Abstract
Among 200 patients with primary myelofibrosis, karyotype at diagnosis was abnormal in 83 (42%). To assess their individual prognostic impact, specific cytogenetic abnormalities with more than or equal to 5 informative cases were identified and the rest grouped separately as “other abnormalities.” Median survival in patients with sole +9 (n = 6), sole 20q− (n = 21), sole 13q− (n = 8), normal karyotype (n = 117), “other abnormalities” (n = 28), complex karyotype (n = 13), and sole +8 (n = 7) were “not reached,” 112, 105, 80, 46, 34, and 28 months, respectively (P = .01). Accordingly, 4 cytogenetic risk groups were considered: (1) favorable (sole +9, 20q−, or 13q−), (2) normal, (3) unfavorable (complex karyotype or sole +8), and (4) “other abnormalities.” Multivariable analysis confirmed the International Prognostic Scoring System (IPSS)–independent prognostic value of both 4-way and 2-way (ie, favorable/normal vs unfavorable/other abnormalities; IPSS-adjusted hazard ratio = 0.37; 95% confidence interval, 0.24-0.58) cytogenetic risk categorization (P < .01). The ability to prognostically dissect a specific IPSS category has major therapeutic implications.
Introduction
Among myeloid malignancies, cytogenetic information has established prognostic value in acute myeloid leukemia and myelodysplastic syndromes and is incorporated into formal prognostic systems.1,2 Similarly, the prognostic value of cytogenetic findings in primary myelofibrosis (PMF) has long been suggested3-5 and recently reiterated in 3 separate studies.6-8 The latter were all in agreement regarding the favorable prognostic impact of sole abnormalities of 20q− and 13q−.4,9 In addition, 1 of the 3 studies7 identified +9 (plus or minus 1 other abnormality) as another favorable prognostic marker in PMF, but the number of informative patients (n = 3 of 131 evaluated at diagnosis) was too small to be certain about the particular observation. Similarly, small sample sizes make it difficult to fully acknowledge observations from previous studies regarding the detrimental effect of −7/7q−,7,10,11 −5/5q−,7 −17 (or other abnormalities of chromosome 17),7 12p−,4 +8,4 +13,12 and complex karyotype.7 In the current study, we expanded our cohort of PMF patients with cytogenetic information at or near the time of diagnosis (n = 200): (1) to validate some of the aforementioned observations and (2) to examine the prognostic interaction between cytogenetic risk categorization and the International Prognostic Scoring System (IPSS) for PMF.13
Methods
After approval by the Mayo Clinic Institutional Review Board, clinical and laboratory data were collected from adult patients (≥ 18 years) with PMF, in whom cytogenetic information at or within 6 months of diagnosis (and before initiation of specific therapy) was available. The diagnosis of PMF was according to the 2001 World Health Organization criteria.14 Bone marrow chromosome and JAK2V617F analysis were according to previously published methods.15,16 Cytogenetic results were interpreted and reported according to the International System for Human Cytogenetic Nomenclature17 ; however, to be consistent with methodology used in recent studies,7 the presence of less than 20 evaluable metaphases did not disqualify patients from study inclusion as long as more than or equal to 10 metaphases were examined in those patients with “normal” reports. Abnormal karyotype was defined by the presence of at least 2 metaphases with structural or 3 metaphases with numerical abnormalities regardless of number of metaphases examined. Beforehand, it was decided that cytogenetic group assignment required the presence of at least 5 informative cases. Cytogenetic abnormalities occurring in less than 5 patients were grouped together and operationally referred to as “other abnormalities.”
All statistical analyses considered parameters at diagnosis. Differences in the distribution of continuous variables between categories were analyzed by either Mann-Whitney or Kruskal-Wallis test. Patients groups with nominal variables were compared by χ2 test. Survival analysis was considered from the date of diagnosis to date of death or last contact; patients who underwent allogeneic hematopoietic cell transplantation (allo-HCT) were censored at the time of transplantation. Survival curves were prepared by the Kaplan-Meier method and compared by the log-rank test. The Cox proportional hazard regression model was used for multivariable analysis. P values less than .05 were considered significant. The Stat View (SAS Institute) statistical package was used for all calculations.
Results and discussion
A total of 200 patients with PMF met the criteria stipulated in “Methods” for study inclusion; an additional 22 patients were excluded because of an inadequate number of metaphases. Clinical and laboratory features of the study population are outlined in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The IPSS risk distributions were low in 66 (33%) patients, intermediate-1 (int-1) in 64 (32%), int-2 in 44 (22%), and high in 26 (13%). JAK2V617F mutation analysis was performed in 167 patients, and 104 (62%) were mutation-positive. Cytogenetic findings were normal in 117 (58%) patients. Among the 83 (42%) patients with abnormal karyotype, 21 displayed sole 20q−, 8 sole 13q−, 7 sole +8, and 6 sole +9. Thirteen patients had complex karyotype (≥ 3 cytogenetic abnormalities) and 28 had cytogenetic abnormalities that individually occurred in less than 5 patients and therefore grouped in the “other abnormalities” category.
