplification of VWF coding exons 2 through 52 by polymerase chain reaction as previously described.7 Polymerase chain reaction products were sequenced by the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit on an ABI Prism 310 or an ABI 377. Candidate mutations were confirmed by sequencing both strands. In addition, if available, further family members of the index patients were studied.
Expression studies
VWF mutagenesis and recombinant expression of 19 mutations identified during this study were performed as described previously.21
In brief, site-directed mutagenesis of full-length VWF cDNA was carried out with the use of the Quick Change kit (Stratagene). Mutagenesis primers carrying the respective base-exchange at a central position were 40 base pairs in length. The expression vector VWF-pcDNA3.1 containing full-length mVWF and wild-type VWF (wtVWF) cDNA, respectively, was used to transform Top10 supercompetent cells (Invitrogen). Plasmids were purified by the Endofree Plasmid Maxi Kit (QIAGEN). 293-EBNA cells (2 × 106; Invitrogen) were transiently transfected with 4 μg of full-length mVWF or wtVWF cDNA by liposomal transfer (Lipofectamine 2000; Invitrogen). The cells were grown for 72 hours (24 hours in Dulbecco modified Eagle medium [Invitrogen] with 10% [vol/vol] fetal bovine serum and 48 hours in serum-free Iscove modified Dulbecco medium [Sigma-Aldrich] + 2% (vol/vol) Ultroser G [Invitrogen]). VWF secreted in the medium was concentrated in Centricon tubes to one-tenth of the original volume before subsequent analysis. To analyze intracellular VWF, the transfected cells were lysed by 3 rounds of freezing (−80°C) and thawing in lysis buffer (0.1M Tris/HCl pH 8.0; 0.6% [vol/vol] Triton X-100).
The same methods were applied for cotransfection with wtVWF and mVWF cDNA, using 2 μg each of wtDNA and mDNA. These experiments were done to reproduce the patients' heterozygous state. All experiments were done in triplicate in comparison to wtVWF cDNA.
Characterization of recombinant VWF
VWF:Ag, VWF:CB, and multimer analysis were determined as described above. In addition we applied a VWF:glycoprotein Ib (GpIb) binding assay with the use of a recombinant GpIbα fragment in the presence of 1 mg/mL Ristocetin essentially as described27 but with the following modifications: a GpIbα fragment expressed in HEK293 cells spanning amino acids 1 to 268 tagged with a carboxy-terminal Flag sequence and bound to Flag antibody (Sigma-Aldrich) coated enzyme-linked immunoabsorbent assay plates captures recombinant VWF from cell culture medium. Bound VWF is detected by a polyclonal rabbit VWF antibody and a second monoclonal goat anti–rabbit peroxidase-labeled antibody (both from Dako).
ADAMTS13 proteolysis
Selected mutants of VWF being characteristic for the spectrum of type 2A/IIE VWF were subjected to ADAMTS13 proteolysis (p.R1121M, p.C1130W, p.C1153Y, p.C1173R, p.C1190R). Full-length recombinant mVWF (rmVWF; 1 μg) was cleaved by 5 mU of recombinant ADAMTS13 in a final volume of 300 μL in comparison to rwtVWF as previously described.13 Susceptibility to proteolysis was assessed by multimer analysis and visual evaluation.
Results
Clinical symptoms
Although most patients were analyzed because of a bleeding tendency, detailed descriptions of bleeding symptoms were not available in many cases. However, from the cases with reports on bleeding symptoms, the bleeding tendency appears mild to moderate, but may also be severe, ranging from simple epistaxis and frequent hematoma to menorrhagia, postsurgery bleeds, occasionally joint bleeding in 2 patients and even central nervous system bleeding in 1 patient (Table 1). One family was tested only because of a prolongation of the activated partial thromboplastin time in the index patient (IC) who had no bleeding symptoms.
Mutations and phenotypic VWF parameters of 57 patients with VWD 2A/IIE
| Exon . | nt . | aa . | Ag 0.5 . | SD . | CB 0.8 . | SD . | Ratio CB . | RCo 0.5 . | SD . | Ratio RCo . | FVIII 0.6 . | SD . | bt < 6′ . | s . | bs* . | IC† . | C‡ . | ref . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | 2867T>C | V956A | 0.31 | 0.37 | 1.19 | 0.25 | 0.81 | 0.62 | — | m | (1) | 1 | 1 | novel | ||||
| 22 | 2926C>T | R976C | 1.29 | 0.46 | 1.04 | 0.25 | 0.81 | — | ND | 0.85 | 0.09 | — | m | (2) | 3 | 5 | novel | |
| 25 | 3271T>C | C1091R | 1.20 | 1.20 | 1.00 | — | ND | — | — | m | (3) | 1 | 1 | novel | ||||
| 25 | 3314C>A | A1105D | 0.14 | 0.13 | 0.93 | 0.11 | 0.79 | — | — | — | — | 1 | 1 | novel | ||||
| 25 | 3359G>C | W1120S | 0.34 | 0.19 | 0.56 | 0.21 | 0.62 | 0.72 | 20 | — | — | 1 | 2 | 28 | ||||
| 25 | 3362G>T | R1121M | 0.23 | 0.20 | 0.87 | — | — | — | — | — | — | 1 | 1 | novel | ||||
