Abstract
The immunomodulatory drugs (IMiDs) lenalidomide and pomalidomide yield high response rates in patients with multiple myeloma, but the use of IMiDs in multiple myeloma is associated with neutropenia and increased risk for venous thromboembolism (VTE) by mechanisms that are unknown. We show that IMiDs down-regulate PU.1, a key transcription factor involved in granulocyte differentiation in vitro and in patients treated with lenalidomide. Loss of PU.1 results in transient maturation arrest with medullary accumulation of immature myeloid precursors and subsequent neutropenia. Accumulation of promyelocytes leads to high levels of the platelet aggregation agonist, cathepsin G stored in the azurophilic granules of promyelocytes. High levels of cathepsin G subsequently may increase the risk of VTE. To our knowledge, this is the first report investigating the underlying mechanism of IMiD-induced neutropenia and increased risk of VTE in multiple myeloma.
Introduction
Thalidomide and its immunomodulatory derivatives (IMiDs), such as lenalidomide (CC-5013) and pomalidomide (CC-4047), represent a novel class of agents with potent activity against multiple myeloma (MM). However, the use of IMiDs has been associated with 2 major adverse effects: (1) higher incidence of neutropenia that is the most common reason for holding or termination of treatment, and (2) increased risk for venous thromboembolism (VTE) that is further amplified with combination therapy.
In 2 multicenter randomized trials including 353 patients with MM, 23% of patients experienced grade 3 or 4 neutropenia.1,2 In patients receiving pomalidomide, the rate of grade 3 or 4 neutropenia was 32% in a phase 2 trial3 and 58% in a phase 1 trial.4 Clinical trials on myelodysplastic syndrome have shown that the frequency of lenalidomide-induced neutropenia is dose dependent and increases with duration of treatment.5,6 The mechanism of induction of neutropenia is unclear, especially because IMiDs are not stem cell toxic and in vitro induce the expansion of CD34+ cells.7
Previous studies have shown that thalidomide/IMiDs increase the risk of VTE in patients with MM, especially if used in combination with dexamethasone. In 2 phase 3 trials comparing lenalidomide plus dexamethasone versus dexamethasone alone, the incidence of VTE in the absence of thromboembolic prophylaxis was 4 times greater in the lenalidomide/dexamethasone group, 16% versus 4%.1,2,8 In a phase 1 trial evaluating pomalidomide alone, in 24 patients with refractory and/or relapsed MM, 4 (17%) developed VTE without thromboembolic prophylaxis during the first year of therapy.4 The observation that aspirin is effective in preventing IMiD-induced VTE,9 however, sharply contrasts with the lack of efficacy of aspirin in VTE prevention, suggesting that the pathogenesis in IMiD-induced VTE is different from that of “standard” VTE and, moreover, that platelet activation might contribute to thalidomide/IMiD-induced VTE.
In this study, we analyzed the effect of IMiDs on granulocytic progenitors. We show that down-regulation of the transcription factor PU.1 by IMiDs leads to maturational arrest of granulocytes with accumulation of myeloid progenitors (promyelocytes) and peripheral blood neutropenia. Azurophilic granules containing platelet aggregation agonists, such as cathepsin G (CG), stored in promyelocytes subsequently increase and may enhance platelet aggregation and risk for VTE.
Methods
Patient bone marrow cells
Bone marrow mononuclear cells were obtained from healthy volunteers and MM patients and isolated by separation on Hypaque-Ficoll gradients as described previously.10 CD34+ cells were obtained from leukaphereses products from patients who were scheduled for autologous transplantation with enriched CD34+ cells. Leukaphereses products were subjected either to positive CD34 cell selection with the Isolex 300 device11 or using CD34+ antibody-specific microbeads followed by magnetic separation column according to the manufacturer's protocol (Miltenyi Biotec). Subset analyses showed that CD34+ selected cells had a predominantly mature phenotype (CD34+/CD38+, 95.5% ± 5.3%; CD34+/CD33+, 79.9% ± 20.1%; CD34+/DR+, 98.9% ± 1.2%).
Patient serum and WBC sampling
All procedures were approved by the University of Pittsburgh Institutional Review Board, and the patients gave informed consent in accordance with the Declaration of Helsinki. Blood samples were collected before treatment with lenalidomide (baseline day −28 to day 1), before start of each cycle, and 1 month and 2 months after discontinuation of lenalidomide treatment. Human plasma was used for the detection of CG by the sandwich-type enzyme-linked immunosorbent assay (ELISA). After lysing the red blood cells with red blood cell lysis buffer (BioLegend), cells were collected by centrifugation followed by extraction of total RNA and then subjected to quantitative reverse-transcribed polymerase chain reaction (RT-PCR) analysis for the mRNA expression of CG.
Colony-forming assay
A total of 1.5 × 103 CD34+ cells were cultured in 2 mL Methocult GF H4434 (Stem Cell Technologies) in the presence of thalidomide, lenalidomide, pomalidomide (0.1, 1, 10, and 100μM), or dimethyl sulfoxide (DMSO; 0.1%, control). Methocult GF H4434 contained the following hematopoietic growth factors: 3 U/mL erythropoietin, 50 ng/mL rh stem cell factor (rhSCF), 10 ng/mL recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF), and 10 ng/mL recombinant human interleukin-3 (rhIL-3). Cells were incubated in 5% CO2 at 37°C for 14 days. At day 7 CFU-erythroid and at day 14 burst-forming units–erythroid, colony-forming units–granulocyte-macrophage (CFU-GM), and colony-forming units–granulocyte erythrocyte macrophage megakaryocyte (CFU-GEMM) were evaluated.
Flow cytometry
CD34+ cells were analyzed for expression of specific surface antigens using the following monoclonal antibodies: anti-CD34, anti-CD11a, anti-CD33 (BD Biosciences), and anti–glycophorin A/B (Sigma-Aldrich). Stained cells were analyzed by FACSCalibur flow cytometer and CellQuest software (BD Biosciences).
Pathologic evaluation of bone marrow aspirate smears and biopsies
Bone marrow aspirate smears were stained with Wright-Giemsa stain. To evaluate a left shift in granulocytopoiesis, a 500-cell count was performed, which included the enumeration of 6 stages of maturing neutrophilic granulocytes (ie, blasts, promyelocytes, myelocytes, metamyelocytes, bands, and segmented neutrophils). The findings were correlated with hematoxylin and eosin–stained histologic sections of the bone marrow biopsy. Review of the biopsy sections included assessment for degree of cellularity, architecture, cellular composition, myeloid/erythroid ratio, and the presence of any abnormal infiltrates.
PU.1/MPO double antigen labeling immunohistochemistry
Double antigen labeling immunohistochemistry studies were performed on marrow biopsy sections, with simultaneous assessment of PU.1 expression and myeloperoxidase expression for granulocytic progenitor cells. In brief, B-Plus–fixed, paraffin-embedded, decalcified bone marrow tissue sections were deparaffinized, rehydrated, and treated with hydrogen peroxide to inactivate endogenous peroxidase activity. Sections were incubated with a primary antibody against PU.1 (Cell Signaling Technology), and positive reactions were visualized using the ImPRESS polymerized reporter enzyme staining system (Vector Laboratories). Incubation with nickel enhanced diaminobenzidine (Vector Laboratories) yields a nuclear black reaction product when positive. Sections were incubated overnight with myeloperoxidase (Dako North America). Positive reactions were developed using the ImPRESS reagent and Vector NovaRED substrate (Vector Laboratories) to yield a red reaction product when positive that was cytoplasmic (myeloperoxidase) for granulocytic precursors. Slides were counterstained with Shandon hematoxylin (Thermo Scientific), dehydrated, and coverslipped for viewing.
Real-time PCR analysis
For the determination of mRNA levels of CG, total RNA was isolated using Mini RNA Isolation II Kit (Zymo Research). Total RNA was converted into cDNA using the Superscript III RT (Invitrogen). Quantitative RT-PCR was performed on an ABI Prism 7700 Sequence Detection System (Applied Biosystems). PCR was carried out with the SYBR Green PCR Master Mix (Bio-Rad) using 1 μL cDNA in a 20-μL final reaction mixture (15 minutes at 95°C, 40 cycles of 15 seconds at 95°, 60 seconds at 60°C, and 10 minutes at 79°C). The average threshold cycle (CT) for each gene was determined from triplicate reactions, and data were analyzed by taking the difference between mean threshold PCR cycles values for target and control gene (ΔCT). Target gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the ΔCT value. This was then calibrated to the control sample in each experiment to give the ΔΔCT value, where the control had a ΔΔCT value of 0. The fold target gene expression, compared with the calibrator value, is given by the formula: 2−ΔΔCT. The following primer sets were used: CG 5′-TGAGAGTGCAGAGGGATAGG-3′ and 5′-CGACTTTCCATAGGAGACGA-3′; β-actin 5′-GGACTTCGAGCAAGAGATGG-3′ and 5′-AGCACTGTGTTGGCGTACAG′-3′.
CG ELISA
Supernatants from CD34+ cell cultures and patient plasma samples were analyzed by sandwich-type ELISA.12 Serial dilution of standard CG was performed and samples were incubated for 1 hour at 37°C on precoated 96-well plates with monoclonal anti-CG antibody, followed by incubation with polyclonal rabbit IgG anti–human CG. Bound polyclonal anti–rabbit IgG CG was detected with goat anti–rabbit IgG conjugated to horseradish peroxidase. A colored product was formed in proportion to the amount of CG present in the sample or standard using the tetramethylbenzidine liquid substrate system (Sigma-Aldrich). The reaction was stopped by addition of stop solution and absorbance was read at 450 nm.
Analysis of cytokine secretion
CD34+ cells (106 cells/mL) were cultured with 50 ng/mL rhSCF, 10 ng/mL rhIL-3, or 10 ng/mL rhIL-6 in the presence of lenalidomide (100μM) or vehicle only (0.1% DMSO). Supernatants were collected on days 1, 3, and 6 and analyzed for granulocyte-colony stimulating factor (G-CSF) by Bio-Plex Cytokine Assay (Bio-Rad) according to the manufacturer's instructions. Intra-assay variability, expressed as coefficient of variation (CV = SD divided by the mean) was calculated based on determining quadruplicates of standards.13
Western blot analyses
Cells were harvested, washed 3 times with phosphate-buffered saline, and lysed with lysis buffer (NP-40 1%, 0.5mM dithiothreitol, 1mM sodium orthovandate, 1 μg/mL aprotinin, 50μM sodium fluoride, 500μM phenylmethylsulfonyl fluoride).14 Cell lysates were subjected to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to Hybond C super filters (GE Healthcare). The blots were probed with anti-PU.1 (Spi-1) antibody (Santa Cruz Biotechnology), granulocyte-colony stimulating factor (G-CSF) receptor antibody (R&D Systems), and β-actin antibody (GE Healthcare). Immune complexes were detected using enhanced chemiluminescence (GE Healthcare).