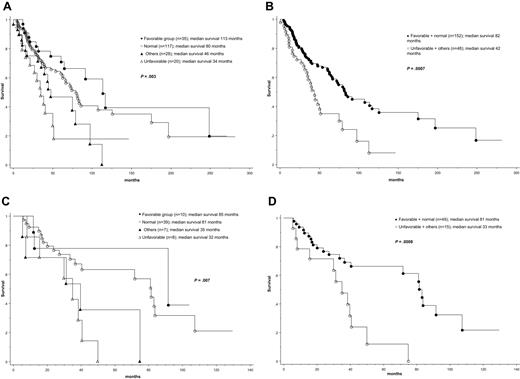
At the time of this report, median follow-up was 41 months and 99 (50%) deaths were recorded. Causes of death were documented in 52 cases, and their distribution among the different cytogenetic groups is outlined in supplemental Table 1. As expected, the IPSS13 effectively delineated patient groups with different prognosis; median survival in low, int-1, int-2, and high IPSS risk categories was 188, 71, 47, and 26 months, respectively (P < .01). Patients also displayed significantly different survival based on cytogenetic findings; median survival was the longest in patients with sole +9 (not reached), sole 20q− (112 months), sole 13q− (105 months), or normal karyotype (80 months) and significantly shorter in patients with “other abnormalities” (46 months), complex karyotype (34 months), or sole +8 (28 months; P = .01; Table 1). Based on these findings, 4 cytogenetic risk groups were considered: favorable (+9, 20q−, 13q−; n = 35), normal (n = 117), “other abnormalities” (n = 28), and unfavorable (+8 or complex karyotype; n = 20); presenting clinical and laboratory features for these 4 cytogenetic risk groups are outlined in supplemental Table 1. The respective median survivals for these 4 groups were 113, 80, 46, and 34 months (P = .003; Figure 1A; Table 1). Consideration of 2 (ie, favorable/normal vs unfavorable/other abnormalities) instead of 4 cytogenetic groups also resulted in significant survival differences (82 vs 42 months; P < .001; Figure 1B).
Cytogenetic risk categories in 200 patients with primary myelofibrosis
| Patient group . | No. of patients (median survival, mo) . | Cytogenetic risk group . | Median survival, mo* . | |
|---|---|---|---|---|
| All patients | 200 (74) | |||
| Sole del(20q) | 21 (112) |  | ||
| Sole del(13q) | 8 (105) | Favorable | ||
| Sole +9 | 6 (not reached) | 113 | ||
| Normal | 117 (80) | Normal | ||
| Other abnormalities | ||||
| Sole −5/del(5q) | 1 (not reached) |  | ||
| Sole −7/7q− | 2 (56) | |||
| 20q− plus one other abnormality | ||||
| 13q− plus one other abnormality | 2 (58) | |||
| +9 plus one other abnormality | 1 (22) | Others | 46 | |
| +8 plus one other abnormality | 1 (4) | |||
| Others with 2 abnormalities† | 7 (29) | |||
| Others with sole abnormalities‡ | 11 (46) | |||
| Complex karyotype | 13 (34) |  | Unfavorable | |
| Sole +8 | 7 (28) | 34 | ||
| Patient group . | No. of patients (median survival, mo) . | Cytogenetic risk group . | Median survival, mo* . | |
|---|---|---|---|---|
| All patients | 200 (74) | |||
| Sole del(20q) | 21 (112) |  | ||
| Sole del(13q) | 8 (105) | Favorable | ||
| Sole +9 | 6 (not reached) | 113 | ||
| Normal | 117 (80) | Normal | ||
| Other abnormalities | ||||
| Sole −5/del(5q) | 1 (not reached) |  | ||
| Sole −7/7q− | 2 (56) | |||
| 20q− plus one other abnormality | ||||
| 13q− plus one other abnormality | 2 (58) | |||
| +9 plus one other abnormality | 1 (22) | Others | 46 | |
| +8 plus one other abnormality | 1 (4) | |||
| Others with 2 abnormalities† | 7 (29) | |||
| Others with sole abnormalities‡ | 11 (46) | |||
| Complex karyotype | 13 (34) |  | Unfavorable | |
| Sole +8 | 7 (28) | 34 | ||
P = .003, favorable versus normal versus other abnormalities versus unfavorable.
47,XY,t(1;9)(p10;q10),+der(1;9)(q10;p10)[14]/46,XY[6]; 46,XY,t(2;11)(p15;q23)?c[17]/46,idem,del(5)(q13q33)[3]; 46,XX,−7,+r[18]/46,XX[2]; 46,XX,−20,+mar[20]; 46,XY, 8p−,9q+; +8 and del(20q−); +1,der(1;13)[2].
+19; 47,XY,+21[20]; 1q+; 46,XY,i(17q)[7]/46,XY[13]; 46,XY,inv(3)(q21q26.2)[12]; 46,XY,+1,der(1;7)(q10;p10)[7]/46,XY[13]; 46,XX,+1,der(1;7)(q10;p10)[8]/46,XX[2]; 46,XY,t(2;12)(p13;q13)?c[20]; 46,XY,add(2)(p13)[6]46,idem,+9,−21[1]/46,XY[13]; add(20)(q13.3); 46,XX,add(9)(q13)[2]/46,XX[28].
Survival data. (A) Survival data of 200 patients with PMF stratified according to 4 different cytogenetic risk categories: (1) favorable (sole abnormalities of 13q−, 20q−, or +9), (2) normal, (3) unfavorable (complex abnormalities or +8), and (4) other cytogenetic abnormalities. (B) Survival data of 200 patients with PMF stratified according to 2 different cytogenetic risk categories: (1) favorable or normal and (2) unfavorable or other cytogenetic abnormalities. (C) Survival data of 64 patients with intermediate-1 risk PMF (according to the IPSS)13 stratified according to 4 different cytogenetic risk categories as in panel A. (D) Survival data of 64 patients with intermediate-1 risk PMF stratified according to 2 different cytogenetic risk categories as in panel B.
Survival data. (A) Survival data of 200 patients with PMF stratified according to 4 different cytogenetic risk categories: (1) favorable (sole abnormalities of 13q−, 20q−, or +9), (2) normal, (3) unfavorable (complex abnormalities or +8), and (4) other cytogenetic abnormalities. (B) Survival data of 200 patients with PMF stratified according to 2 different cytogenetic risk categories: (1) favorable or normal and (2) unfavorable or other cytogenetic abnormalities. (C) Survival data of 64 patients with intermediate-1 risk PMF (according to the IPSS)13 stratified according to 4 different cytogenetic risk categories as in panel A. (D) Survival data of 64 patients with intermediate-1 risk PMF stratified according to 2 different cytogenetic risk categories as in panel B.
Multivariable analysis confirmed the IPSS-independent prognostic value of both 2-way and 4-way cytogenetic risk categorization (P < .01 in both instances); the corresponding IPSS-adjusted hazard ratios and 95% confidence intervals are provided in supplemental Table 2. Additional analysis within specific IPSS categories disclosed that prognostic distinction was most apparent in the IPSS int-1 risk category (Figure 1C-D; supplemental Table 3). JAK2V617F mutational status did not affect survival either in the entire cohort of study patients (P = .27) or when analysis was restricted to the IPSS int-1 risk group (P = .63).
The results of the current study have major therapeutic implications. Allo-HCT is currently the only treatment approach in PMF that is potentially curative.18 However, most patients and physicians alike show limited enthusiasm for such therapy because of the associated high treatment-related mortality and morbidity.19 Similarly, after the wave of recent descriptions of JAK-STAT-relevant mutations in myeloproliferative neoplasms,20 several anti-JAK2 investigational drugs are currently being evaluated in PMF. Such drugs carry unknown risks that are not necessarily less important than those seen with allo-HCT. Therefore, any information that can facilitate treatment decision making for the individual patient is highly desired, and the results of the current study support the value of cytogenetic information in that regard. Accordingly, we recommend application of IPSS risk stratification first followed by second-level prognostication using cytogenetic data.
In conclusion, the current study, by virtue of its larger sample size of informative cases evaluated at the time of diagnosis, validates the recent observations by Tam et al regarding the respective favorable and unfavorable prognostic impact of sole +9 and complex karyotype.7 Similarly, an adequate number of patients were included to also confirm our own previous observations regarding the adverse prognostic impact of sole +8.4 On the other hand, the number of informative patients with −7/7q− was not large enough to help clarify the discrepancy between recent studies regarding its prognostic value.7,8,10 The same can be said regarding abnormalities of chromosomes 5, 17, or noncomplex combinations of good-risk and high-risk abnormalities. A larger number of study patients is needed to resolve these issues and to determine the prospect of including cytogenetic information during future revisions of the IPSS.13
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.H. designed the research, collected and analyzed data, and wrote the manuscript; A.D.P. reviewed the manuscript; D.L.V.D. analyzed and interpreted cytogenetic findings and reviewed the manuscript; C.A.H. reviewed the pathologic findings and reviewed the manuscript; and A.T. designed the research, contributed patients, collected, analyzed, and interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ayalew Tefferi, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: tefferi.ayalew@mayo.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal