Abstract
Acute myeloid leukemias (AMLs) result from multiple genetic alterations in hematopoietic stem cells. We describe a novel t(12;18)(p13;q12) involving ETV6 in a patient with AML. The translocation resulted in overexpression of SETBP1 (18q12), located close to the breakpoint. Overexpression of SETBP1 through retroviral insertion has been reported to confer growth advantage in hematopoietic progenitor cells. We show that SETBP1 overexpression protects SET from protease cleavage, increasing the amount of full-length SET protein and leading to the formation of a SETBP1–SET-PP2A complex that results in PP2A inhibition, promoting proliferation of the leukemic cells. The prevalence of SETBP1 overexpression in AML at diagnosis (n = 192) was 27.6% and was associated with unfavorable cytogenetic prognostic group, monosomy 7, and EVI1 overexpression (P < .01). Patients with SETBP1 overexpression had a significantly shorter overall survival, and the prognosis impact was remarkably poor in patients older than 60 years in both overall survival (P = .015) and event-free survival (P = .015). In summary, our data show a novel leukemogenic mechanism through SETBP1 overexpression; moreover, multivariate analysis confirms the negative prognostic impact of SETBP1 overexpression in AML, especially in elderly patients, where it could be used as a predictive factor in any future clinical trials with PP2A activators.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous clonal malignancy that predominantly affects middle-aged and elderly adults. The disease is characterized by a differentiation block in early progenitors, which leads to the accumulation of immature cells in bone marrow (BM) and peripheral blood. In recent years, several genetic markers with prognostic impact in AML have been identified, leading to a better understanding of the biology of this disease and, in some cases, providing targets for molecular therapies.1 Cytogenetic aberrations have been reported as the most important prognostic factors for survival and response to therapy in AML, allowing the identification of molecular markers that have greatly advanced our understanding of leukemogenesis2 ; nevertheless, the nature of alterations responsible for initiation or progression of the disease is mostly unknown. Recent attempts to identify initiating or progression mutations by extensively resequencing tyrosine kinase genes,3,4 expression profiling studies,5-7 array-based comparative genomic hybridization and/or single nucleotide polymorphism,8-11 and even by unbiased whole-genome sequencing,12 confirm that AML results from multiple genetic alterations in hematopoietic stem cells, and suggest that we have not yet discovered most of the relevant aberrations that contribute to the pathogenesis of this disease.
The ETV6 gene (12p13) encodes a transcription factor frequently rearranged in both myeloid and lymphoid leukemias. Translocation breakpoints are distributed throughout the gene, and ETV6 contributes to the pathogenesis of leukemia by diverse molecular mechanisms that are only partially understood. In most cases, the translocations result in the generation of in-frame fusion genes between different domains of ETV6 and partner genes encoding either kinases or transcription factors.13 However, in some cases involving the 5′ end of ETV6, functionally significant fusions could not be detected and a different leukemogenic mechanism has been described: the deregulation of the expression of oncogenes located close to the breakpoints.13,14 This molecular mechanism, which has been described mainly in lymphoid leukemias and lymphomas, is an uncommon mechanism in myeloid leukemias, although some examples have been reported.13,15
Here, we describe a novel leukemogenic mechanism in a patient with AML and a t(12;18)(p13;q12) involving ETV6. The translocation resulted in overexpression of SETBP1 (18q12), located close to the breakpoint. We show that SETBP1 overexpression protects SET from protease cleavage, increasing the amount of full-length SET protein and leading to the formation of a SETBP1–SET-PP2A complex that results in PP2A inhibition and therefore promotes the proliferation of leukemic cells. Moreover, we show that SETBP1 overexpression is a recurrent molecular event associated with a significantly shorter overall survival (OS) in AML, especially in elderly patients.
Methods
Case reports
A 76-year-old white man was diagnosed with myelodysplastic syndrome. Disease evaluation of the patient 3 years after the diagnosis showed anorexia, perspiration, and loss of 7 kg. Laboratory findings at this moment were: hemoglobin 10.9 g/dL, white blood cell count 38.2 × 109/L with 55% blasts, and platelet count 251 × 109/L; BM aspirate was hypercellular, showing 80% blasts. Flow cytometry showed positivity for CD34, CD13, CD33, CD11b, and HLA-DR. The patient was diagnosed as AML-M5. Karyotype showed 2 clones: 46,XY,t(12,18)(p13;q12)[77%]/47,idem,+19[23%]. The patient received standard induction chemotherapy for 2 months and had partial remission at the next evaluation. Laboratory findings showed hemoglobin 9.9 g/dL, white blood cell count 25.5 × 109/L with 32% blasts, and 260 × 109/L platelets. Karyotype at this time was: 46,XY,t(12,18)(p13;q12)[24%]/46,XY[76%]. The patient relapsed 2 months later and eventually died. Karyotype at relapse showed the 2 clones detected at diagnosis: 46,XY,t(12,18)(p13;q12)[32%]/47,idem,+19[68%]. All samples were taken anonymously, and the study was approved by the Institutional Committee of the University of Navarra.
FISH
Bacterial artificial chromosomes obtained from the Roswell Park Cancer Institute (Buffalo, NY) were used to map the breakpoints in the patient samples. The order of the probes is centromere-418C2-96B19-434C1-telomere (12p13), and centromere-840B16-937P23-252G8-941F5-telomere (18q12). Probes were labeled with SpectrumGreen-dUTP or SpectrumOrange-dUTP. Centromeric probes for chromosomes 12 and 18 were also used in fluorescence in situ hybridization (FISH) experiments. FISH was performed as previously described.14
RACE
3′- and 5′-rapid amplification of cDNA ends (RACE)–polymerase chain reaction (PCR) were performed from total RNA using the GeneRacer kit (Invitrogen). 3′RACE-PCR (94°C 30 seconds, 72°C 2 minutes 30 seconds, 5 cycles; 94°C 30 seconds, 70°C 2 minutes 30 seconds, 5 cycles; 94°C 30 seconds, 64°C 30 seconds, 68°C 2 minutes 30 seconds, 25 cycles) was performed using the primer ETV6-EX1F (exon 1) and a reverse primer from the kit. Nested PCR was performed under the same conditions with ETV6-EX1Fn (exon 1). Products were cloned and sequenced. For 5′RACE, the same PCR conditions were used, with primers ETV6-EX3R and ETV6-EX3Rn (exon 3; supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Nucleic acid isolation and RT-PCR
Total DNA was isolated using the QIAmp DNA minikit, and total RNA using the RNeasy minikit (QIAGEN). cDNA was synthesized with SuperScriptIII Reverse Transcriptase (Invitrogen). Reverse-transcribed (RT)–PCR reactions were carried out with Ampli-Taq Gold DNA Polymerase (Applied Biosystems) after optimizing cycling conditions for each primer pair. To confirm the presence of the fusion products, RT-PCR reactions were performed on patient RNA with primers ETV6-EX1F2, and CR18-B and CR18-E (supplemental Table 1). Products were cloned and sequenced.
Real-time RT-PCR
Quantification of the expression of SETBP1, SET, and ETV6 was performed using TaqMan Gene Expression Assays (Applied Biosystems) specific for each gene. GAPDH was used as internal control. A gene was considered overexpressed if its expression value was higher than the cut-off value established for each gene (mean ± 5 SD), defined by the analysis of 10 normal BM samples.
Cell culture and transfection
HEL, K562, 32Dcl3, and HEK293 cell lines were grown at 37°C in a 5% CO2 atmosphere. HEK293 cells were maintained in Dulbecco's minimum essential medium, HEL and K562 in RPMI-1640 (Invitrogen), and 32Dcl3 in RPMI 1640 + 10% conditioned medium of cell line WEHI-3B (DSM ACC 26). Media were supplemented with 10% fetal bovine serum, penicillin G (100 U/mL), and streptomycin (0.1 mg/mL). For transfection experiments, HEK293 cells were seeded into 10-cm dishes and transfected with 60 μL of Lipofectamine2000 (Invitrogen) and 12 μg of the plasmids expressing SETBP1(SKI homologous region [SHR])–V5, SETBP1-GFP, SETBP1(SETBD)–GFP, SET-GFP, or empty vector as control. HEL and 32Dcl3 cells were seeded in culture flasks and transfected using the Nucleofector System (solution V; protocol X-005 for HEL, and E-032 for 32Dcl3; Amaxa), with 4 μg of plasmidic vectors or 75nM SET siRNA pool designed and synthesized by Dharmacon RNA Technologies.
Plasmids
Human SET cDNA was obtained by RT-PCR from K562 RNA using an upstream primer containing an EcoRI site followed by the first 19 nucleotides of SET cDNA, and a downstream primer containing the last 21 nucleotides of SET linked to a BamHI site. The EcoRI/BamHI digested PCR product was subcloned into the pEGFP-C2 vector leading to the pEGFPC2-SET construct. Human SETBP1 cDNA was obtained by RT-PCR from peripheral blood and subcloned into the vector pEGFP-C2 through XhoI/SalI sites, resulting in the pEGFPC2-SETBP1 construct. The region from amino acids 1167 to the end of SETBP1 was obtained by digestion from the pEGFPC2-SETBP1 construct and subcloned into the vector pEGFP-C2 through HindIII/SacII sites, resulting in the pEGFPC2-SETBP1(SETBD) construct. The region from amino acids 449 to amino acids 857 of SETBP1 was obtained by PCR from the pEGFPC2-SETBP1 and subcloned into the pcDNA3.1V5/His vector, leading to the pcDNA3.1V5/His-SETBP1(SHR). All cloning procedures were verified by sequencing.
Immunoprecipitation and Western blotting
Cells were lysed in 100 μL of lysis buffer containing 1% Triton X-100 and protease inhibitors (Complete Mini; Roche Diagnostics). After incubation on ice (30 minutes), protein extracts were clarified (12 000g, 15 minutes, 4°C), denatured, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot. Antibodies used were goat polyclonal anti-SET (Santa Cruz Biotechnology), mouse monoclonal anti-PP2A (clone 1D6, Upstate), rabbit monoclonal anti-PP2AY307 (Epitomics), rabbit polyclonal anti-GFP (Santa Cruz Biotechnology), and mouse monoclonal anti-βtubulin (Sigma-Aldrich). For immunoprecipitation, lysates were precleared (1 hour, 4°C) on a rotating wheel and immunoprecipitated with antibody-bound protein G-Sepharose (8 hours, 4°C). After washings, immunoprecipitations were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot. Proteins were detected with the appropriate secondary antibodies by chemiluminescence (ECL kit; GE Healthcare).
Proliferation assay, viability, and total cell counts
Cell proliferation was measured in triplicate wells by 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay in 96-well plates using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega). Cell viability and total counts were measured in triplicate with the Nucleocounter (Biogen), and the results confirmed by Trypan Blue method.
PP2A assays
PP2A assays were performed with cell lysates (50 μg) using a PP2A immunoprecipitation phosphatase assay kit (Millipore) as previously described,16 except that we added the protease inhibitor cocktail Complete (Roche Diagnostics) to the protein extraction mix.
Statistical analysis
Statistical analyses were performed using SPSS 15 for windows (SPSS Inc). OS was defined as the time from diagnosis to death of any cause or end of follow-up. Disease-free survival (DFS) was defined as the time from complete remission until relapse or death. Event-free survival (EFS) was defined as the time from diagnosis until first event, in which failure to achieve complete remission, relapse, death, or end of follow-up were considered events. OS, DFS, and EFS were determined according to the Kaplan-Meier method and survival comparisons were done with the log-rank test if proportional hazard assumption was fulfilled, and Breslow otherwise. The Cox proportional hazards model was used to assess patient outcome for patient-stratified groups of age. The hazard model was adjusted taking into consideration relevant parameters that included cytogenetic group, complete remission, and SETBP1 overexpression. Good risk and intermediate risk were analyzed together because of the small number of patients with good-risk cytogenetics. A P value less than .05 was considered statistically significant.
Bioinformatics analysis of the SETBP1 proximal promoter
Analysis of the proximal promoter of SETBP1 was performed with MotifScanner,17-19 which scans DNA sequences with precompiled motif models. This algorithm assumes that motifs are hidden in a noisy background sequence represented by a higher-order Markov model. The motif models of transcriptional factors were obtained from the public version of Jaspar and Transfac databases. The proximal promoter of SETBP1 was defined as 3000 bp upstream the transcription start site and was extracted from Ensembl database release 53.
Results
SETBP1 (18q21) is overexpressed in a patient with a translocation t(12;18) involving ETV6
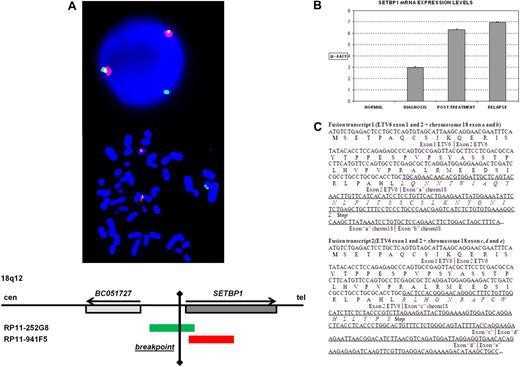
Karyotype at diagnosis of a patient with AML-M5 secondary to myelodysplastic syndrome was 46,XY,t(12,18)(p13;q12)/47,idem,+19. FISH showed that the breakpoint on 12p13 was located between exons 2 and 3 of ETV6 (supplemental Figure 1). To identify the fusion partner of ETV6, RACE-PCR experiments were performed on RNA from BM of the patient. 3′RACE-PCR identified 2 clones containing ETV6 exon 2 followed by sequences that overlapped with the human cDNA clone BC051727 (Mammalian Gene Collection Program Team) located in chromosome 18 (Figure 1). Alignment of these sequences to the human genome did not reveal the presence of a known gene, but the novel sequences were split into segments that were flanked by consensus splice donor and acceptor sites. This indicated that these sequences could be part of a novel gene, whose exact identity and complete cDNA sequence were not further analyzed. Because the complete transcript is currently unknown, the different exons identified in this sequence were arbitrarily named as follows: fusion transcript 1, ETV6-exon-1 + ETV6-exon-2 + exon-“a” + exon-“b”; fusion transcript 2, ETV6-exon-1 + ETV6-exon-2 + exon-“c” + exon-“d” + exon-“e.” Stop codons were found in the 3 different reading frames for exons “b” and “e,” indicating that these exons are part of a noncoding transcript. The presence of these 2 fusions was confirmed by RT-PCR, cloned, and sequenced (data not shown). Fusion transcripts predicted, in both cases, truncated proteins resulting from the presence of premature stop codons (Figure 1). Besides, the predicted new proteins showed no homology with any known proteins. In these novel sequences, we found no microRNAs that could be deregulated as a consequence of the translocation. 5′RACE-PCR failed to find any reciprocal fusion transcript. Taken together, these data suggest that no functionally significant fusion transcripts were generated by the translocation.
Genetic characterization of a patient with AML-M5 and a t(12;18)(p13;q12) involving ETV6. (A) FISH analysis indicating the breakpoint on 18q12: probe RP11-252G8 (green) splits and hybridizes in both der(18) and der(12). (B) Analysis by quantitative RT-PCR of the SETBP1 expression in samples of the patient at diagnosis, posttreatment control, and relapse. (C) RACE results showing the 2 different fusion transcripts detected. Sequences from ETV6 are in italics; the putative exons from chromosome 18 are underlined.
Genetic characterization of a patient with AML-M5 and a t(12;18)(p13;q12) involving ETV6. (A) FISH analysis indicating the breakpoint on 18q12: probe RP11-252G8 (green) splits and hybridizes in both der(18) and der(12). (B) Analysis by quantitative RT-PCR of the SETBP1 expression in samples of the patient at diagnosis, posttreatment control, and relapse. (C) RACE results showing the 2 different fusion transcripts detected. Sequences from ETV6 are in italics; the putative exons from chromosome 18 are underlined.
To confirm the position of the breakpoint on chromosome 18, bacterial artificial chromosomes located at 18q12 were used as probes in FISH experiments. Analysis on BM cells of the patient showed that one signal hybridized to the normal chromosome 18, and the other split and hybridized to both der(18) and der(12) (Figure 1). FISH showed that the breakpoint was located 5′ and close to the SETBP1 gene. Activation of SETBP1 expression by retroviral integration in hematopoietic progenitor cells has been reported to confer a growth advantage leading to clonal expansion.20 Knowing that SETBP1 was close to the breakpoint in the t(12;18)(p13;q12) and taking into account that ectopic expression of oncogenes is a mechanism involved in leukemia, we analyzed the expression of SETBP1 by real-time PCR (quantitative RT-PCR) in this patient, and we found that SETBP1 was overexpressed at diagnosis and in the posttreatment samples (Figure 1).
Ectopic expression of SETBP1 leads to increased full-length SET protein levels
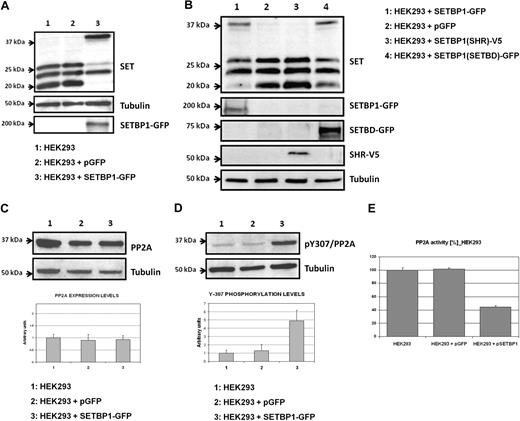
SETBP1 has been reported to specifically interact with the protein SET,21 a potent inhibitor of protein phosphatase 2A (PP2A). To assess whether the ectopic expression of SETBP1 affects SET protein levels, HEK293 cells were transiently transfected with SETBP1-GFP. We found that levels of the 39-kDa full-length form of SET were strongly induced; moreover, an important decrease in the levels of low molecular weight forms of SET was observed. As controls, ectopic expression of the SETBP1-GFP protein was detected by anti-GFP (Figure 2A) and quantitative RT-PCR (supplemental Figure 2A). Analysis by fluorescence microscopy of SETBP1-GFP–transfected HEK293 cells confirmed a predominantly nuclear location of SETBP1 (supplemental Figure 2B).
Ectopic expression of SETBP1 induces increased full-length SET levels and inhibits PP2Ac. (A) Western blot showing endogenous SET expression in HEK293 cells transfected with SETBP1-GFP, an empty vector as a control, or nontransfected cells. (B) Analysis by Western blot showing SET in HEK293 cells transfected with different regions of SETBP1. SHR indicates SKI homologous region; and SETBD, SET binding domain. (C) Western blot and densitometry analysis showing the effect of ectopic SETBP1 over PP2A expression in HEK293 cells. (D) Western blot and densitometry analysis showing the effect of ectopic SETBP1 over PP2A phosphorylation levels of the tyrosine-307. (E) PP2A phosphatase assay in HEK293 cells transfected or not with SETBP1.
Ectopic expression of SETBP1 induces increased full-length SET levels and inhibits PP2Ac. (A) Western blot showing endogenous SET expression in HEK293 cells transfected with SETBP1-GFP, an empty vector as a control, or nontransfected cells. (B) Analysis by Western blot showing SET in HEK293 cells transfected with different regions of SETBP1. SHR indicates SKI homologous region; and SETBD, SET binding domain. (C) Western blot and densitometry analysis showing the effect of ectopic SETBP1 over PP2A expression in HEK293 cells. (D) Western blot and densitometry analysis showing the effect of ectopic SETBP1 over PP2A phosphorylation levels of the tyrosine-307. (E) PP2A phosphatase assay in HEK293 cells transfected or not with SETBP1.
To assess the origin of the proteins of different sizes detected by anti-SET antibodies, HEK293 cells were transfected with SET-GFP and harvested at different times. Full-length SET-GFP protein of approximately 67 kDa could be detected by Western blot 9 hours after transfection, and 2 additional bands of lower molecular weight appeared gradually, together with the full-length form (supplemental Figure 2C). Taking into account the molecular weight of GFP, these bands would correspond to the endogenous short SET forms (Figure 2A). Because SET-GFP is expressed from a cDNA sequence, and alternative splicing cannot take place in the absence of intronic sequences, these results suggest that the endogenous SET protein of 39 kDa is cleaved by proteases, resulting in truncated proteins with distinct sizes.
To analyze the region of SETBP1 that is critical to induce this effect, we transfected HEK293 cells with the vector pcDNA3.1-SETBP1(SHR)-V5, which expresses an internal region of SETBP1 that includes the SHR, and with the vector pEGFPC2-SETBP1(SETBD), expressing the carboxy-terminal region of SETBP1 that includes the SET binding domain (SETBD). We used HEK293 cells transfected with SETBP1-GFP as positive control, with the empty vector as negative control. We found similar results transfecting with SETBP1-GFP and with SETBD-GFP; however, no effects were observed with SETBP1(SHR)-V5, indicating that the region containing SETBD is critical (Figure 2B). Taken together, these results suggest that SETBP1 has a major effect on the 39-kDa form of SET, probably increasing its levels by impairing protease activity on the full-length SET protein.
SETBP1 inhibits PP2A activity through tyrosine phosphorylation of its catalytic subunit
We next investigated the effect of SETBP1 overexpression on the PP2A protein. Expression levels of the catalytic subunit of PP2A (PP2Ac) were not affected in HEK293 cells transfected with SETBP1-GFP, with respect to nontransfected cells (Figure 2C). However, an increase in the phosphorylation of tyrosine-307 was observed (Figure 2D). Of note, phosphorylation of tyrosine-307 is responsible for more than 90% of the phosphatase activity of this protein; indeed, it has been shown that PP2Ac is inactive when tyrosine-307 is phosphorylated.22 These results suggest that ectopic expression of SETBP1 leads to a reduced PP2A activity, which was confirmed by a PP2A phosphatase assay (Figure 2E).
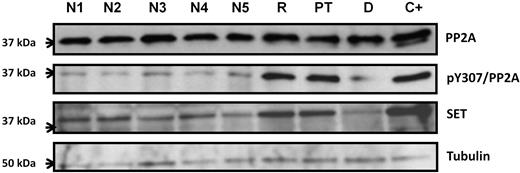
We next assessed whether the observations made in SETBP1-transfected cells could be confirmed in the patient samples. As a positive control for PP2A inactivation and SET overexpression, protein extracts from K562 cells were included.23 The patient showed higher phosphorylation of tyrosine-307 of PP2Ac in both relapse and posttreatment samples, compared with normal donors. We also observed increased SET protein levels, with good correlation between protein and mRNA (Figure 3; supplemental Figure 3).
Comparison of the PP2A and SET protein levels by Western blot between patient samples and normal donors. Phosphorylation on tyrosine-307 of PP2A is also assessed. The K562 cell line was used as a positive control for SET overexpression and hyperphosphorylation of Y-307 of PP2A. N1-N5 indicates normal controls; R, patient sample at relapse; PT, patient sample after treatment; D, patient sample at diagnosis; and C+, K562 cells as positive control.
Comparison of the PP2A and SET protein levels by Western blot between patient samples and normal donors. Phosphorylation on tyrosine-307 of PP2A is also assessed. The K562 cell line was used as a positive control for SET overexpression and hyperphosphorylation of Y-307 of PP2A. N1-N5 indicates normal controls; R, patient sample at relapse; PT, patient sample after treatment; D, patient sample at diagnosis; and C+, K562 cells as positive control.
SETBP1 induces proliferation in HEL and 32Dcl3
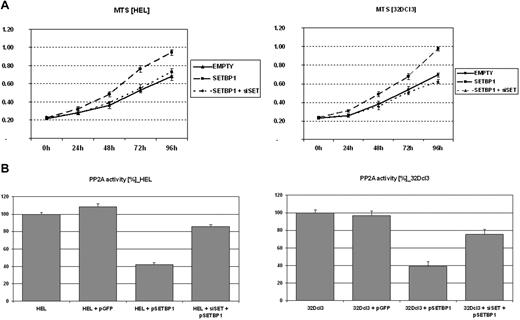
To investigate the effect of SETBP1 on cell growth, the HEL cell line (AML-M6) was chosen as a cellular model. An increased proliferation in cells transfected with SETBP1 compared with mock-transfected cells was observed using the MTS assay (Figure 4A); furthermore, total cell counts and cell viability were determined by Nucleo Counter and confirmed with the Trypan Blue method (data not shown). These results confirmed the higher cell counts and better viability of cells transfected with SETBP1 compared with cells transfected with the empty vector. Similar results were obtained with the 32Dcl3 cell line (Figure 4A). In addition, we observed reduced PP2A activity in both HEL and 32Dcl3 cells transfected with SETBP1.
Effect of SETBP1 on cell proliferation and PP2A activity. (A) MTS assays showing the effect of ectopic SETBP1 expression alone or with SET-specific siRNAs on the growth of HEL and 32Dcl3. (B) PP2A phosphatase assay in HEL and 32Dcl3 cells either not transfected or expressing the empty vector as a control, SETBP1 alone, or SETBP1 with SET down-regulated by SET-specific siRNAs.
Effect of SETBP1 on cell proliferation and PP2A activity. (A) MTS assays showing the effect of ectopic SETBP1 expression alone or with SET-specific siRNAs on the growth of HEL and 32Dcl3. (B) PP2A phosphatase assay in HEL and 32Dcl3 cells either not transfected or expressing the empty vector as a control, SETBP1 alone, or SETBP1 with SET down-regulated by SET-specific siRNAs.
To confirm our hypothesis that SETBP1 inhibits PP2A through SET, SET was down-regulated by SET-specific siRNAs (supplemental Figure 4). We observed that the effects of SETBP1 on cell proliferation and PP2A activity were impaired in both HEL and 32Dcl3 cells after SET knockdown (Figure 4B).
SETBP1 forms a complex including SET and PP2A
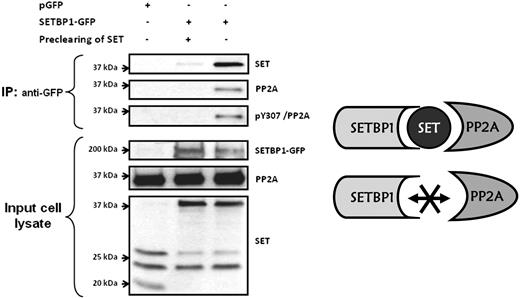
It has been reported that SETBP1 interacts with SET and that the region of SET that binds to SETBP1 is different from the PP2A inhibitory region. Accordingly, it was postulated that SETBP1-SET heterodimers interact with PP2A.21 However, no experimental evidence to confirm this hypothesis has been provided yet. To determine whether SET interacts simultaneously with SETBP1 and PP2A, coimmunoprecipitation assays were carried out in HEK293 cells, transfected or not with SETBP1-GFP. PP2A and SET coimmunoprecipitated with SETBP1, suggesting the existence of a SETBP1–SET-PP2A complex. Preclearing of SET using anti-SET antibodies bound to Sepharose-protein G inhibited the coimmunoprecipitation of PP2A and SET with SETBP1, indicating that PP2A interacts with the SETBP1-SET heterodimer through SET and that SETBP1 and PP2A do not directly interact with each other. Interestingly, we found that the coimmunoprecipitated PP2A was phosphorylated on its tyrosine-307 (Figure 5). Taken together, these results suggest that the SETBP1-SET heterodimer interacts with PP2A forming a SETBP1–SET-PP2A complex in which the presence of SET is critical.
PP2A interacts with SET-SETBP1 heterodimer but not directly with SETBP1. Cell extracts expressing SETBP1-GFP (lanes 2 and 3) were immunoprecipitated with anti-GFP followed by Western blotting with anti-SET and anti-PP2A. Cell extract without SETBP1-GFP expression was used as negative control (lane 1). One of the cell extracts expressing SETBP1-GFP was precleared of SET using anti-SET antibodies bound to Sepharose-protein G before the immunoprecipitation (lane 2).
PP2A interacts with SET-SETBP1 heterodimer but not directly with SETBP1. Cell extracts expressing SETBP1-GFP (lanes 2 and 3) were immunoprecipitated with anti-GFP followed by Western blotting with anti-SET and anti-PP2A. Cell extract without SETBP1-GFP expression was used as negative control (lane 1). One of the cell extracts expressing SETBP1-GFP was precleared of SET using anti-SET antibodies bound to Sepharose-protein G before the immunoprecipitation (lane 2).
Prevalence of SETBP1 overexpression in AML
To study the prevalence of SETBP1 overexpression and its prognostic value in AML, we quantified the expression of SETBP1 in a series of 192 patients with AML at diagnosis, correlated these values with the French-American-British classification, and cytogenetic and molecular markers, and studied the prognostic relevance of this aberration. Patient characteristics are presented in Table 1, and box-plots show the SETBP1 expression levels in supplemental Figure 5. Although patients were treated with different schedules, all received regimens based on anthracycline and cytarabine as induction therapy. High-dose cytarabine, and autologous or allogenic stem cell transplantation when possible, were used as consolidation therapy. All patients provided informed consent. SETBP1 was overexpressed in 53 of 192 patients (27.6%). The prevalence in cytogenetic prognostic groups was as follows: good, 24% (7 of 29); intermediate, 18.5% (17 of 92); and poor, 40.8% (29 of 71). We found genetic aberrations associated with SETBP1 overexpression: monosomy 7 (P = .007), chromosome 18 aberrations (P = .031), and EVI1 overexpression (P = .001). An inverse correlation was observed between SETBP1 overexpression and normal karyotype (P = .010), and NPM1 mutations in patients with wild-type FLT3 (P = .045; Table 2).
Clinical and molecular characteristics of a series of 192 patients with AML
| Characteristic . | No. (%) . |
|---|---|
| Sex | |
| Male | 99 (53) |
| Female | 88 (47) |
| No data | 5 |
| Age | |
| Less than 60 years | 92 (50.3) |
| More than 60 years | 91 (49.7) |
| No data | 9 |
| Complete remission | |
| No | 47 (36.7) |
| Yes | 81 (63.3) |
| No data | 64 |
| Diagnosis | |
| AML-M0 | 21 (10.9) |
| AML-M1 | 35 (18.2) |
| AML-M2 | 38 (19.8) |
| AML-M3 | 7 (3.6) |
| AML-M4 | 33 (17.2) |
| AML-M5 | 36 (18.8) |
| AML-M6 | 12 (6.3) |
| AML-NOS | 10 (5.2) |
| Secondary AML | |
| No | 130 (83.3) |
| Yes | 26 (16.7) |
| No data | 36 |
| Cytogenetic group | |
| Good | 29 (15) |
| Intermediate | 92 (48) |
| Poor | 71 (37) |
| SETBP1 overexpression | |
| No | 139 (72.4) |
| Yes | 53 (27.6) |
| WT1 overexpression | |
| No | 22 (16) |
| Yes | 115 (84) |
| No data | 55 |
| EVI1 overexpression | |
| No | 132 (76.7) |
| Yes | 40 (23.3) |
| No data | 20 |
| NPM1 mutated | |
| No | 18 (37.5) |
| Yes | 30 (62.5) |
| No data | 144 |
| FLT3-ITD | |
| No | 99 (79.2) |
| Yes | 26 (20.8) |
| No data | 67 |
| Characteristic . | No. (%) . |
|---|---|
| Sex | |
| Male | 99 (53) |
| Female | 88 (47) |
| No data | 5 |
| Age | |
| Less than 60 years | 92 (50.3) |
| More than 60 years | 91 (49.7) |
| No data | 9 |
| Complete remission | |
| No | 47 (36.7) |
| Yes | 81 (63.3) |
| No data | 64 |
| Diagnosis | |
| AML-M0 | 21 (10.9) |
| AML-M1 | 35 (18.2) |
| AML-M2 | 38 (19.8) |
| AML-M3 | 7 (3.6) |
| AML-M4 | 33 (17.2) |
| AML-M5 | 36 (18.8) |
| AML-M6 | 12 (6.3) |
| AML-NOS | 10 (5.2) |
| Secondary AML | |
| No | 130 (83.3) |
| Yes | 26 (16.7) |
| No data | 36 |
| Cytogenetic group | |
| Good | 29 (15) |
| Intermediate | 92 (48) |
| Poor | 71 (37) |
| SETBP1 overexpression | |
| No | 139 (72.4) |
| Yes | 53 (27.6) |
| WT1 overexpression | |
| No | 22 (16) |
| Yes | 115 (84) |
| No data | 55 |
| EVI1 overexpression | |
| No | 132 (76.7) |
| Yes | 40 (23.3) |
| No data | 20 |
| NPM1 mutated | |
| No | 18 (37.5) |
| Yes | 30 (62.5) |
| No data | 144 |
| FLT3-ITD | |
| No | 99 (79.2) |
| Yes | 26 (20.8) |
| No data | 67 |
AML indicates acute myeloid leukemia.
Association between SETBP1 overexpression and clinical and genetic parameters in 192 patients with AML at diagnosis
| . | No. of cases . | SETBP1−, no. (%) . | No. SETBP1+, no. (%) . | P . |
|---|---|---|---|---|
| SETBP1 | 192 | 139 (72.4) | 53 (27.6) | |
| Sex | 187 | 136 | 51 | .511 |
| Male | 99 | 70 (70.7) | 29 (29.3) | |
| Female | 88 | 66 (75) | 22 (25) | |
| Age | 183 | 136 | 47 | .219 |
| Less than 60 years | 92 | 72 (78.3) | 20 (21.7) | |
| More than 60 years | 91 | 64 (70.3) | 27 (29.7) | |
| Complete remission | 128 | 180 | 50 | .196 |
| No | 47 | 33 (70.2) | 14 (29.8) | |
| Yes | 81 | 65 (80.2) | 16 (19.8) | |
| Secondary AML | 156 | 247 | 56 | .354 |
| No | 130 | 101 (77.7) | 29 (22.3) | |
| Yes | 26 | 18 (69.2) | 8 (30.8) | |
| Prognostic group | 192 | 139 | 53 | .006* |
| Good | 29 | 22 (76) | 7 (24) | |
| Intermediate | 92 | 75 (82.5) | 17 (18.5) | |
| Poor | 71 | 42 (59.2) | 29 (40.8) | |
| Cytogenetic group | 192 | 139 | 53 | |
| Normal karyotype | .010* | |||
| Yes | 63 | 53 (84) | 10 (16) | |
| No | 128 | 85 (66.4) | 43 (33.6) | |
| Trisomy 8 | .943 | |||
| Yes | 18 | 13 (72.2) | 5 (27.8) | |
| No | 163 | 119 (73) | 44 (27) | |
| Chromosome 18 aberrations | .031* | |||
| Yes | 11 | 5 (45.4) | 6 (54.6) | |
| No | 169 | 127 (75) | 42 (25) | |
| Monosomy 7 | .007* | |||
| Yes | 27 | 14 (51.9) | 13 (48.1) | |
| No | 155 | 119 (76.8) | 36 (23.2) | |
| der(7q) | .318 | |||
| Yes | 16 | 10 (62.5) | 6 (37.5) | |
| No | 166 | 123 (74.1) | 43 (25.9) | |
| WT1 overexpression | 137 | 108 | 29 | .345 |
| No | 22 | 19 (86.7) | 3 (13.6) | |
| Yes | 115 | 89 (77.4) | 26 (22.6) | |
| EVI1 overexpression | 172 | 126 | 46 | .001* |
| No | 132 | 108 (81.8) | 24 (18.2) | |
| Yes | 40 | 18 (45) | 22 (55) | |
| MDS1EVI1 overexpression | 152 | 109 | 43 | .012* |
| No | 138 | 103 (74.6) | 35 (25.4) | |
| Yes | 14 | 6 (42.9) | 8 (57.1) | |
| NPM1 mutated and FLT3 wt | 32 | 28 | 4 | .014* |
| No | 12 | 8 (66.7) | 4 (33.3) | |
| Yes | 20 | 20 (100) | 0 (0.0) | |
| FLT3-ITD | 125 | 100 | 25 | .760 |
| No | 99 | 78 (78.8) | 21 (21.2) | |
| Yes | 26 | 22 (84.6) | 4 (15.4) |
| . | No. of cases . | SETBP1−, no. (%) . | No. SETBP1+, no. (%) . | P . |
|---|---|---|---|---|
| SETBP1 | 192 | 139 (72.4) | 53 (27.6) | |
| Sex | 187 | 136 | 51 | .511 |
| Male | 99 | 70 (70.7) | 29 (29.3) | |
| Female | 88 | 66 (75) | 22 (25) | |
| Age | 183 | 136 | 47 | .219 |
| Less than 60 years | 92 | 72 (78.3) | 20 (21.7) | |
| More than 60 years | 91 | 64 (70.3) | 27 (29.7) | |
| Complete remission | 128 | 180 | 50 | .196 |
| No | 47 | 33 (70.2) | 14 (29.8) | |
| Yes | 81 | 65 (80.2) | 16 (19.8) | |
| Secondary AML | 156 | 247 | 56 | .354 |
| No | 130 | 101 (77.7) | 29 (22.3) | |
| Yes | 26 | 18 (69.2) | 8 (30.8) | |
| Prognostic group | 192 | 139 | 53 | .006* |
| Good | 29 | 22 (76) | 7 (24) | |
| Intermediate | 92 | 75 (82.5) | 17 (18.5) | |
| Poor | 71 | 42 (59.2) | 29 (40.8) | |
| Cytogenetic group | 192 | 139 | 53 | |
| Normal karyotype | .010* | |||
| Yes | 63 | 53 (84) | 10 (16) | |
| No | 128 | 85 (66.4) | 43 (33.6) | |
| Trisomy 8 | .943 | |||
| Yes | 18 | 13 (72.2) | 5 (27.8) | |
| No | 163 | 119 (73) | 44 (27) | |
| Chromosome 18 aberrations | .031* | |||
| Yes | 11 | 5 (45.4) | 6 (54.6) | |
| No | 169 | 127 (75) | 42 (25) | |
| Monosomy 7 | .007* | |||
| Yes | 27 | 14 (51.9) | 13 (48.1) | |
| No | 155 | 119 (76.8) | 36 (23.2) | |
| der(7q) | .318 | |||
| Yes | 16 | 10 (62.5) | 6 (37.5) | |
| No | 166 | 123 (74.1) | 43 (25.9) | |
| WT1 overexpression | 137 | 108 | 29 | .345 |
| No | 22 | 19 (86.7) | 3 (13.6) | |
| Yes | 115 | 89 (77.4) | 26 (22.6) | |
| EVI1 overexpression | 172 | 126 | 46 | .001* |
| No | 132 | 108 (81.8) | 24 (18.2) | |
| Yes | 40 | 18 (45) | 22 (55) | |
| MDS1EVI1 overexpression | 152 | 109 | 43 | .012* |
| No | 138 | 103 (74.6) | 35 (25.4) | |
| Yes | 14 | 6 (42.9) | 8 (57.1) | |
| NPM1 mutated and FLT3 wt | 32 | 28 | 4 | .014* |
| No | 12 | 8 (66.7) | 4 (33.3) | |
| Yes | 20 | 20 (100) | 0 (0.0) | |
| FLT3-ITD | 125 | 100 | 25 | .760 |
| No | 99 | 78 (78.8) | 21 (21.2) | |
| Yes | 26 | 22 (84.6) | 4 (15.4) |
AML indicates acute myeloid leukemia.
Significant values.
Prognostic impact of SETBP1 overexpression in AML
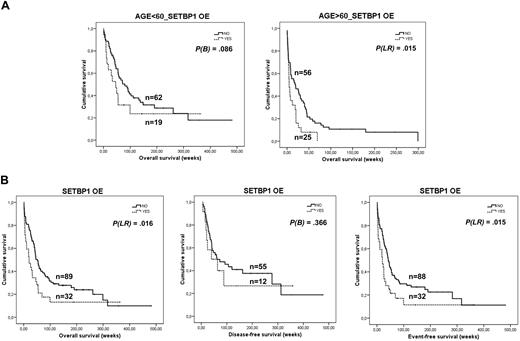
Clinical follow-up data were available for 168 patients (supplemental Table 2), 91 men and 77 women, with a median age of 56 years. Median OS of the global cohort was 31.7 (95% confidence interval, 21.5-41.9 weeks; supplemental Figure 5). As expected, significant differences in OS according to age, cytogenetic group, and complete remission rate were found in this series (P < .01). In addition, we found significant differences in OS between patients with and without SETBP1 overexpression (P < .01; supplemental Figure 6). Interestingly, the prognostic impact of SETBP1 overexpression was particularly evident in patients older than 60 years (Figure 6A). Univariate analysis showed that only complete remission (P < .01) and SETBP1 overexpression (P = .015), but not cytogenetic group (P = .059), were significant in OS in the group of patients older than 60 years. Multivariate analysis demonstrated that SETBP1 overexpression was an unfavorable independent factor associated with OS in elderly patients with AML (Table 3).