| 25 | 3377G>T | C1126F | 0.18 | 0.02 | 0.07 | 0.01 | 0.36 | — | — | — | — | — | — | 1 | 3 | novel | ||
| 26 | 3388T>C | C1130R | 0.17 | 0.10 | 0.59 | — | — | — | — | — | — | 1 | 1 | 29 | ||||
| 26 | 3387C>G | C1130W | 0.22 | 0.08 | 0.36 | — | — | — | — | — | — | 1 | 1 | novel | ||||
| 26 | 3437A>C | Y1146C | 0.20 | 0.07 | 0.16 | 0.06 | 0.79 | 0.07 | 0.03 | 0.35 | 0.31 | 0.08 | 11 | m-s | (4) | 12 | 17 | 29 |
| 26 | 3446G>A | C1149Y | 0.17 | 0.01 | 0.05 | 0.03 | 0.30 | 0.07 | 0.42 | 0.25 | — | m-s | (5) | 1 | 6 | novel | ||
| 26 | 3458G>A | C1153Y | 0.89 | 0.27 | 0.55 | 0.21 | 0.62 | 0.40 | 0.45 | 0.56 | — | — | — | 2 | 2 | novel | ||
| 26 | 3507T>G | C1169W | 0.11 | 0.05 | — | — | 0.06 | 0.01 | 0.52 | 0.17 | 0.05 | — | — | — | 1 | 2 | novel | |
| 26 | 3514G>T | G1172C | 0.25 | 0.14 | 0.56 | 0.15 | 0.60 | 0.45 | — | m | (6) | 1 | 1 | novel | ||||
| 26 | 3517T>C | C1173R | 0.19 | 0.08 | 0.42 | 0.10 | 0.53 | 0.61 | 12 | — | — | 1 | 1 | novel | ||||
| 26 | 3518G>T | C1173F | 0.35 | 0.11 | 0.23 | 0.04 | 0.65 | 0.32 | 0.93 | 0.54 | — | — | — | 3 | 4 | novel | ||
| 27 | 3568T>C | C1190R | 0.41 | 0.24 | 0.59 | 0.14 | 0.34 | 0.56 | 8 | — | — | 1 | 1 | 30 | ||||
| 27 | 3569G>A | C1190Y | 0.94 | 0.18 | 0.19 | — | — | — | — | — | — | 1 | 1 | novel | ||||
| 27 | 3583G>T | D1195Y | 0.43 | 0.38 | 0.88 | — | — | — | — | — | — | 1 | 1 | novel | ||||
| 28 | 3679T>C | C1227R | 0.03 | 0.01 | 0.33 | — | — | — | — | — | — | 1 | 1 | 31 | ||||
| 28 | 3833T>C | L1278P | 0.59 | 0.30 | 0.31 | 0.10 | 0.52 | — | — | — | — | — | — | 1 | 3 | novel | ||
| 28 | 3833T>G | L1278R | 0.20 | 0.21 | 1.05 | 0.02 | 0.10 | 0.26 | 20 | — | — | 1 | 1 | novel |
| Exon . | nt . | aa . | Ag 0.5 . | SD . | CB 0.8 . | SD . | Ratio CB . | RCo 0.5 . | SD . | Ratio RCo . | FVIII 0.6 . | SD . | bt < 6′ . | s . | bs* . | IC† . | C‡ . | ref . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | 2867T>C | V956A | 0.31 | 0.37 | 1.19 | 0.25 | 0.81 | 0.62 | — | m | (1) | 1 | 1 | novel | ||||
| 22 | 2926C>T | R976C | 1.29 | 0.46 | 1.04 | 0.25 | 0.81 | — | ND | 0.85 | 0.09 | — | m | (2) | 3 | 5 | novel | |
| 25 | 3271T>C | C1091R | 1.20 | 1.20 | 1.00 | — | ND | — | — | m | (3) | 1 | 1 | novel | ||||
| 25 | 3314C>A | A1105D | 0.14 | 0.13 | 0.93 | 0.11 | 0.79 | — | — | — | — | 1 | 1 | novel | ||||
| 25 | 3359G>C | W1120S | 0.34 | 0.19 | 0.56 | 0.21 | 0.62 | 0.72 | 20 | — | — | 1 | 2 | 28 | ||||
| 25 | 3362G>T | R1121M | 0.23 | 0.20 | 0.87 | — | — | — | — | — | — | 1 | 1 | novel | ||||
| 25 | 3377G>T | C1126F | 0.18 | 0.02 | 0.07 | 0.01 | 0.36 | — | — | — | — | — | — | 1 | 3 | novel | ||
| 26 | 3388T>C | C1130R | 0.17 | 0.10 | 0.59 | — | — | — | — | — | — | 1 | 1 | 29 | ||||
| 26 | 3387C>G | C1130W | 0.22 | 0.08 | 0.36 | — | — | — | — | — | — | 1 | 1 | novel | ||||
| 26 | 3437A>C | Y1146C | 0.20 | 0.07 | 0.16 | 0.06 | 0.79 | 0.07 | 0.03 | 0.35 | 0.31 | 0.08 | 11 | m-s | (4) | 12 | 17 | 29 |
| 26 | 3446G>A | C1149Y | 0.17 | 0.01 | 0.05 | 0.03 | 0.30 | 0.07 | 0.42 | 0.25 | — | m-s | (5) | 1 | 6 | novel | ||
| 26 | 3458G>A | C1153Y | 0.89 | 0.27 | 0.55 | 0.21 | 0.62 | 0.40 | 0.45 | 0.56 | — | — | — | 2 | 2 | novel | ||
| 26 | 3507T>G | C1169W | 0.11 | 0.05 | — | — | 0.06 | 0.01 | 0.52 | 0.17 | 0.05 | — | — | — | 1 | 2 | novel | |
| 26 | 3514G>T | G1172C | 0.25 | 0.14 | 0.56 | 0.15 | 0.60 | 0.45 | — | m | (6) | 1 | 1 | novel | ||||
| 26 | 3517T>C | C1173R | 0.19 | 0.08 | 0.42 | 0.10 | 0.53 | 0.61 | 12 | — | — | 1 | 1 | novel | ||||
| 26 | 3518G>T | C1173F | 0.35 | 0.11 | 0.23 | 0.04 | 0.65 | 0.32 | 0.93 | 0.54 | — | — | — | 3 | 4 | novel | ||
| 27 | 3568T>C | C1190R | 0.41 | 0.24 | 0.59 | 0.14 | 0.34 | 0.56 | 8 | — | — | 1 | 1 | 30 | ||||
| 27 | 3569G>A | C1190Y | 0.94 | 0.18 | 0.19 | — | — | — | — | — | — | 1 | 1 | novel | ||||
| 27 | 3583G>T | D1195Y | 0.43 | 0.38 | 0.88 | — | — | — | — | — | — | 1 | 1 | novel | ||||
| 28 | 3679T>C | C1227R | 0.03 | 0.01 | 0.33 | — | — | — | — | — | — | 1 | 1 | 31 | ||||
| 28 | 3833T>C | L1278P | 0.59 | 0.30 | 0.31 | 0.10 | 0.52 | — | — | — | — | — | — | 1 | 3 | novel | ||
| 28 | 3833T>G | L1278R | 0.20 | 0.21 | 1.05 | 0.02 | 0.10 | 0.26 | 20 | — | — | 1 | 1 | novel |
Phenotypic parameters are given in U/mL with lower range limit: Ag indicates VWF:Ag; CB, VWF:collagen binding; RCo, VWF:Ristocetin Cofactor; FVIII, FVIII:C; SD, standard deviation. Mean values and SDs are given for mutations that were identified in more than 1 patient. Ratio indicates ratios of VWF functional parameters to VWF:Ag; bt, bleeding time; s, severity of the bleeding phenotype; bs, reported bleeding symptoms; nt, nucleotide change; aa, predicted amino acid change; IC, number of index cases; C, number of cases; —, no data; ref, literature reference; and ND, not done.
Reported bleeding symptoms: (1) menorrhagia; (2) epistaxis; (3) epistaxis, oral mucosal bleeding, hematoma; (4) epistaxis, hematoma, menorrhagia, hematuria, intestinal bleeding, joint bleeding requiring prophylaxis; (5) epistaxis, hematoma, menorrhagia, hematuria, bleeding after adenectomy, intestinal bleeding, joint bleeding, CNS bleeding; and (6) epistaxis, oral mucosal bleeding.
Total number of ICs was 38.
Total number of cases was 57.
Biochemical parameters
We noted considerable variation in the results of the laboratory phenotypic studies even among 17 patients with the same most common mutation Y1146C (Table 1). Unfortunately, phenotypic data are incomplete because only VWF:Ag, VWF:CB, and VWF:multimers were determined centrally. VWF:Ag, VWF:CB, and VWF:RCo were reduced in most cases. Some patients had normal or near normal VWF:Ag (R976C, C1091R, C1190Y), and R976C and C1091R even had normal VWF:CB. In such cases the diagnosis was merely based on an aberrant multimer pattern. The available VWF:RCo values were low in most cases. Low ratios of VWF:CB and VWF:RCo to VWF:Ag, respectively, also indicate additional functional deficits of some mutants.
Multimer analysis
In all cases individual VWF oligomers were aberrant by lacking a normal triplet structure. The common feature was the absence or at least severe reduction in intensity of the VWF outer proteolytic subbands, indicating reduced proteolysis. Instead, in most cases inner subbands were present, flanking the central bands very closely, giving the impression of a broader single band (Figure 1), except for a few mutants (R976C, C1091R, C1173R/F, D1195Y) that show only slightly reduced proteolysis. Furthermore, in most cases we observed at least a relative reduction in intensity of the large VWF multimers, however, with considerable heterogeneity, ranging from their complete absence (C1126F, Y1146C, C1227R) to the presence of even very large (supranormal) multimers (C1130R, C1190R, D1195Y) although with reduced intensity (Figure 2). A good correlation of multimer patterns with data for VWF:CB was seen only in the mutations C1126F and C1190Y, probably because the lack of HMWM is more expressed. In some patients with Y1146C this could also be shown (data not shown) but not by the cumulative dataset of this most common mutation. Mutation L1278R correlates with a normal VWF:CB/VWF:Ag ratio but with a severely reduced ratio for VWF:RCo indicating a possible GpIb binding defect.
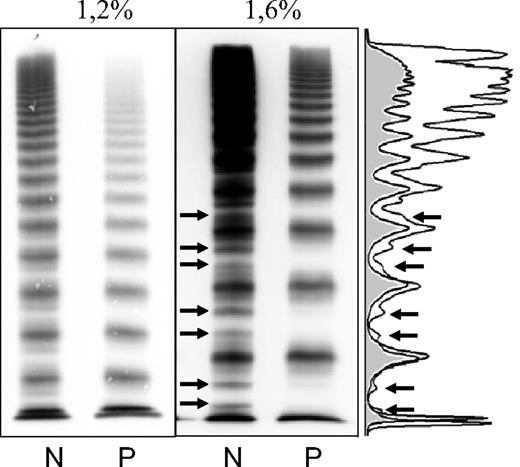
VWF multimers of a patient with VWD subtype 2A/IIE (mutation p.C1173R). Low (1.2%) and medium (1.6%) resolution gel. Densitometric evaluation of the 1.6% gel. Arrows point to the outer proteolytic bands only visible in the normal multimers (N). The patient's multimers (P) lack the outer proteolytic bands, a characteristic feature of this subtype.
VWF multimers of a patient with VWD subtype 2A/IIE (mutation p.C1173R). Low (1.2%) and medium (1.6%) resolution gel. Densitometric evaluation of the 1.6% gel. Arrows point to the outer proteolytic bands only visible in the normal multimers (N). The patient's multimers (P) lack the outer proteolytic bands, a characteristic feature of this subtype.
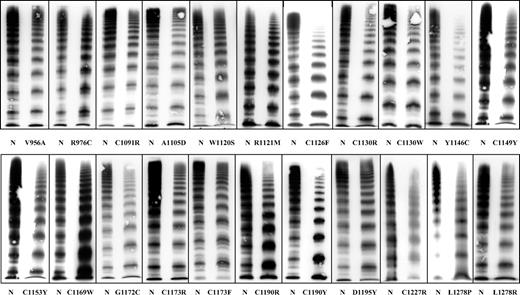
VWF multimers of 22 index patients with VWD subtype 2A/IIE corresponding to 22 different mutations. Although there is some phenotypic heterogeneity, the common feature of a relative loss of large multimers and/or decreased proteolysis is seen in all of them.
VWF multimers of 22 index patients with VWD subtype 2A/IIE corresponding to 22 different mutations. Although there is some phenotypic heterogeneity, the common feature of a relative loss of large multimers and/or decreased proteolysis is seen in all of them.
Identification of candidate mutations
By complete sequencing of the VWF coding exons at least 1 of 22 different candidate missense mutations was identified in 38 ICs (Table 1). Seventeen of them are novel mutations. Twenty are located in the D3 domain, only 2 mutations are located in the adjacent N-terminal region of the VWF A1 domain. Most mutations (15 of 22) involve cysteine codons, either resulting in the loss (12 of 15) or in the gain (3 of 15) of a cysteine codon. One patient was compound-heterozygous for p.W1120S and c.1795delG, another one for p.C1169W and the common VWD type 3 mutation c.2435delC. In one patient with VWF:Ag of only 0.03 U/mL and the mutation p.C1227R, we suspected compound heterozygosity with a second mutation; however, the extended mutation screening remained negative.
One mutation was particularly common: c.3437A>G, predicting the gain of a cysteine (p.Y1146C) was identified in 12 ICs. Other mutations occurring repeatedly were c.2926C>T (p.R976C, 3 ICs), c.3458G>A (p.C1153Y, 2 ICs), and c.3518G>T (p.C1173F, 3 ICs).
Expression studies
By expression of mVWF alone (homozygous) and coexpressed with wtVWF (heterozygous), respectively, we attempted to confirm the phenotype/genotype correlation. With 2 exceptions (p.R976C, p.D1195Y) all homozygous mutants correlate with significantly reduced VWF:Ag in the medium compared with VWF:Ag in cell lysate which was normal except for p.C1149Y. Correspondingly, the ratio of VWF:Ag in medium to lysate was reduced, suggesting a secretion defect in most mutants, including p.C1149Y. In addition, we observed a reduction or lack of HMWM, in all homozygous mutants with severe alterations caused by homozygous expression of p.A1105D, p.W1120S, p.C1130R/W, p.Y1146C, p.C1149Y, p.G1172C, p.C1173F, and p.C1190R and only slightly altered by p.R976C, p.D1195Y, p.C1227R, p.L1278P, and p.L1278R, as well as a corresponding reduction of VWF functional activities in all mutants except for p.D1195Y. (Figure 3; Table 2).
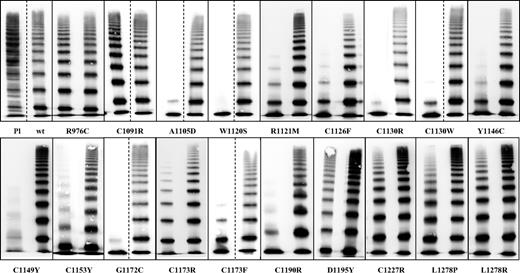
Multimers of 19 recombinant VWF2A/IIE mutants compared with plasma (Pl) and recombinant wtVWF. This figure is a composite of several gels. The first lane in the particular boxes represents the homozygous expression; the second lane represents the coexpression experiment. Most samples were analyzed side by side on the same gel. Dashed lines indicate that the 2 mutants were not analyzed on the same gel. Mutants with C1153Y were analyzed on the same gel but not side by side.
Multimers of 19 recombinant VWF2A/IIE mutants compared with plasma (Pl) and recombinant wtVWF. This figure is a composite of several gels. The first lane in the particular boxes represents the homozygous expression; the second lane represents the coexpression experiment. Most samples were analyzed side by side on the same gel. Dashed lines indicate that the 2 mutants were not analyzed on the same gel. Mutants with C1153Y were analyzed on the same gel but not side by side.
Expression data of 19 mutations located in the D3 and A1 domains
| Mutation . | Homozygous expression . | Coexpression . | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Med . | Lys . | Ratio* . | Med . | Ratio† . | Med . | Ratio‡ . | Med . | Lys . | Ratio* . | Med . | Ratio† . | Med . | Ratio‡ . | |||||||||
| Ag . | SD . | Ag . | SD . | GpIb . | SD . | CB . | SD . | Ag . | SD . | Ag . | SD . | GpIb . | SD . | CB . | SD . | |||||||
| WT | 100.0 | 8.0 | 100.0 | 9.6 | 1.00 | 100.0 | 1.1 | 1.00 | 100.0 | 19.9 | 1.00 | 100.0 | 5.0 | 100.0 | 9.6 | 1.00 | 100.0 | 1.1 | 1.00 | 100.0 | 19.9 | 1.00 |
| R976C | 59.5 | 4.7 | 72.8 | 2.5 | 0.82 | 31.6 | 3.0 | 0.53 | 32.2 | 2.1 | 0.54 | 68.2 | 0.9 | 83.2 | 4.1 | 0.82 | 52.9 | 3.0 | 0.78 | 45.2 | 4.5 | 0.66 |
| C1091R | 48.2 | 4.4 | 65.0 | 3.1 | 0.74 | 42.6 | 3.4 | 0.88 | 28.2 | 1.9 | 0.58 | 68.2 | 3.2 | 89.1 | 12.8 | 0.77 | 51.0 | 1.1 | 0.75 | 34.1 | 4.3 | 0.50 |
| A1105D | 32.3 | 2.7 | 111.7 | 5.8 | 0.29 | 3.9 | 0.0 | 0.12 | 0.7 | 0.2 | 0.02 | 48.7 | 3.6 | 86.4 | 6.5 | 0.56 | 25.2 | 0.0 | 0.52 | 18.7 | 2.0 | 0.38 |
| W1120S | 21.5 | 0.0 | 80.4 | 7.6 | 0.27 | 3.2 | 0.0 | 0.15 | 0.6 | 0.3 | 0.03 | 34.4 | 3.6 | 87.0 | 9.7 | 0.39 | 25.2 | 1.9 | 0.73 | 17.1 | 1.3 | 0.50 |
| R1121M | 19.0 | 3.2 | 90.4 | 3.3 | 0.21 | 3.9 | 0.0 | 0.20 | 1.0 | 0.7 | 0.05 | 44.6 | 7.1 | 88.1 | 7.2 | 0.51 | 31.6 | 3.0 | 0.71 | 19.8 | 1.4 | 0.44 |
| C1126F | 23.1 | 8.1 | 97.5 | 5.0 | 0.24 | 3.9 | 0.0 | 0.17 | 0.8 | 0.4 | 0.04 | 37.4 | 3.9 | 92.4 | 8.4 | 0.40 | 18.7 | 1.1 | 0.50 | 9.5 | 2.9 | 0.25 |
| C1130R | 25.1 | 6.2 | 85.1 | 16.9 | 0.30 | 4.5 | 1.1 | 0.18 | 0.7 | 0.2 | 0.03 | 61.0 | 7.7 | 97.4 | 15.9 | 0.63 | 36.1 | 2.2 | 0.59 | 17.3 | 5.4 | 0.28 |
| C1130W | 39.5 | 4.9 | 103.1 | 16.3 | 0.38 | 5.8 | 1.9 | 0.15 | 1.1 | 0.2 | 0.03 | 92.8 | 10.0 | 81.3 | 5.2 | 1.14 | 57.4 | 1.1 | 0.62 | 51.0 | 2.7 | 0.55 |
| Y1146C | 24.6 | 7.7 | 103.6 | 5.5 | 0.24 | 3.9 | 0.0 | 0.16 | 1.0 | 0.4 | 0.04 | 80.0 | 6.2 | 113.2 | 28.7 | 0.71 | 49.7 | 1.1 | 0.62 | 44.6 | 6.7 | 0.56 |
| C1149Y | 10.3 | 0.9 | 35.4 | 2.2 | 0.29 | 5.8 | 0.0 | 0.57 | 0.3 | 0.0 | 0.03 | 46.2 | 1.5 | 57.3 | 6.0 | 0.81 | 36.1 | 3.0 | 0.78 | 30.1 | 0.0 | 0.65 |
| C1153Y | 33.3 | 5.8 | 120.0 | 15.2 | 0.28 | 11.6 | 1.9 | 0.35 | 1.3 | 0.5 | 0.04 | 47.7 | 2.7 | 85.0 | 5.8 | 0.56 | 25.8 | 1.1 | 0.54 | 15.0 | 1.0 | 0.31 |
| G1172C | 26.2 | 1.5 | 106.5 | 6.1 | 0.25 | 5.8 | 0.0 | 0.22 | 0.9 | 0.3 | 0.04 | 55.4 | 4.1 | 92.8 | 16.7 | 0.60 | 25.2 | 3.4 | 0.45 | 18.1 | 0.6 | 0.33 |
| C1173F | 36.9 | 5.5 | 131.8 | 12.1 | 0.28 | 5.8 | 0.0 | 0.16 | 1.1 | 0.2 | 0.03 | 51.8 | 4.9 | 97.2 | 2.4 | 0.53 | 28.4 | 1.1 | 0.55 | 18.2 | 1.8 | 0.35 |
| C1173R | 25.1 | 7.6 | 119.7 | 15.0 | 0.21 | 3.9 | 0.0 | 0.15 | 0.7 | 0.5 | 0.03 | 45.6 | 10.2 | 92.7 | 13.9 | 0.49 | 27.7 | 4.9 | 0.61 | 19.6 | 3.1 | 0.43 |
| C1190R | 24.1 | 3.2 | 97.5 | 6.3 | 0.25 | 2.6 | 0.0 | 0.11 | 0.4 | 0.4 | 0.02 | 31.3 | 4.7 | 99.2 | 9.7 | 0.32 | 14.8 | 1.1 | 0.47 | 10.1 | 0.9 | 0.32 |
| D1195Y | 83.1 | 5.5 | 74.5 | 4.4 | 1.12 | 107.1 | 4.0 | 1.29 | 66.9 | 5.1 | 0.80 | 102.6 | 5.4 | 68.4 | 9.7 | 1.50 | 88.4 | 4.0 | 0.86 | 74.6 | 14.0 | 0.73 |
| C1227R | 22.1 | 3.6 | 94.9 | 8.2 | 0.23 | 9.7 | 1.9 | 0.44 | 0.4 | 0.5 | 0.02 | 44.1 | 3.2 | 89.8 | 13.7 | 0.49 | 27.7 | 4.0 | 0.63 | 20.0 | 1.9 | 0.45 |
| L1278P | 22.1 | 1.8 | 90.9 | 6.9 | 0.24 | 0.0 | 0.0 | 0.00 | 0.0 | 0.0 | 0.00 | 49.7 | 3.9 | 82.1 | 16.7 | 0.61 | 18.7 | 3.0 | 0.38 | 22.5 | 0.9 | 0.45 |
| L1278R | 19.0 | 2.4 | 93.9 | 11.6 | 0.20 | 0.0 | 0.0 | 0.00 | 0.4 | 0.4 | 0.02 | 56.4 | 4.4 | 96.6 | 8.0 | 0.58 | 21.9 | 0.0 | 0.39 | 26.2 | 0.0 | 0.46 |
| Mutation . | Homozygous expression . | Coexpression . | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Med . | Lys . | Ratio* . | Med . | Ratio† . | Med . | Ratio‡ . | Med . | Lys . | Ratio* . | Med . | Ratio† . | Med . | Ratio‡ . | |||||||||
| Ag . | SD . | Ag . | SD . | GpIb . | SD . | CB . | SD . | Ag . | SD . | Ag . | SD . | GpIb . | SD . | CB . | SD . | |||||||
| WT | 100.0 | 8.0 | 100.0 | 9.6 | 1.00 | 100.0 | 1.1 | 1.00 | 100.0 | 19.9 | 1.00 | 100.0 | 5.0 | 100.0 | 9.6 | 1.00 | 100.0 | 1.1 | 1.00 | 100.0 | 19.9 | 1.00 |
| R976C | 59.5 | 4.7 | 72.8 | 2.5 | 0.82 | 31.6 | 3.0 | 0.53 | 32.2 | 2.1 | 0.54 | 68.2 | 0.9 | 83.2 | 4.1 | 0.82 | 52.9 | 3.0 | 0.78 | 45.2 | 4.5 | 0.66 |
| C1091R | 48.2 | 4.4 | 65.0 | 3.1 | 0.74 | 42.6 | 3.4 | 0.88 | 28.2 | 1.9 | 0.58 | 68.2 | 3.2 | 89.1 | 12.8 | 0.77 | 51.0 | 1.1 | 0.75 | 34.1 | 4.3 | 0.50 |
| A1105D | 32.3 | 2.7 | 111.7 | 5.8 | 0.29 | 3.9 | 0.0 | 0.12 | 0.7 | 0.2 | 0.02 | 48.7 | 3.6 | 86.4 | 6.5 | 0.56 | 25.2 | 0.0 | 0.52 | 18.7 | 2.0 | 0.38 |
| W1120S | 21.5 | 0.0 | 80.4 | 7.6 | 0.27 | 3.2 | 0.0 | 0.15 | 0.6 | 0.3 | 0.03 | 34.4 | 3.6 | 87.0 | 9.7 | 0.39 | 25.2 | 1.9 | 0.73 | 17.1 | 1.3 | 0.50 |
| R1121M | 19.0 | 3.2 | 90.4 | 3.3 | 0.21 | 3.9 | 0.0 | 0.20 | 1.0 | 0.7 | 0.05 | 44.6 | 7.1 | 88.1 | 7.2 | 0.51 | 31.6 | 3.0 | 0.71 | 19.8 | 1.4 | 0.44 |
| C1126F | 23.1 | 8.1 | 97.5 | 5.0 | 0.24 | 3.9 | 0.0 | 0.17 | 0.8 | 0.4 | 0.04 | 37.4 | 3.9 | 92.4 | 8.4 | 0.40 | 18.7 | 1.1 | 0.50 | 9.5 | 2.9 | 0.25 |
| C1130R | 25.1 | 6.2 | 85.1 | 16.9 | 0.30 | 4.5 | 1.1 | 0.18 | 0.7 | 0.2 | 0.03 | 61.0 | 7.7 | 97.4 | 15.9 | 0.63 | 36.1 | 2.2 | 0.59 | 17.3 | 5.4 | 0.28 |
| C1130W | 39.5 | 4.9 | 103.1 | 16.3 | 0.38 | 5.8 | 1.9 | 0.15 | 1.1 | 0.2 | 0.03 | 92.8 | 10.0 | 81.3 | 5.2 | 1.14 | 57.4 | 1.1 | 0.62 | 51.0 | 2.7 | 0.55 |
| Y1146C | 24.6 | 7.7 | 103.6 | 5.5 | 0.24 | 3.9 | 0.0 | 0.16 | 1.0 | 0.4 | 0.04 | 80.0 | 6.2 | 113.2 | 28.7 | 0.71 | 49.7 | 1.1 | 0.62 | 44.6 | 6.7 | 0.56 |
| C1149Y | 10.3 | 0.9 | 35.4 | 2.2 | 0.29 | 5.8 | 0.0 | 0.57 | 0.3 | 0.0 | 0.03 | 46.2 | 1.5 | 57.3 | 6.0 | 0.81 | 36.1 | 3.0 | 0.78 | 30.1 | 0.0 | 0.65 |
| C1153Y | 33.3 | 5.8 | 120.0 | 15.2 | 0.28 | 11.6 | 1.9 | 0.35 | 1.3 | 0.5 | 0.04 | 47.7 | 2.7 | 85.0 | 5.8 | 0.56 | 25.8 | 1.1 | 0.54 | 15.0 | 1.0 | 0.31 |
| G1172C | 26.2 | 1.5 | 106.5 | 6.1 | 0.25 | 5.8 | 0.0 | 0.22 | 0.9 | 0.3 | 0.04 | 55.4 | 4.1 | 92.8 | 16.7 | 0.60 | 25.2 | 3.4 | 0.45 | 18.1 | 0.6 | 0.33 |
| C1173F | 36.9 | 5.5 | 131.8 | 12.1 | 0.28 | 5.8 | 0.0 | 0.16 | 1.1 | 0.2 | 0.03 | 51.8 | 4.9 | 97.2 | 2.4 | 0.53 | 28.4 | 1.1 | 0.55 | 18.2 | 1.8 | 0.35 |
| C1173R | 25.1 | 7.6 | 119.7 | 15.0 | 0.21 | 3.9 | 0.0 | 0.15 | 0.7 | 0.5 | 0.03 | 45.6 | 10.2 | 92.7 | 13.9 | 0.49 | 27.7 | 4.9 | 0.61 | 19.6 | 3.1 | 0.43 |
| C1190R | 24.1 | 3.2 | 97.5 | 6.3 | 0.25 | 2.6 | 0.0 | 0.11 | 0.4 | 0.4 | 0.02 | 31.3 | 4.7 | 99.2 | 9.7 | 0.32 | 14.8 | 1.1 | 0.47 | 10.1 | 0.9 | 0.32 |
| D1195Y | 83.1 | 5.5 | 74.5 | 4.4 | 1.12 | 107.1 | 4.0 | 1.29 | 66.9 | 5.1 | 0.80 | 102.6 | 5.4 | 68.4 | 9.7 | 1.50 | 88.4 | 4.0 | 0.86 | 74.6 | 14.0 | 0.73 |
| C1227R | 22.1 | 3.6 | 94.9 | 8.2 | 0.23 | 9.7 | 1.9 | 0.44 | 0.4 | 0.5 | 0.02 | 44.1 | 3.2 | 89.8 | 13.7 | 0.49 | 27.7 | 4.0 | 0.63 | 20.0 | 1.9 | 0.45 |
| L1278P | 22.1 | 1.8 | 90.9 | 6.9 | 0.24 | 0.0 | 0.0 | 0.00 | 0.0 | 0.0 | 0.00 | 49.7 | 3.9 | 82.1 | 16.7 | 0.61 | 18.7 | 3.0 | 0.38 | 22.5 | 0.9 | 0.45 |
| L1278R | 19.0 | 2.4 | 93.9 | 11.6 | 0.20 | 0.0 | 0.0 | 0.00 | 0.4 | 0.4 | 0.02 | 56.4 | 4.4 | 96.6 | 8.0 | 0.58 | 21.9 | 0.0 | 0.39 | 26.2 | 0.0 | 0.46 |
Med indicates VWF:Ag secreted into the medium; Lys, VWF:Ag in cell lysate; and SD, standard deviation.
Ratio of VWF:Ag in medium to VWF:Ag in lysate; GpIb, VWF:GpIb binding.
Ratio of VWFGpIb binding to VWF:Ag; CB, VWF:CB.
Ratio of VWF:CB to VWF:Ag.
Heterozygous mutants displayed a much broader spectrum of VWF multimer patterns, ranging from the presence of all multimer sizes at normal concentrations to relative reduction of HMWM, thus reflecting the situation in the patients. However, VWF:CB and VWF:GpIb binding did not correlate well with the multimer phenotype. This can partly be explained by the observation that standardized high-quality multimer analysis is superior in detecting a relative reduction of HMWMs rather than VWF:RCo or VWF:CB.25,28 Coexpression of wtVWF and mVWF resulted only in a relative decrease of HMWM, similar to the pattern observed in the patients. The ratios of VWF:GpIb and VWF:CB to VWF:Ag of almost all mutants were also decreased, although to a lesser extent.
ADAMTS13 proteolysis of VWF
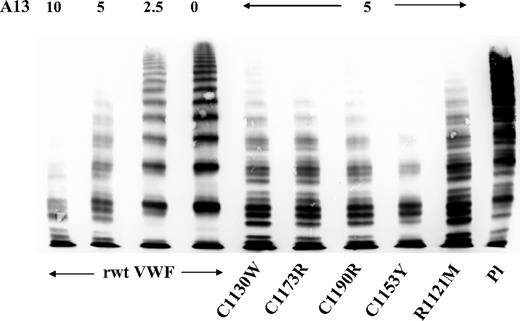
Proteolytic susceptibility of 5 representative rVWF mutants to ADAMTS13 measured by a static assay and visualized by multimer analysis was not principally different in comparison to rwtVWF. mVWF and wtVWF both displayed a similar multimer pattern and a comparable oligomer quintuplet structure as the result of the specific ADAMTS13 action on VWF (Figure 4).
VWF:multimers of recombinant wtVWF and mVWF proteolyzed by rhuADAMTS13 (A13) in a static assay. Similar proteolytic susceptibility of rwtVWF and mVWF is evident. Pl indicates plasma.
VWF:multimers of recombinant wtVWF and mVWF proteolyzed by rhuADAMTS13 (A13) in a static assay. Similar proteolytic susceptibility of rwtVWF and mVWF is evident. Pl indicates plasma.
Discussion
By VWF multimer analysis we could identify a distinct VWD phenotype with a remarkably high prevalence of 29% among patients with VWD2A.20 This phenotype, which closely resembles the previously described VWD type IIE, is primarily characterized by a relative reduction of HMWM, although in some cases even ultralarge multimers can be detected by the highly sensitive luminescent visualization method and photo imaging. The phenotype differs from the classical type 2A (phenotype IIA) by the lack of a defined triplet structure.
In all 57 patients we could identify a particular mutation, confirming a previous observation that aberrant VWF multimers are highly predictive for mutations at the VWF gene.25,29 Of the 22 different mutations only 5 had been described before by different investigators.29-32 Despite the common multimer phenotype, VWF:Ag and VWF:functional parameters were rather heterogeneous, ranging from even normal VWF:Ag and normal functional parameters, which would question the diagnosis of VWD, to values below 0.1 U/mL. Accordingly, clinical symptoms were ranging from no bleeding symptoms in 1 of 3 ICs with the mutation p.R976C, who was only tested because of a prolonged activated partial thromboplastin time, to severe bleeding symptoms in patients with the mutation p.Y1146C and p.C1149Y. Even 17 patients with the same common mutation p.Y1146C presented with heterogeneous VWF parameters, for example, normal and abnormal ratios of VWF:CB to VWF:Ag, respectively, and a range of clinical symptoms from mild to severe. Interestingly, p.Y1146C has previously also been identified in patients with VWD type 1.29,31
The nature of most of the identified mutations, their clustering in the D3 domain, and the similar features of the phenotype suggest a common underlying pathomechanism. However, several possible mechanisms may act in concert:
Secretion defect
By analyzing secreted and intracellular VWF we could show that most mutations correlate with a moderate-to-severe secretion defect that could explain the decreased VWF:Ag observed in the patients' plasma. Similar results for other mutations in the D3 domain, for example, p.C1130F, p.C1149R, have previously been reported.10,11 In addition to a secretion defect, the mutation p.C1149R also causes intracellular degradation of VWF,11 which suggests that our observation of low intracellular rmVWF p.C1149Y may also be the result of increased intracellular degradation or, alternatively, decreased VWF synthesis.
Multimerization defect
Because most mutations affect cysteines, it is suggestive that they might also impair the multimerization of VWF by affecting the polymerization process of the primary VWF dimers at their VWF D3 domains. Two such mutations (p. C1130F and p.C1149R) have been reported previously under this aspect.33 Cysteine residues C1099, C1142, C1222, C1225, and C1227 were suggested to play a direct role in amino-terminal intersubunit disulfide bonding of VWF.17-19 Mutations affecting such residues should directly impair the multimerization process. This is supported by the example of a naturally occurring mutation (p.C1225G) in a patient lacking HMWM34 and also by experimental evidence after recombinant expression of the artificial mutations C1222A, C1225A, and C1227A, respectively, which also correlated with a relative decrease of large VWF multimers.18 This is in line with the mutation p.C1227R in one of our cases: both, patient's plasma and rmVWF were lacking the largest multimers. Most mutations, however, are involving other cysteine residues in the D3 domain. Some of them are repeatedly found in several unrelated patients, in particular p.Y1146C, which we identified in 12 ICs (32%). Whether the respective cysteines are directly involved in intersubunit bonding or whether their mutation indirectly impairs the multimerization process, as can be assumed for the noncysteine mutants, cannot be answered by our study.
Decreased half-life
Some mutations in the VWF D3 domain cause a considerably shortened half-life. In an animal model it has been shown that the most severe defect, R1205H, which correlates with VWF:Ag less than 0.1 U/mL is caused by enhanced clearance of mVWF.35 Other mutations in D3 with decreased half-life are C1130F/G/R, C1149R, and W1144G.36,37 C1130R was also identified in our study. In addition, we found 2 novel mutations at the same respective codons (C1130W, C1149Y), suggesting decreased half-life as well. The decreased half-life may not only affect the steady state level of VWF:Ag but probably also the proteolytic pattern of individual VWF oligomers. A reduced residence time of secreted VWF HMWM will also reduce the time of exposure to the VWF protease ADAMTS13, resulting in limited proteolysis which could be an explanation for the reduced proteolytic pattern of the IIE phenotype but also for the persistence of supranormal multimers in some mutants (C1130R, C1190R, D1195Y).38
Decreased ADAMTS13-mediated proteolysis
The reduction of VWF proteolysis in patients with VWD2A/IIE3 is indicated by the lack of the characteristic triplet pattern as the result of ADAMTS13-mediated proteolysis.13 We performed proteolysis studies with 5 representative rmVWF constructs and rADAMTS13 and found no significant difference between the proteolytic pattern of rmVWF and rwtVWF in our in vitro static system (Figure 4). However, the assay, attempted to show a possible decreased intrinsic proteolytic susceptibility of VWF2A/IIE, does not sufficiently reproduce the conditions in the circulation where shear stress facilitates VWF proteolysis by ADAMTS13. We, therefore, speculate that in contrast to VWF2A/IIA with enhanced proteolytic susceptibility, the reduced susceptibility of VWF2A/IIE is not an intrinsic property of the mutant protein but rather the result of its functional deficits. In this model, decreased binding of mVWF to its receptors in the circulation may prevent sufficient unfolding of its A2 domain, thereby reducing the accessibility for ADAMTS13, which would sufficiently explain the observed typical multimer structure with decreased proteolytic subbands. Proteolysis experiments in flow-based assays should confirm our hypothesis.
Issues of VWD classification
The particular phenotype described in our study closely resembles the multimer pattern seen in patients with VWD2A/IIE.3 However, such patients have previously been found only in single families. Unfortunately, there are no molecular studies on these historical patients which would allow a direct comparison with our cohort.
The detection of a mutation cluster mainly in the D3 domain of VWF emphasizes the validity of our grouping of these patients into a distinct phenotype by multimer analysis. However, according to the current classification of VWD6 subtle aberrations in multimer structure do not justify the diagnosis of VWD type 2A. Therefore, the phenotypic expression spectrum of these mutants is rather a continuum from VWD type 1 to VWD type 2A, depending on the underlying defect. This is further underlined by earlier publications on cysteine mutations in the D3 domain that were regarded as VWD type 1 mutations, although the phenotype was identical to the phenotype described here.10 Similar mutations were also identified by a recent European study on patients historically diagnosed with VWD type 129 and in a comparable Canadian study.31
Diagnostic issues
The observed high prevalence of the VWD 2A/IIE phenotype of 29% among patients with VWD 2A is remarkable. We think that the abundance of this phenotype in Germany is not due to a particular population structure but is rather a matter of underdiagnosis in the past and may be due to technical problems with multimer analysis. The relative reduction of HMWM can only be detected by standardized sensitive multimer analysis requiring homogeneous transfer of smaller and HMWM during the blotting procedure. Additional densitometric evaluation can be helpful (Figure 1). However, although HMWM can be better differentiated in low-resolution gels, the typical IIE structure of individual oligomers with a lack of the outer bands of the triplet and pronounced inner subbands instead requires a medium resolution gel that, although extremely useful in detecting functional deficits, is not commonly used in other laboratories.25 The observation that ratios of VWF:CB or VWF:RCo to VWF:Ag were normal in several patients emphasizes the usefulness of multimer analysis in identifying patients with this particular VWD phenotype.
In conclusion, we could show that the particular phenotype described here is frequent and despite some heterogeneity an entity of its own, with a common molecular and pathogenic background most probably involving the multimerization process, intracellular transport, and secretion and/or decreased half-life of mVWF. Although these mechanisms may act in concert, a different individual contribution of single mechanisms may be responsible for the observed heterogeneity and the resulting problem of classification of this particular phenotype.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the following colleagues who have contributed with material and clinical data to our study: L. Balleisen, M. Barthels, F. Bergmann, W. Eberl, D. Franke, V. Freitag, H. Gazda, R. Grosse, H. Hauch, U. Harbrecht, K. Hofmann, H. Kiesewetter, H. Lenk, G. Marx, E. Mondorf, H.H. Wolf, I. Martinez-Saguer, R. Scharf, I. Scharrer, W. Streif, M. van der Planken, M. von Depka-Prondzinski, K. Welte, C. Wermes, and J. Wendisch.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG; grants Schn 325/4-1, Schn 325/4-2) and the German National Genome Research Network NGFN2 (Cardiovascular Diseases Network, grant NHK-S17T22).
Authorship
Contribution: R.S. designed the study; collected, evaluated, and interpreted the phenotypic, genotypic, and expression data; and wrote the paper; J.J.M. and B.Z. contributed significantly with patients' data and reviewed the manuscript; A.P. and K.W. characterized the patients phenotypically by VWF parameters and multimer analysis; F.O. and T.O. carried out genotyping and the expression studies; S.S. also carried out genotyping and evaluated and interpreted VWF multimers; and U.B. contributed to the design of the study, evaluated and interpreted phenotypic data and VWF multimers, characterized recombinant VWF mutants phenotypically, and reviewed the manuscript.
Conflict-of-interest disclosure: R.S. has received research funding from CSL Behring. The remaining authors declare no competing financial interests.
Correspondence: Reinhard Schneppenheim, University Medical Center Hamburg-Eppendorf, Department of Pediatric Hematology and Oncology, Martinistr 52, D-20246 Hamburg, Germany; e-mail: schneppenheim@uke.de.