Platelet aggregation measurement
Blood was collected from healthy human volunteers for platelet preparation. Platelet-rich plasma was prepared by centrifugation of blood at 180g for 20 minutes. A total of 250 counts of platelets (250 μL of platelet-rich plasma) were incubated with 25 μL of patient plasma (1:10 dilution) at 37°C in an aggregometer (Dual Aggrometer, Chrono-Log). For the inhibition study, the CG inhibitor, α-1-antichymotrypsin (EMD Biosciences), was added at a final concentration of 300nM to platelet-rich plasma 1 minute before incubation with patient plasma. The resulting aggregation was expressed as a percentage of light transmission.
Statistical analyses
Each experiment was repeated at least 3 times, and all quantitative data are presented as mean plus or minus SD. Statistical differences were determined by Student t test. The results were considered significantly different at P less than .05.
Results
Lenalidomide and pomalidomide induce development of granulocytic progenitors with a concomitant block of maturation
We analyzed the effect of thalidomide, lenalidomide, and pomalidomide on colony formation of unstimulated and stimulated CD34+ cells. Stimulation of CD34+ cells was performed by culturing CD34+ cells in a cocktail of rhSCF, rhIL-3, and rhIL-6 for 12 hours after thawing. In both unstimulated and stimulated CD34+ cells, IMiDs significantly increased numbers of myeloid colonies (CFU-G, CFU-GM, and CFU-GEMM) in all tested concentrations. The up-regulation of myeloid colonies was especially evident in stimulated CD34+ cells, with an increase from mean 77 colonies (control) up to 247 colonies (pomalidomide).13 Thalidomide had only marginal stimulatory effect on the induction of myeloid precursor colonies. In accordance with our previous results, formation of erythroid colonies was strongly inhibited by IMiDs in cultures of stimulated and unstimulated CD34+ cells. Again, thalidomide had no significant effects on erythroid colonies (Figure 1A-B). Next we determined whether a particular subset of myeloid colonies was induced by IMiDs. As shown in Figure 1C, in cultures of bone marrow mononuclear cells from healthy donors and MM patients, pomalidomide and lenalidomide significantly induced development of CFU-G (P < .001) with a mean of 92 for lenalidomide and 121 for pomalidomide compared with control with 8 CFU-G. The number of CFU-GM and CFU-GEMM was unchanged.
Lenalidomide and pomalidomide induce development of granulocytic progenitors. Purified CD34+ cells were cultured with increasing concentrations of thalidomide, pomalidomide, lenalidomide, or 0.1% DMSO as vehicle control. (A) Erythroid colonies (burst-forming units-erythroid/CFU-erythroid) and (B) myeloid colonies (CFU-G, CFU-GM, and CFU-GEMM) were evaluated as described in “Colony-forming assay.” (C) Bone marrow mononuclear cells from healthy donors and MM patients were cultured with lenalidomide (left panel), pomalidomide (right panel), or DMSO as described in “Colony-forming assay.” Results are shown as mean ± SD from triplicates. *P ≤ .001. **P ≤ .01. ***P ≤ .05.
Lenalidomide and pomalidomide induce development of granulocytic progenitors. Purified CD34+ cells were cultured with increasing concentrations of thalidomide, pomalidomide, lenalidomide, or 0.1% DMSO as vehicle control. (A) Erythroid colonies (burst-forming units-erythroid/CFU-erythroid) and (B) myeloid colonies (CFU-G, CFU-GM, and CFU-GEMM) were evaluated as described in “Colony-forming assay.” (C) Bone marrow mononuclear cells from healthy donors and MM patients were cultured with lenalidomide (left panel), pomalidomide (right panel), or DMSO as described in “Colony-forming assay.” Results are shown as mean ± SD from triplicates. *P ≤ .001. **P ≤ .01. ***P ≤ .05.
The finding that IMiDs induce development of granulocytic precursors is in contrast to the clinical finding that IMiDs cause neutropenia. Because our previous studies showed that IMiDs are not directly toxic to bone marrow hematopoietic cells,15 we concluded that these effects might be induced by a maturational arrest of granulopoiesis. We therefore analyzed the marker expression profile of CD34+ cells cultured with and without IMiDs or thalidomide. CD33, a differentiation antigen whose expression is largely restricted to cells of the myeloid lineage,16 increased from 60% (control) to 74% and 78% (P < .05) in cells treated with lenalidomide and pomalidomide, respectively (Figure 2A), suggesting an increase in myeloid precursors. There was no significant change of CD11a+ cells with IMiD treatment. As expected, CD34+, a marker for early hematopoietic progenitors, decreased uniformly in all treatment groups. Glycophorin A, a marker for red cells, stayed at a low level on days 3, 6, and 9 (Figure 2B-D).
IMiDs induce a maturational block of granulocytic progenitors. Purified CD34+ cells were cultured with thalidomide, lenalidomide, or pomalidomide and analyzed for (A) CD33, (B) CDIIa, (C) CD34, and (D) glycophorin A expression at day 0, 3, 6, and 9 by fluorescence-activated cell sorter analysis. Results are shown as mean ± SD. (E) Table represents the ratio of the myeloid precursors/erythroid precursors in 6 patient bone marrow aspirates, before treatment and at neutrophil nadir induced by lenalidomide. (F) Representative bone marrow aspirates before treatment and at neutrophil nadir from a patient receiving lenalidomide. Arrows indicate immature myeloid cells. Note: during grade 4 neutropenia, the aspirate remains cellular, with a predominant population of neutrophilic precursors. Slides were evaluated using an Olympus BX45 microscope equipped with a 100×/1.35 numeric aperture (Olympus; original magnification ×1000).
IMiDs induce a maturational block of granulocytic progenitors. Purified CD34+ cells were cultured with thalidomide, lenalidomide, or pomalidomide and analyzed for (A) CD33, (B) CDIIa, (C) CD34, and (D) glycophorin A expression at day 0, 3, 6, and 9 by fluorescence-activated cell sorter analysis. Results are shown as mean ± SD. (E) Table represents the ratio of the myeloid precursors/erythroid precursors in 6 patient bone marrow aspirates, before treatment and at neutrophil nadir induced by lenalidomide. (F) Representative bone marrow aspirates before treatment and at neutrophil nadir from a patient receiving lenalidomide. Arrows indicate immature myeloid cells. Note: during grade 4 neutropenia, the aspirate remains cellular, with a predominant population of neutrophilic precursors. Slides were evaluated using an Olympus BX45 microscope equipped with a 100×/1.35 numeric aperture (Olympus; original magnification ×1000).
To evaluate medullary changes in granulopoiesis in human subjects treated with lenalidomide, we evaluated sequential bone marrows from 6 patients comparing bone marrow features before treatment, and at the time of grade 4 neutropenia. Marrows at the time of neutropenia showed an accumulation of myeloid precursors (myelocytes, metamyelocytes), supporting our hypothesis of impairment of myeloid maturation by IMiDs. In 5 of 6 patients, the myeloid/erythroid ratio increased from a median of 1.95 before treatment to 9.2 at the time of the neutrophil nadir, whereas bone marrow cellularity was unchanged (Figure 2E). A representative bone marrow aspirate at the time of the neutrophil nadir of a patient treated with lenalidomide is shown in Figure 2F.
Lenalidomide and pomalidomide decrease PU.1, which is critical for the maturation of neutrophils
After finding evidence that peripheral blood neutropenia might be induced by a maturational block of neutrophils, we focused on the identification of transcription factors controlling the maturation of myeloid progenitors. Western blot analysis revealed a down-regulation of PU.1 protein after pomalidomide and lenalidomide treatment (Figure 3A-B). This was further confirmed by quantitative RT-PCR. In CD34+ cells cultured for 3 days with either control vehicle or IMiD, mRNA of PU.1 was significantly (P ≥ .05) down-regulated by both pomalidomide and lenalidomide compared with control as shown in Figure 3C.
IMiDs down-regulate PU.1 in myeloid progenitors in vitro and in patients. Purified CD34+ cells were cultured in the presence of DMSO (control), (A) pomalidomide, or (B) lenalidomide for 6 and 8 days, and PU.1 protein expression was determined. β-Actin served as loading control. (C) CD34+ cells were cultured in the presence of DMSO (control), pomalidomide, or lenalidomide for 3 days. Total RNA was extracted and then subjected to quantitative RT-PCR. Data were analyzed according to the ΔCT method. Results are shown as mean CG mRNA fold increase by pomalidomide or lenalidomide compared with baseline (control). The level of mRNA was normalized to GAPDH expression. (D) Double labeling immunohistochemistry on paraffin-embedded bone marrow biopsy sections for PU.1 expression (black nuclear staining) and myeloperoxidase (red cytoplasmic staining) before and during treatment with either lenalidomide or dexamethasone (control). Slides were evaluated using an Olympus BX45 microscope equipped with a 100×/1.35 numeric aperture (Olympus; original magnification ×1000).
IMiDs down-regulate PU.1 in myeloid progenitors in vitro and in patients. Purified CD34+ cells were cultured in the presence of DMSO (control), (A) pomalidomide, or (B) lenalidomide for 6 and 8 days, and PU.1 protein expression was determined. β-Actin served as loading control. (C) CD34+ cells were cultured in the presence of DMSO (control), pomalidomide, or lenalidomide for 3 days. Total RNA was extracted and then subjected to quantitative RT-PCR. Data were analyzed according to the ΔCT method. Results are shown as mean CG mRNA fold increase by pomalidomide or lenalidomide compared with baseline (control). The level of mRNA was normalized to GAPDH expression. (D) Double labeling immunohistochemistry on paraffin-embedded bone marrow biopsy sections for PU.1 expression (black nuclear staining) and myeloperoxidase (red cytoplasmic staining) before and during treatment with either lenalidomide or dexamethasone (control). Slides were evaluated using an Olympus BX45 microscope equipped with a 100×/1.35 numeric aperture (Olympus; original magnification ×1000).
To confirm our in vitro findings, we analyzed PU.1 expression by double labeling immunohistochemistry of maturing neutrophils on bone marrow biopsy samples of myeloma patients treated with lenalidomide/dexamethasone or dexamethasone as control (non-IMiD treatment) for their expression of PU.1 before treatment and at the time of neutropenia. Before treatment, PU.1 (black nuclear staining) showed intense staining in myeloperoxidase-positive (red cytoplasmic staining) neutrophils and precursors (Figure 3D left panel). During therapy, the maturing neutrophils exhibited weak staining for PU.1 in patients treated with lenalidomide. In contrast, intense staining for PU.1 persisted in maturing neutrophils in patients receiving non–IMiD-containing treatment (Figure 3D right panel). These results show that PU.1, which is a critical transcription factor for the maturation of neutrophils, is down-regulated by IMiDs, resulting in their maturational arrest.
IMiDs enhance secretion of G-CSF
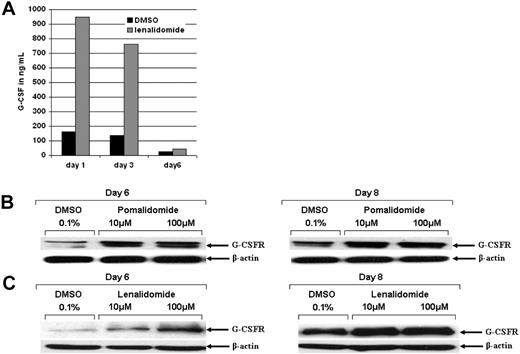
To investigate whether the maturational arrest of neutrophils by IMiDs is a result of a lack of granulocyte-colony stimulating factor G-CSF secretion or down-regulation of the G-CSF receptor, we analyzed G-CSF receptor expression and secretion of its ligand under IMiD treatment. CD34+ cells were treated with DMSO or lenalidomide for 1, 3, and 6 days, and supernatants were analyzed using the Bio-Plex Cytokine Assay. G-CSF was effectively increased by lenalidomide at day 1 (946 ng/mL vs 162 ng/mL control) and day 3 (763 ng/mL vs 138 ng/mL control) of treatment (Figure 4A). We observed a marginal increase of the G-CSF receptor expression on day 6 in IMiD-treated cells compared with control (Figure 4B-C).
Lenalidomide induces G-CSF and G-CSF receptor expression. Purified CD34+ cells were treated with 0.1% DMSO (control), pomalidomide, or lenalidomide. (A) Supernatants of CD34+ cells treated with pomalidomide, lenalidomide, or 0.1% DMSO were analyzed for cytokine secretion (pg/mL) after 1, 3, and 6 days. Black bars represent control; gray bars, lenalidomide treatment. Intra-assay variability, expressed as coefficient of variation (CV = SD divided by the mean) was calculated based on determining quadruplicates of standards. The SD was calculated based on quadruplicates and was less than 10%. (B-C) CD34+ cells were analyzed for G-CSF receptor protein expression by Western blot. β-Actin served as loading control.
Lenalidomide induces G-CSF and G-CSF receptor expression. Purified CD34+ cells were treated with 0.1% DMSO (control), pomalidomide, or lenalidomide. (A) Supernatants of CD34+ cells treated with pomalidomide, lenalidomide, or 0.1% DMSO were analyzed for cytokine secretion (pg/mL) after 1, 3, and 6 days. Black bars represent control; gray bars, lenalidomide treatment. Intra-assay variability, expressed as coefficient of variation (CV = SD divided by the mean) was calculated based on determining quadruplicates of standards. The SD was calculated based on quadruplicates and was less than 10%. (B-C) CD34+ cells were analyzed for G-CSF receptor protein expression by Western blot. β-Actin served as loading control.
IMiDs up-regulate the platelet aggregation agonist CG, released by promyelocytes in vitro and in patients
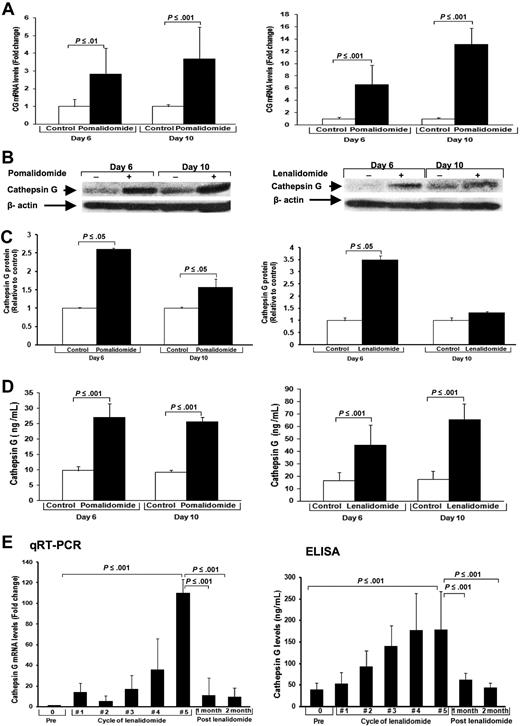
To further support our findings that IMiDs lead to maturational arrest of hematopoietic progenitors, we analyzed the gene expression profile. High density oligonucleotide microarrays were performed after 48 and 72 hours of treatment with pomalidomide. Compared with control, the 3 major serine proteases expressed in azurophilic granules of immature neutrophils were highly up-regulated after 48 hours: CG 3.4-fold, nuclear elastase 2 (NE2) 4.5-fold, and proteinase3 2.5-fold. After 72 hours, up-regulation of CG was 7.6-fold, NE2 11.8-fold, and proteinase3 6.5-fold compared with control. The results of our gene array analyses were confirmed by quantitative RT-PCR and Western blot. By quantitative RT-PCR, we found that after 6 days (2.8-fold, P ≤ .01) and 10 days (3.7-fold, P ≤ .001) CG mRNA expression increased significantly in CD34+ cells treated with pomalidomide compared with controls (Figure 5A left). Treatment with lenalidomide increased CG mRNA expression 6.6-fold on day 6 (P ≤ .001) and 13.1-fold on day 10 (P ≤ .001; Figure 5A right). Western blot analysis confirmed a strong intracellular up-regulation of CG protein in cells treated with lenalidomide (days 6 and 10) and pomalidomide (day 6; P < .05) compared with control (Figure 5B-C).
IMiDs up-regulate the platelet aggregation agonist CG released by promyelocytes. Purified CD34+ cells were cultured with pomalidomide, lenalidomide, or DMSO (control) for different time periods. (A) Quantitative RT-PCR of CD34+ cells. Results are shown as mRNA fold increase by pomalidomide or lenalidomide, compared with control (DMSO). mRNA levels were normalized with β-actin. (B) CG protein expression of CD34+ cells. β-Actin was used as loading control. (C) Densitometric analyses of immunoreactive bands for the protein levels of CG. (D) CG levels of supernatants of cultured CD34+ cells. Results are shown as mean ± SD of CG (ng/mL). (E) White blood cells and plasma from peripheral blood of 14 patients before lenalidomide treatment, during each cycle, and 1 and 2 months after discontinuation of lenalidomide were obtained. (Left panel) Quantitative RT-PCR of white blood cells of patients. CG mRNA fold induction by lenalidomide was compared with baseline (pretreatment). Results are shown as mean CG mRNA fold increase by lenalidomide compared with baseline (pretreatment). The level of mRNA was normalized to GAPDH. Results are shown as mean ± SD. (Right panel) Results of ELISA of plasma of patients are shown as mean ± SD of CG ng/mL.
IMiDs up-regulate the platelet aggregation agonist CG released by promyelocytes. Purified CD34+ cells were cultured with pomalidomide, lenalidomide, or DMSO (control) for different time periods. (A) Quantitative RT-PCR of CD34+ cells. Results are shown as mRNA fold increase by pomalidomide or lenalidomide, compared with control (DMSO). mRNA levels were normalized with β-actin. (B) CG protein expression of CD34+ cells. β-Actin was used as loading control. (C) Densitometric analyses of immunoreactive bands for the protein levels of CG. (D) CG levels of supernatants of cultured CD34+ cells. Results are shown as mean ± SD of CG (ng/mL). (E) White blood cells and plasma from peripheral blood of 14 patients before lenalidomide treatment, during each cycle, and 1 and 2 months after discontinuation of lenalidomide were obtained. (Left panel) Quantitative RT-PCR of white blood cells of patients. CG mRNA fold induction by lenalidomide was compared with baseline (pretreatment). Results are shown as mean CG mRNA fold increase by lenalidomide compared with baseline (pretreatment). The level of mRNA was normalized to GAPDH. Results are shown as mean ± SD. (Right panel) Results of ELISA of plasma of patients are shown as mean ± SD of CG ng/mL.
Secreted CG protein measured by ELISA in supernatants of CD34+ cell cultures treated with pomalidomide was also significantly increased on day 6 (27 ng/mL vs 9.8 ng/mL; P ≤ .001) and day 10 (25.6 ng/mL vs 9.2 ng/mL; P ≤ .001) compared with control (Figure 5D left). Lenalidomide significantly increased CG secretion on day 6 (44.9 ng/mL vs 16.4 ng/mL; P ≤ .001) and on day 10 (65.5 ng/mL vs 17.6 ng/mL; P ≤ .001) compared with control (Figure 5D right).
To confirm our in vitro findings, we evaluated the CG levels in patients treated with lenalidomide. Seven healthy volunteers had CG levels of mean 32.9 plus or minus 0.6 ng/mL (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). We then analyzed CG levels in relapsed/refractory MM patients (n = 14) before treatment with lenalidomide, during each cycle between day 15 until start of the next cycle, and 1 and 2 months after discontinuation of lenalidomide. All patients received 25 mg/day lenalidomide on days 1 to 21 in combination with 40 mg pulse dexamethasone. MM patients per se did not show elevated CG levels (Figure 5E prelenalidomide). But starting after cycle 1, we observed a progressive increase of CG mRNA levels in peripheral white blood cells measured by quantitative RT-PCR over the course of treatment, up to a median of 102.3-fold (cycle 5) compared with pretreatment levels (P ≤ .001; Figure 5E left panel). The most significant up-regulation of CG protein levels compared with pretreatment levels occurred during cycles 4 and 5 (cycle 5 to 224 ng/mL vs 40.7 ng/mL before the start of treatment; P < .001; Figure 5E right panel). One month after stopping lenalidomide, CG levels returned to baseline levels. The posttreatment decrease of CG was significant in patient plasma by ELISA assays (P ≤ .001) and in peripheral white blood cells by quantitative RT-PCR (P ≤ .001; Figure 5E right panel).
The characteristic pattern of CG up-regulation in individual patients, in patients with thromboembolic events, and lenalidomide dose reduction are shown in supplemental Figure 2A-C. To confirm that CG is the critical factor for platelet aggregation, we performed platelet aggregation studies using plasma samples from patients obtained pretreatment, during treatment, and posttreatment with lenalidomide. Plasma from patients treated with lenalidomide significantly increased platelet aggregation compared with plasma from the pretreatment controls, as demonstrated in supplemental Figure 2D (pretreatment: 7% vs cycle 3: 29% vs posttreatment: 9%) and supplemental Figure 2E (pretreatment: 2% vs cycle 5: 18%, vs cycle 8: 24%, vs cycle 10: 19%, vs posttreatment: 2%). Addition of a specific CG inhibitor (α-1-antichymotrypsin) decreased the enhanced platelet aggregation significantly. Plasma from patients off treatment for 1 month did not increase the platelet aggregation (supplemental Figure 2D-E).
Discussion
Without growth factor support, 28% and 58% of relapsed/refractory MM patients treated with lenalidomide or pomalidomide, respectively, developed grade 3 or 4 neutropenia in clinical trials.1-4 Previously, we found that IMiDs induce a shift in lineage commitment by down-regulation of GATA1, resulting in suppression of erythropoiesis and induction of myelopoiesis.15 The lineage bias effects of lenalidomide is concentration dependent, promoting the expansion of immature erythroid progenitors at low nanomolar concentrations.17,18 We, along with others, showed that the induction of myelopoiesis by IMiDs is accompanied by a maturational arrest of osteoclasts.19,20
Here, we show that lenalidomide and pomalidomide had no toxic effects on CD34+ cells, and indeed increased total colony numbers. We observed a shift in lineage commitment toward myelopoiesis associated with an induction of CFU-G formation. Further, CD34+ cells cultured with IMiDs showed a significant increase in expression of the early myeloid antigen, CD33. This is in accordance with previous findings from our laboratory that CD33 is significantly up-regulated in CD34+ cells cultured with pomalidomide under different conditions, such as SCF+ erythropoietin or SCF+ IL-3+ IL-6/soluble IL-6 receptor fusion protein (hyper–IL-6).15 CD33 expression is detected on myeloblasts, promyelocytes, myelocytes, a proportion of metamyelocytes, and also monocytes. CD33 is largely absent from band forms and mature granulocytes, suggesting that terminal maturation was impaired in IMiD-treated cells and may account for myelosuppression in the clinical setting.16,21 Our findings that sequential bone marrow samples from patients treated with lenalidomide show a significant expansion in myeloid progenitors accompanied by an increase in promyelocytes and other immature myeloid precursors at the time of grade 4 neutropenia support this notion.
We further characterized the effects of IMiDs on transcriptional regulation of granulocytic maturation and found that PU.1 protein expression in hematopoietic progenitors was down-regulated in vitro in a dose-dependent manner by pomalidomide and lenalidomide. Immunohistochemical staining of bone marrow samples of patients treated with lenalidomide showed decreased PU.1 expression. PU.1 is known to be a critical transcription factor regulating granulopoiesis.22 PU.1-deficient hematopoietic progenitors show a perturbation of neutrophil maturation.22 Anderson et al verified that PU.1−/− mice develop neutrophils, which fail to attain functional competence.22 Dakic et al showed that PU.1−/− granulocytes, in contrast to wild-type controls, maintained c-kit expression and displayed some, but not complete, maturation as measured by morphologic criteria.23 These data indicate that PU.1 is not essential for specification of granulocytic precursors; however, it is required for terminal differentiation of these precursors into mature neutrophils. Down-regulation of PU.1 by IMiDs in hematopoietic stem cells therefore may underlie the medullary maturation impairment and consequent peripheral blood neutropenia. This is further supported by the findings that CD64+ neutrophils are increased in patients treated with pomalidomide despite peripheral blood neutropenia.24 CD64 is the high affinity receptor for IgG that is expressed on neutrophil precursors during early myelopoiesis but is down-regulated with terminal differentiation and is minimally detectable in healthy subjects.25
Our findings of induction of G-CSF in CD34+ cells treated with IMiDs (Figure 4A) are consistent with previous results generated with pomalidomide15 and may represent a self-regulatory mechanism intended to overcome the drug-induced maturation impairment. After 3 days of IMiD exposure, G-CSF concentration in culture supernatants increased from 140 pg/mL in controls, to 767 pg/mL and 1500 pg/mL after lenalidomide and pomalidomide treatment, respectively.13 Myeloid precursor cells express G-CSFR, and in response to stimulation by G-CSF, mature into neutrophilic granulocytes.26 In our experiments, expression of G-CSFR was stable or slightly up-regulated on hematopoietic cells treated with IMiDs, consistent with a persistent immature myeloid cell population.
In accordance with these results are findings from DeKoter et al who showed that PU.1−/− myeloid cells were not responsive to G-CSF–dependent differentiation of granulocytes.27
Gene expression analysis of hematopoietic progenitors treated with IMiDs revealed a striking up-regulation of the serine proteases CG, NE2, and proteinase3. CG and NE2 are major serine proteases contained in the granules of immature neutrophils.28 CG is a potent platelet agonist and triggers platelet aggregation similar to that observed with thrombin.29-31 Like thrombin, CG potently stimulates platelet aggregation through structurally related protease-activated receptors.32,33 Although NE2 does not activate platelets by itself,34-37 it significantly enhances the platelet aggregating property of CG, especially at lower concentrations.30 CG and NE2 can proteolytically cleave the integrin αIIbβ3, the mediator of platelet aggregation.38 This induces a conformational change, which leads to increased binding of fibrinogen to platelets and increased fibrin-induced cross-linking of platelets. Expression of CG is highest and restricted to an early stage of neutrophil differentiation during the promyelocyte stage and declines with terminal maturation.39-41 Levels of serine proteases therefore correlate with the number of promyelocytes and are highest in acute promyelocytic leukemia patients42 who have a high risk for intravascular disseminated coagulation.43 In our studies, CG was rapidly and highly up-regulated in vitro and in patients treated with IMiDs, suggesting an increased number of immature myeloid cells, further supporting our findings on the maturational arrest of myeloid cells by IMiDs. We observed an increase of the CG/absolute neutrophil count ratio over the time of treatment, suggesting that the relative higher amount of CG reflects more immature granulocytes during treatment with lenalidomide.
In addition, increased levels of CG might enhance the risk of VTE.9 In our studies, CG normalizes shortly after termination of treatment with a subsequent decreased VTE risk corresponding with the short period of time needed for the maturation of promyelocytes and normalization of CG levels[b]. Further, the fact that aspirin is effective in prevention of IMiD-induced VTE9 suggests that platelet activation is a major contributor to IMiD-induced VTE.
Interestingly, we found a report from Yamazaki and Aoki showing that CG released from granulocytes binds to human NK cells and caused enhancement of NK cytotoxicity through its protease activity.44 This is especially interesting in light of reports that increased NK-cell activity is one of the antimyeloma mechanisms of IMiDs.45,46
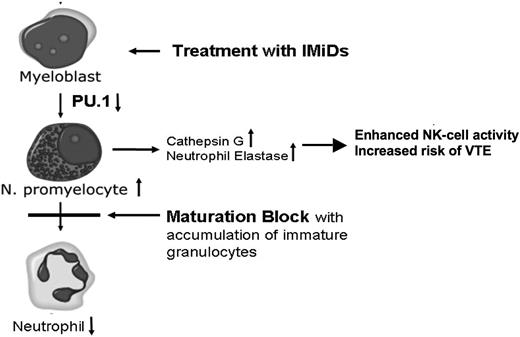
In conclusion, we provide evidence that IMiDs induce a dependent shift in lineage commitment toward myeloid lineage and impair neutrophil differentiation, arising at least in part from suppression of PU.1. Further, we postulate that IMiD-induced up-regulation of CG and NE2 contributes to the increased risk of VTE in patients treated with IMiDs (Figure 6).
Proposed mechanism of IMiD-induced CG up-regulation. Treatment of hematopoietic progenitors leads to a down-regulation of PU.1, which results in a maturational block of neutrophil granulocytes with peripheral blood neutropenia. The block of maturation leads to an accumulation of neutrophil promyelocytes. The azurophilic granules of promyelocytes store high amounts of serine proteases, such as CG and NE2. Subsequently, levels of CG and NE2 are up-regulated, contributing to the increased risk of VTE.
Proposed mechanism of IMiD-induced CG up-regulation. Treatment of hematopoietic progenitors leads to a down-regulation of PU.1, which results in a maturational block of neutrophil granulocytes with peripheral blood neutropenia. The block of maturation leads to an accumulation of neutrophil promyelocytes. The azurophilic granules of promyelocytes store high amounts of serine proteases, such as CG and NE2. Subsequently, levels of CG and NE2 are up-regulated, contributing to the increased risk of VTE.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Rita Bhutta for excellent assistance in preparing this manuscript.
This work was supported by Celgene Corporation.
Authorship
Contribution: R.P. performed research and wrote manuscript; S.A.M. performed and analyzed research; A.C.H. and M.V.R. performed and analyzed platelet aggregation studies; M.Y.M., P.S., and G.D.R. analyzed data and wrote manuscript; P.S. was involved in discussing and interpreting the data; L.M. and A.L. performed research; and S.L. designed and analyzed research and wrote the manuscript.
Conflict-of-interest disclosure: P.S. is an employee of the company that manufactures the tested drugs (Celgene Corporation). S.L. received support for this research from Celgene Corporation. The remaining authors declare no competing financial interests.
Correspondence: Suzanne Lentzsch, Division of Hematology/Oncology, University of Pittsburgh Cancer Institute, 5150 Centre Ave, #568, Pittsburgh, PA 15232; e-mail: lentzschs@upmc.edu.