SETBP1 overexpression and cumulative survival in AML. (A) Kaplan-Meier analyses of OS for SETBP1 in the series of age-stratified 168 patients with AML, and clinical follow-up data available. Patients older than 60 years with SETBP1 overexpression showed a poorer outcome compared with patients with no SETBP1 overexpression. (B) Kaplan-Meier analyses of OS, DFS, and EFS for SETBP1 overexpression in a series of 121 patients with AML and clinical follow-up data available who received induction therapy (note that this group of 121 patients is included in the previous series of 168 patients in panel A).
SETBP1 overexpression and cumulative survival in AML. (A) Kaplan-Meier analyses of OS for SETBP1 in the series of age-stratified 168 patients with AML, and clinical follow-up data available. Patients older than 60 years with SETBP1 overexpression showed a poorer outcome compared with patients with no SETBP1 overexpression. (B) Kaplan-Meier analyses of OS, DFS, and EFS for SETBP1 overexpression in a series of 121 patients with AML and clinical follow-up data available who received induction therapy (note that this group of 121 patients is included in the previous series of 168 patients in panel A).
Multivariate analysis of clinical and biologic variables in the group of patients with AML older than 60 years
| Variable . | Subset . | Hazard ratio (95% confidence interval) . | P . |
|---|---|---|---|
| Complete remission | No/yes | 1/0.218 (0.110-0.430) | .001 |
| SETBP1 overexpression | No/yes | 1/2.311 (1.173-4.553) | .015 |
| Variable . | Subset . | Hazard ratio (95% confidence interval) . | P . |
|---|---|---|---|
| Complete remission | No/yes | 1/0.218 (0.110-0.430) | .001 |
| SETBP1 overexpression | No/yes | 1/2.311 (1.173-4.553) | .015 |
AML indicates acute myeloid leukemia.
Only 121 of this series of 168 patients received induction therapy. We also found significant differences in age, cytogenetic group, and complete remission (data not shown) in this series. When we investigated the prognostic impact of SETBP1 overexpression in this cohort, we found that patients with SETBP1 overexpression had significantly worse OS (P = .016) and EFS (P = .015; Figure 6B). We found no differences in DFS (P = .366). Sixty-six patients of this series were included in the intermediate cytogenetic risk group, and 47 had normal karyotype. Patients with SETBP1 overexpression had a worse OS than those without (median, 26 vs 45.9 weeks); however, differences were not statistically significant, probably because of the small number of SETBP1+ cases. A worse outcome in DFS (median, 20.9 vs 61.4 weeks) and EFS (median, 23.3 vs 40 weeks) was also observed.
To analyze the molecular events that could lead to SETBP1 overexpression in the AML cases, we performed a bioinformatic analysis of the proximal promoter of SETBP1. We identified several hypothetical binding sites for transcription factors previously implicated in leukemia, such as GATA1, GATA2, EVI1, HMG-1, or AP-1 (supplemental Table 3). Moreover, there is a CpG island within the putative proximal promoter of SETBP1 that spans 1812 bp and could lead to the hypothetical epigenetic deregulation of SETBP1 in cases with overexpression of this gene. Interestingly, 6 of the 7 retroviral insertion sites described by Ott et al20 are located in a region of 200 bp (40513701-40513912), where this CpG island is located (40512982-40514793).
Discussion
We report here a novel mechanism of transformation in acute myeloid leukemia. We first investigated a t(12;18)(p13;q12) involving ETV6 in a patient with AML. Functionally significant fusions could not be detected. Knowing that SETBP1 (18q12) was close to the translocation breakpoint, and taking into account that ectopic expression of oncogenes is a mechanism involved in leukemia, we postulated that overexpression of SETBP1 could be the major event in this case. Indeed, SETBP1 was overexpressed at diagnosis and in the posttreatment samples of the patient (Figure 1). As activation of SETBP1 expression by retroviral integration in hematopoietic progenitor cells had been reported to confer a growth advantage leading to clonal expansion20 and we could establish that SETBP1 overexpression was a recurrent molecular event with poor prognostic impact in AML, we decided to investigate the leukemogenic mechanism of overexpression of this gene. In this paper, we show that SETBP1 overexpression protects SET from protease cleavage, increasing the amount of full-length SET protein, and leading to the formation of a SETBP1–SET-PP2A complex that results in PP2A inhibition, and therefore promotes the proliferation and expansion of leukemic cells.
SETBP1 encodes a protein of 1542 amino acids and 170 kDa, localized predominantly in the nucleus, as we have confirmed in this study (supplemental Figure 2B). The physiologic function as well as the molecular mechanism by which SETBP1 acts remains unknown. The protein contains a region homologous to the dimerization domain of SKI, and a SET-binding region, although the functional significance of these interactions has not been determined.21 The protein SET (I2PP2A/TAF-Iβ) inhibits PP2A through the phosphorylation of the PP2Ac tyrosine-307.24 Indeed, SET is overexpressed in multiple solid tumors25 and in chronic myeloid leukemia where its correlation has been demonstrated with the expression and oncogenic activity of BCR-ABL, leading to PP2A inhibition.23
Here, we demonstrate that SETBP1 overexpression increased the 39-kDa full-length SET, probably because the SETBP1-SET interaction protects SET from protease-mediated cleavage. As a consequence, we detected a decrease in the 27-, 24-, and 20-kDa SET forms (Figure 2A). It has been reported that SET inhibits the DNase activity of the tumor suppressor NM23-H1 and that the cleavage of SET by Granzyme A during the cytotoxic T lymphocyte-induced apoptosis releases NM23-H1 from inhibition and triggers NM23-H1 to translocate into the nucleus, where it nicks the DNA.26,27 Interestingly, the cleavage of SET by Granzyme A generates 3 polypeptides with similar molecular weight to those we detected. Although more studies are needed to clarify this point, our results suggest a possible role for SETBP1 impairing the cleavage of SET by Granzyme A in the apoptosis caspase-independent pathway induced by cytotoxic T lymphocytes. This could point to a novel defense mechanism in leukemic cells.
PP2A is a major protein phosphatase implicated in many cell processes,28-32 and its loss of function has been associated with cell transformation.33,34 Because SET is a potent inhibitor of PP2A and SETBP1 overexpression alters the level of SET, we analyzed the effect of SETBP1 overexpression on PP2A. In addition to the previously reported SETBP1-SET and SET-PP2A complexes, we demonstrated the formation of a SETBP1–SET-PP2A heterotrimeric complex, in which PP2A is phosphorylated and, therefore, inhibited (Figure 5). Our results suggest that this is the molecular mechanism by which PP2A is inhibited in patients with SETBP1 overexpression and AML because PP2A and SETBP1 did not interact in the absence of SET (Figure 5). Interestingly, we only detected the full-length SET form in the immunoprecipitate, indicating that the shorter processed forms are not present in the SETBP1–SET-PP2A complex. It has been reported that these forms conserve their inhibitory activity of PP2A, but it remains unclear whether this activity is similar to that of full-length SET35 ; our results suggest that the processed forms may have lower PP2A inhibitory activity and that SETBP1 may increase the inhibition of PP2A by protecting the full-length SET protein, and permitting its interaction with PP2A.
Moreover, SET is a key modulator of DNA replication, chromatin remodeling and gene transcription,36,37 differentiation,38 and cell-cycle regulation.39 We postulate that the protecting role that SETBP1 overexpression plays on SET might reflect changes in the expression patterns of genes whose acetylation depends on the presence of the full-length SET protein because 39 kDa SET, but not the processed forms, has been reported to inhibit histone acetylation and to bind to HuR, which stabilizes immediate early gene mRNAs.25,36,40 Our results showed that the SETBD region is critical for this function (Figure 2B); therefore, the interaction of SETBP1 and SET could have other effects apart from the inhibition of PP2A.
To assess the prevalence and the prognostic significance of SETBP1 overexpression, we analyzed the expression of this gene in 192 patients with AML at diagnosis, and we found that SETBP1 overexpression was a recurrent event in AML, accounting for 27.6% of all cases. Our results show that SETBP1 overexpression predicts shorter OS and that the impact on prognosis is especially significant in patients older than 60 years (Figure 6A). AML is a disease of the elderly, with a median age at diagnosis more than 60 years. However, the gradual improvements achieved in the last 2 decades have been mainly focused on the group of patients younger than 60 years, whereas there has been no change in the OS of patients older than 60 years, probably because of both patient- and disease-specific factors, although this subgroup represents two-thirds of the total number of cases.41 It is therefore important to identify genetic markers that could predict prognosis in this subgroup of patients, as well as to advance our knowledge of disease biology to develop novel targeted therapies. Our data suggest that SETBP1 overexpression could distinguish 2 subgroups of AML elderly patients; furthermore, it could be a predictive factor of response to PP2A activators, such as FTY720, which has been proposed as a future alternative for treating blast-crisis chronic myeloid leukemia and Philadelphia-positive acute lymphocytic leukemia.42
Although multivariate analysis confirmed the negative prognostic impact of SETBP1 overexpression in our series, it was associated with other poor prognostic markers, such as monosomy 7 and EVI1 overexpression. AML is a multistep genetic disease, and the activation of SETBP1 may cooperate with additional mutations to induce leukemia.20 In the patient with t(12;18), we found no FLT3, NPM1, or JAK2 mutations, although trisomy 19 was observed in the blast cells of the patient as a secondary event that would cooperate in the progression of the disease. Moreover, posttreatment samples showed overexpression of SET and ETV6, which could act synergistically with SETBP1 overexpression (supplemental Figure 3). The mechanisms by which SETBP1 is overexpressed in the series analyzed remain unknown. However, bioinformatic analysis of the putative promoter region of SETBP1 allowed us to identify several transcription factors previously implicated in leukemia (supplemental Table 3); so we postulate that their deregulation could promote SETBP1 overexpression. FISH analysis suggests that the breakpoint in the patient with the t(12;18) is located upstream to the transcription start site of SETBP1, close to the retroviral insertion sites described by Ott et al.20 Moreover, 6 of the 7 retroviral insertion sites described in that paper are located in a region of 200 bp (40513701-40513912), where we have found a CpG island (40512982-40514793). Taken together, this suggests that this is an important region for the transcriptional regulation of the SETBP1 gene and that epigenetic aberrations could be another mechanism of SETBP1 overexpression in patients with AML without 18q aberrations. Further studies to confirm this and to analyze the promoter region of SETBP1 are in progress.
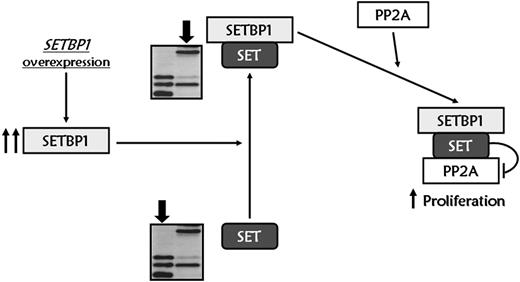
In conclusion, we report a novel mechanism of leukemic transformation. Overexpression of SETBP1 promotes an increase in full-length SET levels, impairing the phosphatase activity of the tumor suppressor PP2A through the formation of a SETBP1–SET-PP2A complex, and promoting the proliferation of cells (Figure 7). Besides, we demonstrate that SETBP1 overexpression protects SET from protease cleavage, which could have important effects on both the histone acetylation inhibitory activity of SET and Granzyme A–mediated caspase-independent apoptosis induced by cytotoxic T lymphocytes. Furthermore, deregulation of SETBP1 by translocations or other unknown mechanisms seems to play a crucial role in the leukemic transformation of AML. We have shown that SETBP1 overexpression is a recurrent molecular event with independent prognostic value in AML, especially in the subgroup of elderly patients. Further research into the physiologic function of this gene will contribute to a better understanding of the multiple steps that give rise to AML.
Proposed molecular model for SETBP1 signaling pathway. SETBP1 overexpression protects SET from protease cleavage, increasing the amount of full-length SET protein, and leading to the formation of a SETBP1–SET-PP2A complex that results in PP2A inhibition and therefore promotes the proliferation of leukemic cells.
Proposed molecular model for SETBP1 signaling pathway. SETBP1 overexpression protects SET from protease cleavage, increasing the amount of full-length SET protein, and leading to the formation of a SETBP1–SET-PP2A complex that results in PP2A inhibition and therefore promotes the proliferation of leukemic cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Elizabeth Guruceaga, of the Unit of Proteomics, Genomics and Bioinformatics of Center for Applied Medical Research, for the bioinformatics analysis, and Enrique Andreu for useful discussion.
This work was supported by the Ministerio de Educación y Ciencia, Ministerio de Ciencia e Innovación (PI081687, M.D.O.; and SAF2007-61827, C.B.), Departamento de Salud del Gobierno de Navarra (14/2008), ISCIII-RTICC (RD06/0020/0078), and Fundación para la Investigación Médica Aplicada y UTE (Spain).
Authorship
Contribution: I.C., F.J.B., L.G.-O., and N.M. performed research; C.V. performed FISH analyses; F.J.N. and E.B. contributed to bioinformatics and statistical analyses; J.R. and M.J.C. made available clinical histories and patient samples; and I.C., C.B., and M.D.O. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: María D. Odero, Division of Oncology, Center for Applied Medical Research, University of Navarra, Pio XII-55 31008-Pamplona, Spain; e-mail: modero@unav.es.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal