Abstract
In megakaryocytes, the maturation process and oxidative stress response appear to be closely related. It has been suggested that increased oxygen tension and reactive oxygen species (ROS) promote megakaryopoiesis and that the expression of stress-responsive genes responsible for ROS elimination declines during megakaryocytic maturation. NF-E2 p45 is an essential regulator of megakaryopoiesis, whereas Nrf2 is a key activator of stress-responsive genes. Because p45 and Nrf2 have similar DNA-binding specificities, we hypothesized that p45 competes with Nrf2 to repress stress-responsive genes and achieves favorable intracellular conditions to allow ROS to be efficiently used as signaling molecules. We conducted comprehensive gene expression profiling with wild-type and p45-null megakaryocytes and examined the functional relationship between p45 and Nrf2. We found that 2 characteristic gene clusters are defined within p45 target genes: platelet genes and cytoprotective genes. The former are unique targets activated by p45, whereas the latter are common targets of p45 and Nrf2. Further analysis suggested that, as a less efficacious activator, p45 maintains moderate expression of cytoprotective genes through competing with Nrf2 and promotes ROS accumulation. Increased ROS enhanced platelet gene expression. These results suggest that p45 dominates over Nrf2 to enhance megakaryocytic maturation by promoting ROS accumulation.
Introduction
Reactive oxygen species (ROS) cause cellular damage by oxidizing nucleic acids, proteins, and lipids. To avoid the toxic effects of ROS, our cells are equipped with several protective systems. However, at times, achieving minimal levels of ROS may not be preferable because ROS have been suggested to serve as signaling molecules under certain circumstances. For instance, megakaryocytes have been reported to use ROS for differentiation signals. Megakaryocytes are closely associated with bone marrow sinusoids or lung capillaries during maturation, where the cells are exposed to oxidative stress.1-3 The increase in oxygen tension or ROS has been shown to promote megakaryocytic maturation.4,5 Based on these observations, it has been suggested that the maturation process of megakaryocytes is closely related to the cellular response to oxidative stress. However, it remains largely unknown how the intracellular ROS level and response to oxidative stress affect megakaryocytic maturation.
NF-E2 is a heterodimeric transcription factor composed of Cap′N′Collar (CNC) transcription factor p45 and small Maf proteins.6,7 NF-E2 plays a key role in megakaryocytic differentiation and platelet production through binding to the Maf recognition element (MARE).8-11 As small Maf proteins lack activation domains, transcriptional activation capacity of NF-E2 depends on the N-terminal region of p45.12,13 p45-null mice have neonatal hemorrhage because of severe thrombocytopenia.8 Whereas megakaryocytes proliferate in response to thrombopoietin (TPO) and increase their ploidy through endomitosis, even in the absence of p45,8 proplatelet formation, the terminal stage of megakaryocytic differentiation, is completely defective in p45-null megakaryocytes.14 The expression of Thromboxane synthetase (Txas) and Rab27b, 2 direct target genes of p45, is decreased in p45-null megakaryocytes.15,16 In addition, p45 depletion impairs proliferation of megakaryocytes,17 suggesting that p45 also contributes to the proliferation of megakaryocytes.
Nrf2 also belongs to the CNC family. Through heterodimerization with small Maf proteins, Nrf2 confers cytoprotection against oxidative stress.18-20 The transactivation domains of Nrf2 determine the heterodimer activity of Nrf2 and small Mafs.21 Under basal conditions, Nrf2 is ubiquitinated by the Keap1-based ubiquitin E3 complex and is degraded by the proteasome. However, in response to increased ROS, Nrf2 is stabilized and activates the transcription of numerous cytoprotective genes.22 The induced cytoprotective enzymes/proteins act to eliminate ROS and maintain homeostasis of intracellular ROS levels.
Recently, a sophisticated fractionation of megakaryocytes revealed the maturation stage-specific gene expression profiles.23 According to this work, genes involved in the stress response, many of which are established Nrf2 targets, are highly expressed in immature megakaryocytes, but their expression declines as megakaryocytes mature. This study implies that megakaryocytes reduce the concentration of antioxidant proteins during differentiation, which favors megakaryocytes, using ROS accumulation as a maturation signal. p45-null megakaryocytes showed elevated expression of certain stress-responsive genes, which is interesting considering that p45-null megakaryocytes are stalled at an earlier stage of maturation.23
Because p45 and Nrf2 possess similar DNA-binding specificities, we hypothesized that p45 competes with Nrf2 and reduces stress-responsive gene expression in mature megakaryocytes. To test this hypothesis, we conducted gene expression profiling analysis of p45-null megakaryocytes and examined the functional relationship between p45 and Nrf2 in megakaryocytes. We found that 2 characteristic gene clusters are defined within p45 target genes: platelet genes and cytoprotective genes. The platelet genes are exclusively activated by p45, whereas the cytoprotective genes, such as NAD(P)H:quinone oxidoreductase (Nqo1), are competitively regulated by p45 and Nrf2. Our analysis suggested that, being a weaker activator than Nrf2, p45 maintains expression of cytoprotective genes at moderate levels by competing with Nrf2 in mature megakaryocytes. This competitive transcriptional mechanism promotes ROS accumulation and platelet gene expression. By contrast, in immature megakaryocytes, p45 and Nrf2 cooperatively promote proliferation. Increased p45 levels during megakaryocyte maturation are associated with a transition from Nrf2-p45 cooperation to competition. Importantly, this shift in the balance of 2 CNC factors promotes ROS accumulation and drives megakaryocyte maturation.
Methods
Mice
Together with control wild-type (WT) littermates, p45-null, Nrf2-null, and Keap1-null embryos were obtained from p45 heterozygous mating pairs,8 Nrf2-heterozygous mating pairs,18 and Keap1-heterozygous mating pairs,24 respectively. The polymerase chain reaction (PCR) conditions and primers for genotype determination have been previously described. All mice experiments were approved by the Animal Care and Use Committee of Tohoku University.
Primary culture of megakaryocytes
Whole livers were recovered from mouse embryos at 14.5-day (E14.5), and single-cell suspensions were prepared by successive passage through 25-gauge needles. Fetal liver cells were maintained in RPMI 1640 (Wako) supplemented with 20% charcoal-stripped fetal bovine serum, 50 U/mL penicillin, 50 μg/mL streptomycin, and 50 ng/mL of recombinant human TPO (generously provided by Kirin); 10mM N-acetyl cysteine (NAC; Sigma-Aldrich) was added just after plating. A total of 100μM-diethylmaleate (DEM; Sigma-Aldrich) was added just after plating and at every 24 hours. Megakaryocytes were harvested from day 1 or day 3 cultures for RNA purification, the preparation of nuclear extracts, chromatin immunoprecipitation (ChIP) assay, and flow cytometric analysis. Megakaryocytes were harvested on day 3 unless otherwise noted. RNA was prepared from a day 4 culture for microarray analysis. The frequency of megakaryocytes generated in the primary culture and their ROS levels varied in each experiment, probably because of subtle differences in the embryonic stages when the fetal livers were collected. Therefore, all samples from primary megakaryocytes were compared within the same litter and processed at the same time.
Microarray analysis
CD41+ cells were selected from a day 4 primary culture of E14.5 fetal liver cells using a MACS magnetic system (Miltenyi Biotec). The fetal liver culture with TPO described in “Primary culture of megakaryocytes” was incubated with FITC-conjugated anti-CD41 antibody (BD Biosciences PharMingen, clone MWReg30) followed by incubation with anti–fluorescein isothiocyanate (FITC) microbeads. Subsequently, the microbeads were selected magnetically through MACS large-cell columns (Miltenyi Biotec). Total RNA was extracted from the CD41+ cells using Isogen (Nippon Gene). RNA was further purified using an RNeasy Mini Kit (QIAGEN), processed and hybridized to a mouse expression array Affymetrix Mouse Genome 430 2.0 Array (Affymetrix). The experimental procedures for the GeneChip were performed according to the Affymetrix technical manual.
Quantitative real time RT-PCR
CD41+ cells were selected by biotinylated anti-CD41 antibody (Serotec; clone MWReg30) and streptavidin-coupled Dynabeads (Veritas) from a culture of E14.5 fetal liver cells. Total RNA was purified from the CD41+ cells, and cDNA was synthesized from the RNA with random hexamers. Real-time reverse-transcription (RT) PCR was performed using an ABI7300 Sequence Detection System. The reaction was carried out for 40 to 60 cycles of 15 seconds at 95°C and 1 minute at 60°C in qPCR Mastermix (Eurogentec). Ribosomal RNA control reagents (Applied Biosystems) were used as an internal control. The probes and primers for detecting Ho-1, Nqo1, Gsta4, Txnrd1, and Gpx1 have been previously described.20,25 The probes and primers for detecting the expression of other genes are described in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Flow cytometry
The primary culture of fetal liver cells with TPO was stained with PE-conjugated anti-CD61 (BD Biosciences Pharmingen, 2C9.G2 subclone of HMβ3-11 ) and biotinylated anti-CD41 that was subsequently complexed with APC-conjugated streptavidin (BD Biosciences Pharmingen). The fluoroprobe 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA; 5μM; Invitrogen) was added, and cells were incubated at 37°C and 5% CO2 for 2 hours. Cells were analyzed using a FACSCalibur (BD Biosciences). The DNA content was analyzed, as previously described.26 For cell sorting, megakaryocytes were identified after labeling with phycoerythrin (PE)–conjugated anti-CD41 antibody and incubated with 5μM DCFDA for 2 hours. CD41+ cells were sorted according to the intensity of DCFDA using a FACS Vantage (BD Biosciences). The standard line for fractionating the cells into 2 populations intersects the midpoint between the maximum and the minimum levels of DCFDA signal intensity of the CD41+ population on a log scale. The gates were set for CD41+DCFDAhigh and CD41+DCFDAlow populations on the right and left sides of the standard line. The sorted cells were used for RNA extraction to examine gene expression profiles.
ChIP assays
ChIP assays were performed using a day 3 primary culture of megakaryocytes from E14.5 fetal livers. Immunoprecipitation was performed using control rabbit IgG, anti-Nrf2 (Santa Cruz Biotechnology; sc-13032), anti-p45,13 anti–trimethyl-histone H3 Lys4 (Upstate; 07-473), anti–dimethyl-histone H3 Lys4 (Upstate; 07-030), anti–acetyl-histone H3 (Upstate; 06-599), and anti–acetyl-histone H4 (Upstate; 06-866) antibodies. The sequences of the PCR primer sets for the Txas promoter, Nqo1 promoter, and Txas intron are described in supplemental Table 1. Chromatin DNA was quantitatively analyzed by real-time PCR.
Immunoblot analysis
Nuclear extracts were prepared from primary megakaryocytes as described before,27 and immunoblot analysis was performed using anti-Nrf2,28 anti-p45 (Santa Cruz Biotechnology; sc-291), and anti-LaminB (Santa Cruz Biotechnology; sc-6217). Whole-cell extracts were prepared from transiently transfected 293T cells, and immunoblot analysis was performed with anti-Flag antibody (Sigma-Aldrich; F3165).
Transient transfection and reporter assay
The 293T cells were maintained in Dulbecco modified Eagle medium (Sigma-Aldrich) with 10% fetal bovine serum, 50 U/mL penicillin, and 50 μg/mL streptomycin. NQO1 MARE-Luc, a previously described reporter plasmid,29 was introduced into 293T cells along with Nrf2 and p45 expression vectors using FuGENE6 transfection reagent (Roche Diagnostics) or Lipofectamine 2000 (Invitrogen). Luciferase activity was measured 24 hours after transfection using a Dual Reporter Assay System (Promega) and a luminometer (Berthold Japan). All samples were prepared in triplicate, and mean and SD were calculated.
Plasmid construction
The DNA fragment encoding the N-terminal half of Nrf2 (amino acids 1-426) connected to the C-terminal half of p45 (amino acids 206-377) was generated by PCR. The fragment was cloned between NotI and XbaI sites of p3xFLAG-CMV-10 (Sigma-Aldrich).
Results
Expression profiling revealed 2 characteristic gene clusters within p45 target genes
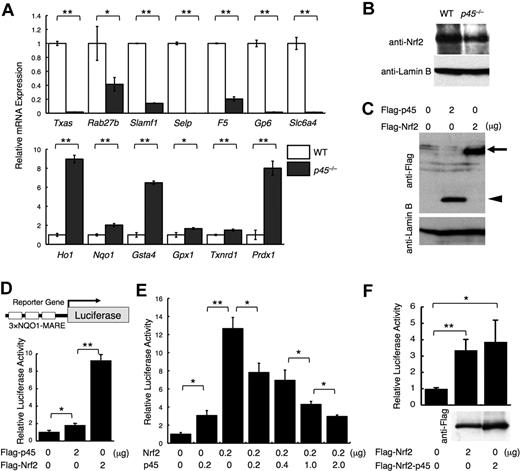
To determine the downstream targets of p45 in megakaryocytes, we conducted an expression array analysis of p45-null megakaryocytes cultured from livers of E14.5 embryos (GEO accession no. GSE15581). The expression of genes encoding platelet enzymes and membrane proteins, including Txas, Rab27b, Signaling lymphocytic activation molecule family member 1 (Slamf1), Selectin P (Selp), Factor 5 (F5), Glycoprotein 6 (Gp6), and Solute carrier family 6, member 4 (Slc6a4), was decreased in the absence of p45 (Figure 1A top panel; supplemental Table 2). We also examined the expression of cytoprotective genes, which are normally regulated by Nrf2. Supporting the previous report,23 expression of detoxifying enzymes and stress-responsive genes, such as Heme oxygenase 1 (Ho-1), NADP(H):quinone oxidoreductase (Nqo1), Glutathione-S-transferase a4 (Gsta4), Glutathione peroxidase 1 (Gpx1), Thioredoxin reductase 1 (Txnrd1), and Peroxiredoxin 1 (Prdx1), was increased in the absence of p45 (Figure 1A bottom panel; supplemental Table 3). Many of these genes were well-characterized Nrf2 target genes.27,30
p45 competitively inhibits Nrf2-mediated transcriptional activation. (A) Quantitative RT-PCR of platelet genes and cytoprotective genes in p45-null megakaryocytes. The relative values were calculated against the values of WT megakaryocytes. (B) Immunoblot analysis of nuclear extracts of p45-null and WT megakaryocytes cultured from fetal livers. Anti-Nrf2 and anti–lamin B antibodies were used. (C) Comparison of protein abundance of p45 and Nrf2. FLAG-tagged p45 and FLAG-tagged Nrf2 were transiently overexpressed in 293T cells. Whole-cell extracts were prepared and subjected to immunoblot analysis with anti-FLAG or anti–lamin B antibodies.  and ◀ indicate FLAG-Nrf2 and FLAG-p45, respectively. (D) Comparison of transcriptional activation abilities of p45 and Nrf2. Equal amount of the expression vectors were introduced into 293T cells with a luciferase gene driven by MARE of the Nqo1 promoter in triplicate as a reporter gene. (E) Reporter assay in 293T cells with the same reporter gene used in panel D. Expression vectors of Nrf2 and/or p45 without tags were added as effector molecules. (F) Comparison of transcriptional activation abilities of FLAG-tagged Nrf2 and FLAG-tagged fusion protein of the N-terminal half of Nrf2 and C-terminal half of p45 with the same reporter gene used in panel D. The protein expression detected by anti-FLAG antibody is shown below. The relative luciferase activities were calculated against the activity generated by the reporter gene alone (D-F). The average values of triplicate experiments are presented, and the error bars represent SD (A,D-F). The Student t test was used to calculate statistical significance. *P < .05; **P < .005 (A,D-F).
and ◀ indicate FLAG-Nrf2 and FLAG-p45, respectively. (D) Comparison of transcriptional activation abilities of p45 and Nrf2. Equal amount of the expression vectors were introduced into 293T cells with a luciferase gene driven by MARE of the Nqo1 promoter in triplicate as a reporter gene. (E) Reporter assay in 293T cells with the same reporter gene used in panel D. Expression vectors of Nrf2 and/or p45 without tags were added as effector molecules. (F) Comparison of transcriptional activation abilities of FLAG-tagged Nrf2 and FLAG-tagged fusion protein of the N-terminal half of Nrf2 and C-terminal half of p45 with the same reporter gene used in panel D. The protein expression detected by anti-FLAG antibody is shown below. The relative luciferase activities were calculated against the activity generated by the reporter gene alone (D-F). The average values of triplicate experiments are presented, and the error bars represent SD (A,D-F). The Student t test was used to calculate statistical significance. *P < .05; **P < .005 (A,D-F).
p45 competitively inhibits Nrf2-mediated transcriptional activation. (A) Quantitative RT-PCR of platelet genes and cytoprotective genes in p45-null megakaryocytes. The relative values were calculated against the values of WT megakaryocytes. (B) Immunoblot analysis of nuclear extracts of p45-null and WT megakaryocytes cultured from fetal livers. Anti-Nrf2 and anti–lamin B antibodies were used. (C) Comparison of protein abundance of p45 and Nrf2. FLAG-tagged p45 and FLAG-tagged Nrf2 were transiently overexpressed in 293T cells. Whole-cell extracts were prepared and subjected to immunoblot analysis with anti-FLAG or anti–lamin B antibodies.  and ◀ indicate FLAG-Nrf2 and FLAG-p45, respectively. (D) Comparison of transcriptional activation abilities of p45 and Nrf2. Equal amount of the expression vectors were introduced into 293T cells with a luciferase gene driven by MARE of the Nqo1 promoter in triplicate as a reporter gene. (E) Reporter assay in 293T cells with the same reporter gene used in panel D. Expression vectors of Nrf2 and/or p45 without tags were added as effector molecules. (F) Comparison of transcriptional activation abilities of FLAG-tagged Nrf2 and FLAG-tagged fusion protein of the N-terminal half of Nrf2 and C-terminal half of p45 with the same reporter gene used in panel D. The protein expression detected by anti-FLAG antibody is shown below. The relative luciferase activities were calculated against the activity generated by the reporter gene alone (D-F). The average values of triplicate experiments are presented, and the error bars represent SD (A,D-F). The Student t test was used to calculate statistical significance. *P < .05; **P < .005 (A,D-F).
and ◀ indicate FLAG-Nrf2 and FLAG-p45, respectively. (D) Comparison of transcriptional activation abilities of p45 and Nrf2. Equal amount of the expression vectors were introduced into 293T cells with a luciferase gene driven by MARE of the Nqo1 promoter in triplicate as a reporter gene. (E) Reporter assay in 293T cells with the same reporter gene used in panel D. Expression vectors of Nrf2 and/or p45 without tags were added as effector molecules. (F) Comparison of transcriptional activation abilities of FLAG-tagged Nrf2 and FLAG-tagged fusion protein of the N-terminal half of Nrf2 and C-terminal half of p45 with the same reporter gene used in panel D. The protein expression detected by anti-FLAG antibody is shown below. The relative luciferase activities were calculated against the activity generated by the reporter gene alone (D-F). The average values of triplicate experiments are presented, and the error bars represent SD (A,D-F). The Student t test was used to calculate statistical significance. *P < .05; **P < .005 (A,D-F).
These results indicate that 2 characteristic gene clusters are defined within p45 target genes: platelet genes and cytoprotective genes. In p45-null megakaryocytes, the former genes were repressed, whereas the latter genes that substantially overlap with the Nrf2 target genes were activated. Importantly, Nrf2 nuclear accumulation was similar between p45-null and WT megakaryocytes (Figure 1B). Thus, p45 deficiency elevated the expression of cytoprotective genes without increasing Nrf2 abundance in nuclei.
p45 inhibits Nrf2-mediated transcriptional activation
Because Nrf2 and p45 bind to a similar DNA sequence in vitro, the relative concentrations and activities of these factors are important determinants of target gene expression. To assess whether Nrf2 and p45 have similar or distinct transactivation activities, we compared their transactivation potentials in a transient transfection assay.
Transfection of equal amounts of FLAG-Nrf2 and FLAG-p45 expression vectors into 293T cells yielded similar levels of protein expression, as detected with an anti-FLAG antibody (Figure 1C). An equal amount of the expression vectors was then transfected with the luciferase reporter gene driven by MARE from the Nqo1 gene29 (Figure 1D). Whereas both Nrf2 and p45 induced the reporter, p45-mediated activation was considerably weaker than that of Nrf2. Similar results were obtained when p45 and Nrf2 expression vectors without FLAG tags were used (Figure 1E). On coexpression of an increasing amount of p45 with a constant amount of Nrf2, the reporter activity decreased incrementally with the increase of p45 (Figure 1E), indicating that p45 competitively inhibits Nrf2-mediated activation. When the C-terminal half of p45-containing CNC and bZip motifs was fused to the N-terminal half of Nrf2 without CNC or bZip motifs, the fusion protein acquired a transcriptional activity similar to that of Nrf2 (Figure 1F), implying that a property of the transactivation domain, but not the DNA-binding specificity of Nrf2, is responsible for the potent transcriptional activity.
Competitive binding of p45 and Nrf2 to Nqo1 MARE
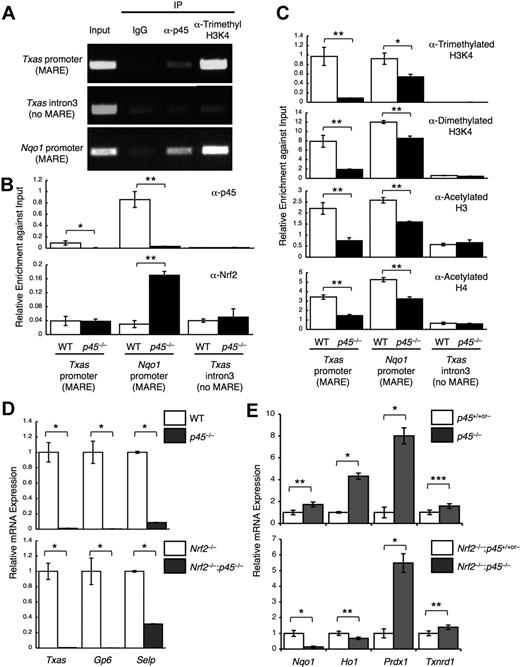
To examine whether the competition between Nrf2 and p45 for common target genes occurs in megakaryocytes in vivo, we investigated the competitive binding of Nrf2 and p45 by ChIP analysis using cultured primary megakaryocytes. Chromatin complexes were immunoprecipitated with an anti-p45 antibody.13 The results indicated that p45 occupies regions containing the Nqo1 MARE and Txas MARE, but not the non-MARE control region (Figure 2A). In addition, trimethylated histone H3K4 was also highly enriched at both MAREs, which is an active chromatin mark (Figure 2A). ChIP analysis with anti-Nrf2 antibody revealed that Nrf2 occupancy at the Nqo1 MARE was higher in p45-null megakaryocytes (Figure 2B). However, little to no Nrf2 occupancy was detected at the Txas MARE in either WT or p45-null megakaryocytes (Figure 2B). Consistent with this result, the active chromatin marks were considerably reduced at the Txas promoter but were mostly retained at the Nqo1 promoter (Figure 2C). We considered this to be a probable consequence of Nrf2 recruitment to the Nqo1 MARE in the absence of p45. Because nuclear accumulation of Nrf2 was indistinguishable between WT and p45-null megakaryocytes (Figure 1B), the increase in Nrf2 occupancy at the Nqo1 MARE can be explained by a lack of competition with p45 for the chromatin site. These results indicate that cytoprotective gene expression is determined by competition of p45 and Nrf2 for common chromatin target sites.
Competitive regulation of cytoprotective genes by p45 and Nrf2. (A) ChIP assays were performed with primary megakaryocytes derived from WT fetal livers using anti-p45 and anti–trimethyl-histone H3 (Lys4) antibodies. (B-C) ChIP assays were performed with primary megakaryocytes derived from p45-null and control fetal livers using anti-p45 and anti-Nrf2 antibodies (B) and anti–trimethyl-histone H3K4, anti–dimethyl-histone H3K4, anti–acetyl-histone H3, and anti–acetyl-histone H4 antibodies (C). Enrichment of the promoter regions containing MAREs of the Txas and Nqo1 genes is shown. The third intron of the Txas gene was used as a negative control. Quantitative analysis was performed, and the average values and SD were calculated from the triplicate samples (B-C). The Student t test was used to calculate statistical significance. *P < .02; **P < .002 (B-C). (D-E) Effect of p45 deletion on the expression of platelet genes (D) and cytoprotective genes (E) in the presence (top panels) and in the absence (bottom panels) of Nrf2. Quantitative RT-PCR was performed, and the relative values were calculated against the values of WT (D top panel), Nrf2−/− (D bottom panel), the mixture of WT and p45+/− (E top panel), and the mixture of Nrf2−/− and Nrf2−/−:p45+/− (E bottom panel) megakaryocytes. The Student t test was used to calculate statistical significance. *P < .002; **P < .05; ***P < .1 (D-E).
Competitive regulation of cytoprotective genes by p45 and Nrf2. (A) ChIP assays were performed with primary megakaryocytes derived from WT fetal livers using anti-p45 and anti–trimethyl-histone H3 (Lys4) antibodies. (B-C) ChIP assays were performed with primary megakaryocytes derived from p45-null and control fetal livers using anti-p45 and anti-Nrf2 antibodies (B) and anti–trimethyl-histone H3K4, anti–dimethyl-histone H3K4, anti–acetyl-histone H3, and anti–acetyl-histone H4 antibodies (C). Enrichment of the promoter regions containing MAREs of the Txas and Nqo1 genes is shown. The third intron of the Txas gene was used as a negative control. Quantitative analysis was performed, and the average values and SD were calculated from the triplicate samples (B-C). The Student t test was used to calculate statistical significance. *P < .02; **P < .002 (B-C). (D-E) Effect of p45 deletion on the expression of platelet genes (D) and cytoprotective genes (E) in the presence (top panels) and in the absence (bottom panels) of Nrf2. Quantitative RT-PCR was performed, and the relative values were calculated against the values of WT (D top panel), Nrf2−/− (D bottom panel), the mixture of WT and p45+/− (E top panel), and the mixture of Nrf2−/− and Nrf2−/−:p45+/− (E bottom panel) megakaryocytes. The Student t test was used to calculate statistical significance. *P < .002; **P < .05; ***P < .1 (D-E).
Nrf2 activates cytoprotective genes in p45-null megakaryocytes
To further verify that Nrf2 is the factor that induces cytoprotective genes in the absence of p45, we measured Nqo1 expression in Nrf2 and p45 double-knockout megakaryocytes. For this purpose, we first generated Nrf2−/−:p45+/− compound mutant mice, which were subsequently intercrossed to obtain Nrf2−/−:p45−/− embryos. Fetal liver cells were prepared from the embryos and cultured with TPO. Comparisons were made within a litter exploiting the fetal liver cell culture from Nrf2−/− embryos as a control.
Platelet genes were decreased by p45 deletion, irrespective of Nrf2 genotype (Figure 2D). Among the cytoprotective genes up-regulated by p45 deletion in the presence of Nrf2 (Figure 2E top panel), Nqo1 and Ho1 were not increased but rather were decreased by p45 deletion in the absence of Nrf2 (Figure 2E bottom panel), indicating a requirement of Nrf2 for increased expression of these genes. In contrast, Prdx1 and Txnrd1 expressions were increased by p45 deletion, irrespective of Nrf2 genotype (Figure 2E). We surmise that a compensatory activating factor, possibly Nrf1, is involved in the regulation of these genes because Nrf1 is also known to participate in the regulation of cytoprotective genes through MARE.31
These results support the idea that increased expression of cytoprotective genes in p45-null megakaryocytes depends, at least in part, on Nrf2. Thus, p45 counteracts, at least in part, Nrf2-mediated activation of cytoprotective genes in normal megakaryocytes.
Reduction of intracellular ROS level in p45-null megakaryocytes
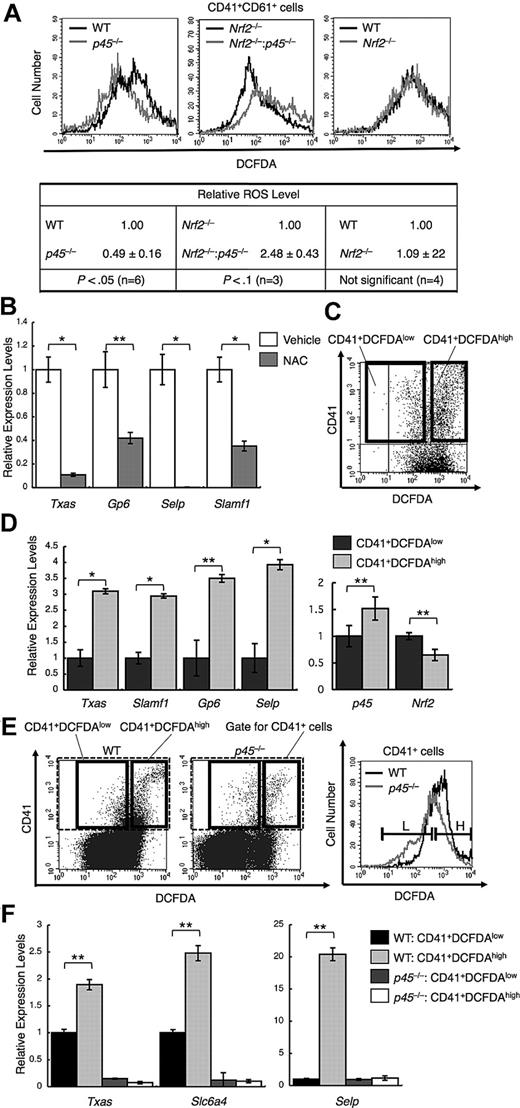
Because p45 is a weaker activator than Nrf2 and limits the expression of cytoprotective genes, it is reasonable to expect that p45 is more permissive for ROS accumulation than Nrf2 and that the intracellular ROS level is decreased in p45-null megakaryocytes. We measured intracellular ROS using DCFDA and flow cytometry. The intensity of DCFDA was lower in CD41+CD61+ cells from p45-null embryos than in the cells from WT embryos (Figure 3A left panel), indicating that ROS accumulation is reduced in the p45-null megakaryocytes.
p45 promotes ROS accumulation and increased ROS enhances platelet gene activation. (A) Flow cytometric analysis of intracellular ROS levels in primary megakaryocytes from p45-null and WT livers (top left panel), Nrf2−/− and Nrf2−/−:p45−/− fetal livers (top middle panel), and Nrf2-null and WT livers (top right panel). The intensities of DCFDA in the CD41+CD61+ cells are displayed in histograms. Average ROS levels obtained from the histograms of p45-null, Nrf2−/−:p45−/−, and Nrf2-null cells were changed to relative values against those from the WT, Nrf2-null, and WT cells, respectively (bottom panel). Average values are shown with SD. Statistical significance of the relative ROS levels was calculated using paired t test. (B) Quantitative RT-PCR of platelet genes in primary megakaryocytes after supplementation with 10mM NAC for scavenging ROS. The relative values were calculated against the values of the vehicle-treated samples. (C) Primary megakaryocytes were fractionated into either DCFDA high or DCFDA low populations according to the fluoroprobe intensity in flow cytometry. The gates are depicted in bold rectangles. (D) Quantitative RT-PCR of platelet genes (left panel) and of p45 and Nrf2 (right panel). The CD41+DCFDAhigh and CD41+DCFDAlow populations were compared. The relative values were calculated against the values of the CD41+DCFDAlow fraction. (E) Primary megakaryocytes cultured from p45-null and WT fetal livers were fractionated as in panel C. The intensities of DCFDA of CD41+ cells contained in the gate indicated with broken lines in the left panel are displayed in histograms (right panel). L and H in the right panel correspond to the gates depicted in bold rectangles in the left panel. (F) Quantitative RT-PCR of platelet genes. The CD41+DCFDAhigh and CD41+DCFDAlow populations from p45-null and WT fetal livers were compared. The relative values were calculated against the values of the CD41+DCFDAlow fraction of WT cells. The average values of triplicate experiments are presented, and the error bars represent SD (B,D,F). The Student t test was used to calculate statistical significance. *P < .001 (B,D). **P < .05 (B,D). **P < .005 (F).
p45 promotes ROS accumulation and increased ROS enhances platelet gene activation. (A) Flow cytometric analysis of intracellular ROS levels in primary megakaryocytes from p45-null and WT livers (top left panel), Nrf2−/− and Nrf2−/−:p45−/− fetal livers (top middle panel), and Nrf2-null and WT livers (top right panel). The intensities of DCFDA in the CD41+CD61+ cells are displayed in histograms. Average ROS levels obtained from the histograms of p45-null, Nrf2−/−:p45−/−, and Nrf2-null cells were changed to relative values against those from the WT, Nrf2-null, and WT cells, respectively (bottom panel). Average values are shown with SD. Statistical significance of the relative ROS levels was calculated using paired t test. (B) Quantitative RT-PCR of platelet genes in primary megakaryocytes after supplementation with 10mM NAC for scavenging ROS. The relative values were calculated against the values of the vehicle-treated samples. (C) Primary megakaryocytes were fractionated into either DCFDA high or DCFDA low populations according to the fluoroprobe intensity in flow cytometry. The gates are depicted in bold rectangles. (D) Quantitative RT-PCR of platelet genes (left panel) and of p45 and Nrf2 (right panel). The CD41+DCFDAhigh and CD41+DCFDAlow populations were compared. The relative values were calculated against the values of the CD41+DCFDAlow fraction. (E) Primary megakaryocytes cultured from p45-null and WT fetal livers were fractionated as in panel C. The intensities of DCFDA of CD41+ cells contained in the gate indicated with broken lines in the left panel are displayed in histograms (right panel). L and H in the right panel correspond to the gates depicted in bold rectangles in the left panel. (F) Quantitative RT-PCR of platelet genes. The CD41+DCFDAhigh and CD41+DCFDAlow populations from p45-null and WT fetal livers were compared. The relative values were calculated against the values of the CD41+DCFDAlow fraction of WT cells. The average values of triplicate experiments are presented, and the error bars represent SD (B,D,F). The Student t test was used to calculate statistical significance. *P < .001 (B,D). **P < .05 (B,D). **P < .005 (F).
We also found that the ROS level was increased in the Nrf2 and p45 double-knockout megakaryocytes compared with that in the Nrf2 single-knockout cells (Figure 3A middle panel). Thus, Nrf2 was required for the decrease of ROS level in the absence of p45, implying that Nrf2-dependent cytoprotective genes substantially contribute to the elimination of ROS. It should be noted that the intensity of DCFDA in Nrf2-null megakaryocytes was almost identical to that of WT megakaryocytes (Figure 3A right panel), indicating that Nrf2 is not a major determinant of ROS level in megakaryocytes in the presence of p45, probably because Nrf2 is inhibited by p45 under normal conditions. These results suggest that p45 enhances ROS accumulation in megakaryocytes as a consequence of limiting cytoprotective gene expression.
ROS accumulation promotes platelet gene expression
We next examined the functional importance of the reduced expression of cytoprotective genes and ROS accumulation during megakaryocytic maturation. We examined the effect of ROS reduction by NAC, a ROS-scavenging reagent, on the expression of platelet genes. Megakaryocytes were isolated using anti-CD41 antibody, and their patterns of gene expression were examined. NAC repressed platelet gene expression, including Txas, Gp6, Selp, and Slamf1 (Figure 3B), suggesting that ROS accumulation is indeed required for full activation of platelet genes in mature megakaryocytes.
We tested whether ROS accumulation and platelet gene expression correlate with each other in megakaryocytes. Megakaryocytes in the primary culture were fractionated into 2 populations depending on their ROS levels. The megakaryocytes in culture were incubated with anti-CD41 antibody and stained with DCFDA, and CD41+DCFDAlow cells and CD41+DCFDAhigh cells were then sorted using flow cytometry (Figure 3C). The expression of platelet genes was markedly higher in CD41+DCFDAhigh than in CD41+DCFDAlow cells (Figure 3D left panel). The accumulation of ROS to high levels correlated with the higher level of platelet gene expression, which is consistent with the notion that ROS is an important signal to confer high-level platelet gene expression.
We also examined the expression levels of Nrf2 and p45 and found that the expression levels did not change much between the 2 fractions. However, albeit the difference was modest, p45 expression was higher, whereas Nrf2 expression was lower, in CD41+DCFDAhigh cells than in CD41+DCFDAlow cells (Figure 3D right panel), implying that the increased p45 expression induced ROS accumulation in megakaryocytes.
We further examined p45-null megakaryocytes by a similar fractionation (Figure 3E). Although CD41+DCFDAhigh cells were generated in p45-null culture, the frequency was found to be lower than that in the WT culture. The platelet gene expression was repressed in p45-null megakaryocytes irrespective of the intracellular ROS levels (Figure 3F), suggesting that p45 is primarily required for platelet gene activation.
Nrf2 activation reduces ROS levels and platelet gene expression
Our results support a model in which competition between Nrf2 and p45 is an important determinant of megakaryocyte maturation. We demonstrated that p45 competes with Nrf2 in megakaryocytes, resulting in the enhancement of ROS accumulation and platelet gene expression. We reasoned that any context that creates an imbalance between Nrf2 and p45 would alter ROS levels and platelet gene expression. Thus, we examined the functional consequences of genetically and pharmacologically modulating Nrf2 levels.
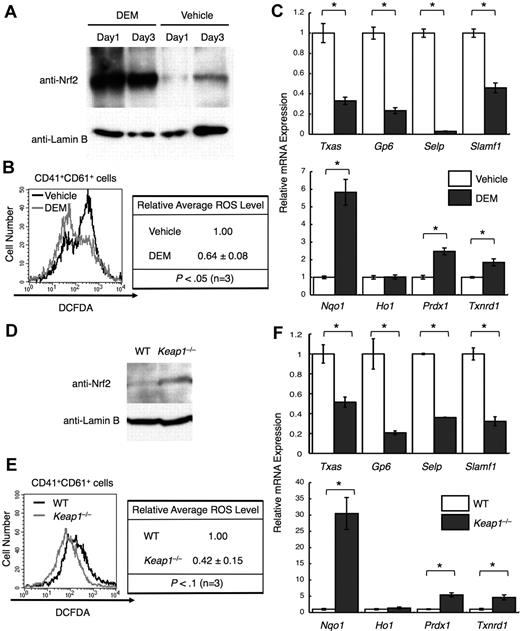
The well-known electrophile DEM induced Nrf220 (Figure 4A). Deletion of Keap1, a ubiquitin ligase that down-regulates Nrf2, also induced Nrf222 (Figure 4D). In both cases, cytoprotective genes were induced, except for Ho-1 (Figure 4C,F bottom panels). As expected from the decreased cytoprotective gene expression, ROS levels declined (Figure 4B,E). Furthermore, platelet gene expression declined (Figure 4C,F top panels), supporting the notion that ROS is an important signal for maximizing platelet gene expression.
Nrf2 activation reduces ROS levels and platelet gene expression. (A) Nuclear accumulation of Nrf2 in primary megakaryocytes by DEM treatment. DEM was added to a final concentration of 100 μM every 24 hours. CD41+ cells were isolated from day 1 and day 3 cultures, and nuclear extracts were prepared. Anti-Nrf2 and anti–lamin B antibodies were used. (B) Flow cytometric analysis of intracellular ROS levels in primary megakaryocytes treated with DEM. The intensities of DCFDA in the CD41+CD61+ cells are displayed in histograms. Average ROS levels obtained from the histograms of DEM-treated cells were changed to relative values against those from vehicle-treated cells. (C) Quantitative RT-PCR of platelet genes (top panel) and of cytoprotective genes (bottom panel) in DEM-treated megakaryocytes. The relative values were calculated against the values of vehicle-treated megakaryocytes. (D) Nuclear accumulation of Nrf2 in primary megakaryocytes cultured from Keap1-null fetal livers. CD41+ cells were isolated, and nuclear extracts were prepared. Anti-Nrf2 and anti–lamin B antibodies were used. (E) Flow cytometric analysis of intracellular ROS levels in Keap1-null megakaryocytes. The intensities of DCFDA in the CD41+CD61+ cells are displayed in histograms. Average ROS levels obtained from the histograms of Keap1-null megakaryocytes were changed to relative values of those from WT cells. Average values are shown with SD (B,E). Statistical significance of the relative ROS levels was calculated using paired t test (B,E). (F) Quantitative RT-PCR of platelet genes (top panel) and of cytoprotective genes (bottom panel) in Keap1-null megakaryocytes. The relative values were calculated against the values of WT megakaryocytes. The average values of triplicate experiments are presented, and the error bars represent SD (C,F). The Student t test was used to calculate statistical significance. *P < .002 (C,F).
Nrf2 activation reduces ROS levels and platelet gene expression. (A) Nuclear accumulation of Nrf2 in primary megakaryocytes by DEM treatment. DEM was added to a final concentration of 100 μM every 24 hours. CD41+ cells were isolated from day 1 and day 3 cultures, and nuclear extracts were prepared. Anti-Nrf2 and anti–lamin B antibodies were used. (B) Flow cytometric analysis of intracellular ROS levels in primary megakaryocytes treated with DEM. The intensities of DCFDA in the CD41+CD61+ cells are displayed in histograms. Average ROS levels obtained from the histograms of DEM-treated cells were changed to relative values against those from vehicle-treated cells. (C) Quantitative RT-PCR of platelet genes (top panel) and of cytoprotective genes (bottom panel) in DEM-treated megakaryocytes. The relative values were calculated against the values of vehicle-treated megakaryocytes. (D) Nuclear accumulation of Nrf2 in primary megakaryocytes cultured from Keap1-null fetal livers. CD41+ cells were isolated, and nuclear extracts were prepared. Anti-Nrf2 and anti–lamin B antibodies were used. (E) Flow cytometric analysis of intracellular ROS levels in Keap1-null megakaryocytes. The intensities of DCFDA in the CD41+CD61+ cells are displayed in histograms. Average ROS levels obtained from the histograms of Keap1-null megakaryocytes were changed to relative values of those from WT cells. Average values are shown with SD (B,E). Statistical significance of the relative ROS levels was calculated using paired t test (B,E). (F) Quantitative RT-PCR of platelet genes (top panel) and of cytoprotective genes (bottom panel) in Keap1-null megakaryocytes. The relative values were calculated against the values of WT megakaryocytes. The average values of triplicate experiments are presented, and the error bars represent SD (C,F). The Student t test was used to calculate statistical significance. *P < .002 (C,F).
p45 dominates over Nrf2 during megakaryocyte differentiation
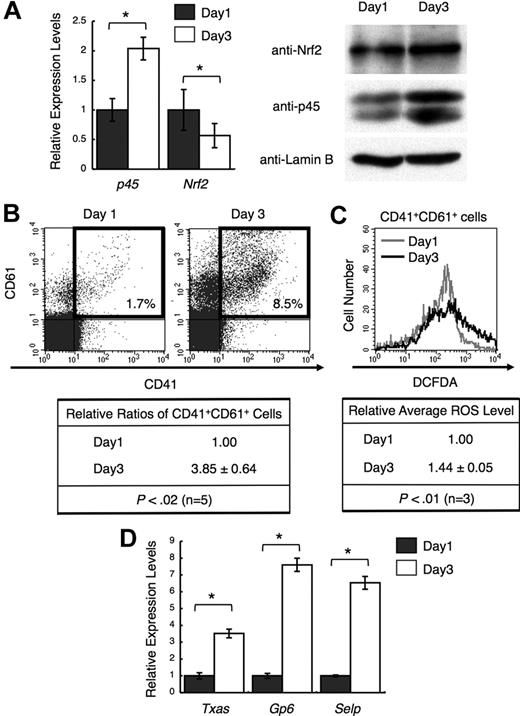
We examined whether the ratio between Nrf2 and p45 changes during megakaryocytic maturation. Because the prior analyses used day 3 cultures, we compared p45 and Nrf2 levels on days 1 and 3. p45 expression was higher at both the mRNA and protein levels in day 3 megakaryocytes, whereas Nrf2 expression was nearly constant, especially at the protein level (Figure 5A). In addition, the number of megakaryocytes with higher levels of ROS was increased in day 3 cultures (Figure 5B-C). The expression of platelet genes was consistently elevated in day 3 megakaryocytes (Figure 5D). Thus, the increase of ROS and the induction of platelet genes were accompanied by an increase in p45 during megakaryocytic maturation.
Increased p45 and ROS levels correlate with megakaryocytic maturation. (A) Expression of p45 and Nrf2 during megakaryocytic maturation. mRNA levels (left panel) and protein levels in nuclei (right panel) are shown. CD41+ cells were isolated from day 1 and day 3 cultures, and total RNA and nuclear extracts were prepared. The relative values were calculated against the values of day 1 megakaryocytes (left panel). Anti-Nrf2, anti-p45, and anti–lamin B antibodies were used (right panel). (B) Flow cytometric analysis showing megakaryocytic differentiation in day 1 and day 3 cultures. Frequencies of CD41+CD61+ cells are shown in the figure (top panel). Relative frequencies of CD41+CD61+ cells were calculated against the value of day 1 culture (bottom panel). Average values are shown with SD. Statistical significance of the relative ratio of CD41+CD61+ cells was calculated using paired t test. (C) Flow cytometric analysis of intracellular ROS levels in primary megakaryocytes at day 1 and day 3. The intensities of DCFDA of CD41+CD61+ cells in panel B (gates depicted in bold rectangles) are shown in the histogram. Average ROS levels obtained from the histograms of day 3 megakaryocytes were changed to relative values against those from day 1 cells. Average values are shown with SD. Statistical significance of the relative ROS levels was calculated using paired t test. (D) Quantitative RT-PCR of platelet genes in day 1 and day 3 megakaryocytes. The relative values were calculated against the values of the day 1 sample. The average values of triplicate experiments are presented, and the error bars represent SD (A,D). The Student t test was used to calculate statistical significance. *P < .05 (A). *P < .001 (D).
Increased p45 and ROS levels correlate with megakaryocytic maturation. (A) Expression of p45 and Nrf2 during megakaryocytic maturation. mRNA levels (left panel) and protein levels in nuclei (right panel) are shown. CD41+ cells were isolated from day 1 and day 3 cultures, and total RNA and nuclear extracts were prepared. The relative values were calculated against the values of day 1 megakaryocytes (left panel). Anti-Nrf2, anti-p45, and anti–lamin B antibodies were used (right panel). (B) Flow cytometric analysis showing megakaryocytic differentiation in day 1 and day 3 cultures. Frequencies of CD41+CD61+ cells are shown in the figure (top panel). Relative frequencies of CD41+CD61+ cells were calculated against the value of day 1 culture (bottom panel). Average values are shown with SD. Statistical significance of the relative ratio of CD41+CD61+ cells was calculated using paired t test. (C) Flow cytometric analysis of intracellular ROS levels in primary megakaryocytes at day 1 and day 3. The intensities of DCFDA of CD41+CD61+ cells in panel B (gates depicted in bold rectangles) are shown in the histogram. Average ROS levels obtained from the histograms of day 3 megakaryocytes were changed to relative values against those from day 1 cells. Average values are shown with SD. Statistical significance of the relative ROS levels was calculated using paired t test. (D) Quantitative RT-PCR of platelet genes in day 1 and day 3 megakaryocytes. The relative values were calculated against the values of the day 1 sample. The average values of triplicate experiments are presented, and the error bars represent SD (A,D). The Student t test was used to calculate statistical significance. *P < .05 (A). *P < .001 (D).
Nrf2 cooperates with p45 in megakaryocytes at the proliferation stage
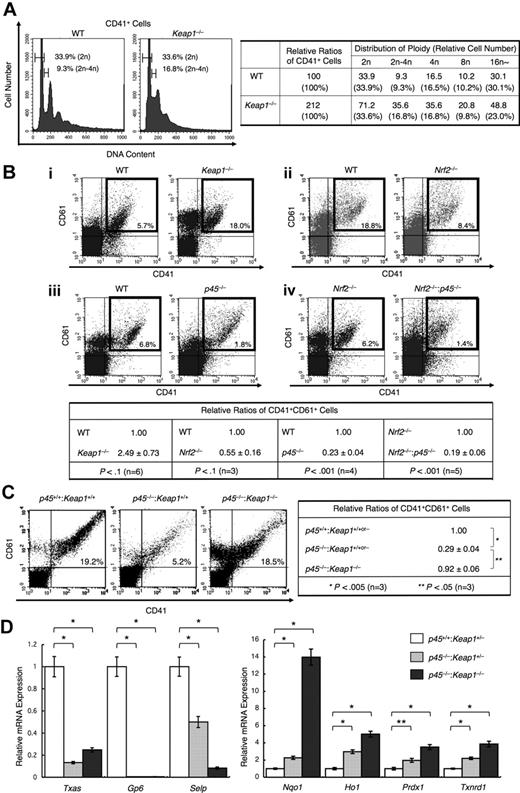
We found that mature megakaryocytes contain increased p45, which indicates that relative levels of Nrf2 to p45 are higher in immature megakaryocytes. Expecting that Nrf2 also plays a role in an earlier stage of megakaryocyte differentiation, we examined the DNA ploidy of Keap1-null megakaryocytes. We found that Keap1 deletion increased the population of megakaryocytes possessing intermediate DNA content between 2n and 4n, ie, cells in S-phase (Figure 6A left panel). Thus, constitutive Nrf2 activation promotes the proliferation of megakaryocytes before the endomitosis stage. The frequency of CD41+CD61+ megakaryocytes generated in the primary culture of Keap1-null fetal liver cells was indeed increased compared with that in WT cultures (Figure 6A right panel, 6Bi).
Nrf2 cooperates with p45 to promote megakaryocytic proliferation but does not activate platelet genes. (A) DNA content of primary megakaryocytes cultured from Keap1-null and control fetal livers (left panel). Frequencies of CD41+ cells containing DNA contents of 2n and between 2n and 4n are indicated. A relative number of CD41+ cells of each ploidy against the total CD41+ cells generated in the WT culture is shown in the right panel. Representative data from 2 independent experiments are shown. (B) Flow cytometric analysis of fetal liver cultures. Cells were stained with FITC-conjugated CD41 and PE-conjugated CD61. Keap1-null cells (i), Nrf2-null cells (ii), and p45-null cells (iii) were compared with control WT cells derived from their corresponding littermates. Nrf2−/−:p45−/− cells were compared with Nrf2-null cells (iv). Representative data are presented from more than 3 independent experiments. Frequencies of CD41+CD61+ cells are shown in the figure. Relative frequencies of CD41+CD61+ cells were calculated against the value of the WT culture (for Keap1-null, Nrf2-null, and p45-null cells) and Nrf2-null culture (for Nrf2−/−:p45−/− cells) (bottom panel). (C) Flow cytometric analysis showing megakaryocytic differentiation in fetal liver cultures from WT, p45-null, and p45−/−:Keap1−/− mice obtained from the same litter. Cells were stained with FITC-conjugated CD41 and PE-conjugated CD61. Frequencies of CD41+CD61+ cells are shown in the figure (left panel). Relative frequencies of CD41+CD61+ cells were calculated against the value of the mixture of WT and Keap1+/− culture (right panel). Average values are shown with SD (B-C). Statistical significance of the relative ratio of CD41+CD61+ cells was calculated using paired t test (B-C). (D) Quantitative RT-PCR of platelet genes and cytoprotective genes in primary megakaryocytes cultured from Keap1+/−, p45−/−:Keap1+/−, and p45−/−:Keap1−/− fetal livers obtained in the same litter. The relative values were calculated against the values of Keap1+/− megakaryocytes. The average values of triplicate experiments are presented, and the error bars represent SD. The Student t test was used to calculate statistical significance. *P < .001. **P < .005.
Nrf2 cooperates with p45 to promote megakaryocytic proliferation but does not activate platelet genes. (A) DNA content of primary megakaryocytes cultured from Keap1-null and control fetal livers (left panel). Frequencies of CD41+ cells containing DNA contents of 2n and between 2n and 4n are indicated. A relative number of CD41+ cells of each ploidy against the total CD41+ cells generated in the WT culture is shown in the right panel. Representative data from 2 independent experiments are shown. (B) Flow cytometric analysis of fetal liver cultures. Cells were stained with FITC-conjugated CD41 and PE-conjugated CD61. Keap1-null cells (i), Nrf2-null cells (ii), and p45-null cells (iii) were compared with control WT cells derived from their corresponding littermates. Nrf2−/−:p45−/− cells were compared with Nrf2-null cells (iv). Representative data are presented from more than 3 independent experiments. Frequencies of CD41+CD61+ cells are shown in the figure. Relative frequencies of CD41+CD61+ cells were calculated against the value of the WT culture (for Keap1-null, Nrf2-null, and p45-null cells) and Nrf2-null culture (for Nrf2−/−:p45−/− cells) (bottom panel). (C) Flow cytometric analysis showing megakaryocytic differentiation in fetal liver cultures from WT, p45-null, and p45−/−:Keap1−/− mice obtained from the same litter. Cells were stained with FITC-conjugated CD41 and PE-conjugated CD61. Frequencies of CD41+CD61+ cells are shown in the figure (left panel). Relative frequencies of CD41+CD61+ cells were calculated against the value of the mixture of WT and Keap1+/− culture (right panel). Average values are shown with SD (B-C). Statistical significance of the relative ratio of CD41+CD61+ cells was calculated using paired t test (B-C). (D) Quantitative RT-PCR of platelet genes and cytoprotective genes in primary megakaryocytes cultured from Keap1+/−, p45−/−:Keap1+/−, and p45−/−:Keap1−/− fetal livers obtained in the same litter. The relative values were calculated against the values of Keap1+/− megakaryocytes. The average values of triplicate experiments are presented, and the error bars represent SD. The Student t test was used to calculate statistical significance. *P < .001. **P < .005.
Conversely, we found that Nrf2 deletion reduced the frequency of CD41+CD61+ megakaryocytes generated in the fetal liver cell culture (Figure 6Bii). The number of CD41+CD61+ megakaryocytes was reduced in the p45-null fetal liver cell culture compared with that of the WT cell culture (Figure 6Biii), which is consistent with a previous report.17 We assumed that p45 and Nrf2 might promote megakaryocytic proliferation cooperatively. Indeed, in our analysis of the primary culture of fetal liver cells, a further reduction in the frequency of CD41+CD61+ megakaryocytes was observed in the Nrf2 and p45 double-knockout cell culture compared with that in the single Nrf2 knockout cell culture (Figure 6Biv). These results indicate that, in immature stages, Nrf2 contributes to enhanced megakaryocytic proliferation in cooperation with p45.
Requirement of p45 for platelet gene expression
We then asked whether Nrf2 compensates for the function of p45 when Nrf2 is constitutively activated in the absence of Keap1. We generated p45−/−:Keap1−/− mice by intercrossing p45+/−:Keap1+/− mice. Fetal liver cells prepared from p45−/−:Keap1−/− and WT littermates were cultured in vitro. Importantly, although the number of CD41+CD61+ cells was reduced by p45 deletion (Figure 6C left and middle panels), this reduction was substantially rescued by the simultaneous deletion of Keap1 (Figure 6C right panel). This result indicates that Nrf2 can compensate for the impaired megakaryocytic proliferation observed in p45-null cells. In contrast, although the platelet gene expression was reduced in p45-null megakaryocytes, this reduction was not rescued by the simultaneous depletion of Keap1 (Figure 6D left panel). Elevated expression of cytoprotective genes resulting from the absence of p45 was further increased by the Keap1 depletion (Figure 6D right panel). Thus, Nrf2 compensated for the loss of p45 at the immature stages, specifically to enhance proliferation. However, at mature stages, Nrf2 was not capable of substituting for p45 to induce platelet gene expression.
Discussion
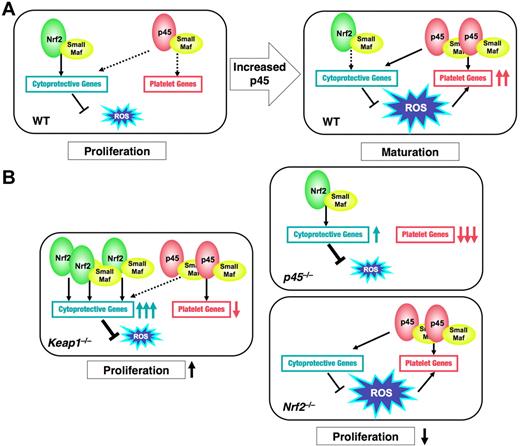
In this study, we identified 2 categories of NF-E2 p45 target genes: platelet genes and cytoprotection genes. Our analyses revealed that the platelet genes are uniquely regulated by p45, whereas the cytoprotective genes are common targets of p45 and Nrf2. The functional domination by p45 over Nrf2 favors ROS accumulation by limiting the expression level of the cytoprotective genes. This ROS accumulation promotes the full activation of platelet genes. Whereas both p45 and Nrf2 function as transcriptional activators, their activities, as well as their contributions to the oxidative stress response in cells, differ considerably. Nrf2 strongly activates the expression of cytoprotective genes in response to ROS and maintains cellular ROS at low levels. By contrast, p45 maintains moderate expression of the cytoprotective genes irrespective of ROS levels, which underlies the ROS accumulation in mature megakaryocytes. Nrf2 increased the expression of cytoprotective genes and reduced cellular ROS levels in the absence of p45. This novel mechanism, involving balancing the activities of 2 CNC factors, is summarized in Figure 7.
Model for p45 and Nrf2-regulated megakaryocytic maturation. (A) In WT megakaryocytes at an immature stage (WT, left panel), Nrf2 seems to make a major contribution to cytoprotective gene regulation, which could promote cell proliferation. In mature megakaryocytes (WT, right panel), p45, as a modest activator, seems to maintain the moderate expression of cytoprotective genes by competing with Nrf2 and to enhance ROS accumulation. Increased ROS cooperates with p45 to promote platelet gene expression. (B) In p45-null cells, platelet genes are dramatically repressed and cytoprotective genes are induced by Nrf2, resulting in the reduction of ROS levels. In Nrf2-null cells, neither gene expressions nor ROS levels are changed. In both cases, proliferation of megakaryocytes is impaired (right panels). In Keap1-null cells, constitutively activated Nrf2 strongly activates cytoprotective genes but not platelet genes. ROS accumulation is inhibited, and proliferation of immature megakaryocytes is promoted (left panel). Solid and broken black arrows indicate explicit and ineffective interaction, respectively.
Model for p45 and Nrf2-regulated megakaryocytic maturation. (A) In WT megakaryocytes at an immature stage (WT, left panel), Nrf2 seems to make a major contribution to cytoprotective gene regulation, which could promote cell proliferation. In mature megakaryocytes (WT, right panel), p45, as a modest activator, seems to maintain the moderate expression of cytoprotective genes by competing with Nrf2 and to enhance ROS accumulation. Increased ROS cooperates with p45 to promote platelet gene expression. (B) In p45-null cells, platelet genes are dramatically repressed and cytoprotective genes are induced by Nrf2, resulting in the reduction of ROS levels. In Nrf2-null cells, neither gene expressions nor ROS levels are changed. In both cases, proliferation of megakaryocytes is impaired (right panels). In Keap1-null cells, constitutively activated Nrf2 strongly activates cytoprotective genes but not platelet genes. ROS accumulation is inhibited, and proliferation of immature megakaryocytes is promoted (left panel). Solid and broken black arrows indicate explicit and ineffective interaction, respectively.
The gene expression profiling analysis described herein revealed that multiple cytoprotective genes, which are well-established Nrf2 target genes, are up-regulated in the p45-null megakaryocytes. Because we detected p45 occupancy at the regulatory regions of these genes in our genome-wide ChIP-sequence analysis of p45 in megakaryocytes (R.F., H.M., E.H.B., H.A., and M.Y., unpublished observation, June 2009), we surmise that p45 limits the expression of these cytoprotective genes by competing with Nrf2. In contrast, the platelet genes were dramatically repressed in p45-null megakaryocytes. Because p45 was occupied the presumptive regulatory regions of the platelet genes as well (R.F., H.M., E.H.B., H.A., and M.Y., unpublished observation, June 2009), we conclude that p45 directly activates the platelet genes. Thus, it is interesting that Nrf2 could not rescue platelet gene expression even if the nuclear concentration of Nrf2 was increased in the absence of Keap1. Although Nrf2 and p45 bind similar MARE sequences, it is formally possible that the distinct contributions of p45 and Nrf2 to platelet gene expression are attributable to distinct binding affinities to specific MARE-related sequences in chromatin. Alternatively, different cofactors may be recruited by the 2 proteins resulting from the structural differences between p45 and Nrf2, consistent with the important capacity of the N-terminal region of Nrf2 to confer strong transcriptional activity (Figure 1F). Another possible explanation is that Nrf2 may not be able to establish the requisite chromatin environment to permit a sufficient level of platelet gene activation.
We demonstrated that the ratio of p45 to Nrf2 increases during megakaryocytic maturation, and p45 competes with Nrf2 for the regulation of cytoprotective genes in mature megakaryocytes. This is in contrast to the situation in immature megakaryocytes, in which p45 appears to cooperate with Nrf2 to promote megakaryocytic proliferation. We suggest that increased p45 expression triggers the switching from Nrf2-p45 cooperation to competition (Figure 7A). Alternatively, certain differentiation-dependent signaling cascades may change p45 activity, as the transcriptional activity of NF-E2 is enhanced by the activation of Ras-MAP kinase32 and cyclic adenosine monophosphate-dependent protein kinase.33,34 These phosphorylation signals may modify p45 or its interacting cofactors, leading to increased p45-dependent transcription of platelet genes.
We demonstrated that Keap1 deletion induces the nuclear accumulation of Nrf2 and increases the number of megakaryocytes in a fetal liver cell culture. These results indicate that Nrf2 promotes the proliferation of immature megakaryocytes. It is interesting to note that Nrf2 enhances cell proliferation in cancer cells.35 Point mutations in Keap136 and Nrf237 characterize various human cancers, which give rise to Keap1 inactivation and Nrf2 stabilization. In both cases, Nrf2 is constitutively activated in the cancer cells, rendering cancer cells resistant to oxidative stress, which correlates with a poor prognosis.37 These results further suggest that transcriptional activation through MARE may promote the proliferation of immature megakaryocytes.
Transcription factors that share DNA-binding proteins compete for occupancy of common cis-elements. For example, a switching of GATA transcription factors occurs during erythroid differentiation. This GATA switch involves GATA1-mediated displacement of GATA2 from chromatin during the differentiation from the early progenitors to erythroblasts.38,39 Furthermore, hemin stimulation of NIH3T3 cells and DMSO treatment of MEL cells induce a switch from Bach1 to Nrf2 and from Bach1 to p45 at the MARE motifs of Ho-1 gene and β-Globin gene, respectively.40,41 These latter cases illustrate competitive regulation between a repressor and an activator because Bach1 is a repressive member of the CNC family. The regulation of cytoprotective genes by p45 and Nrf2 in megakaryocytes is unique, as the domination of a potent activator Nrf2 by a less efficacious activator p45 establishes a permissive environment for ROS accumulation without completely shutting off the stress response pathway.
An important challenge that remains is the elucidation of the primary source of ROS in megakaryocytes. Several hematopoietic growth factors, including TPO, interleukin-3, and granulocyte-macrophage colony stimulating factor, induce ROS and promote cell proliferation.42 In addition to intrinsic ROS, megakaryocytic maturation might be influenced by exogenous ROS, such as lipid peroxides in hyperlipidemia and hyperglycemia. Platelets produced from megakaryocytes in an oxidative environment may acquire hyperaggregability because of elevated expression of platelet gene products that determine platelet reactivity. We think that the present study provides the first step into this new area of research.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Kazuhiko Igarashi for critical reading of the manuscript, Ms Hiroko Meguro and Dr Shogo Yamamoto for assistance with the microarray analysis, Kirin Pharma Co Ltd for providing recombinant TPO, and Ms Eriko Naganuma and the Biomedical Research Core of Tohoku University Graduate School of Medicine for technical support.
This work was supported by the National Institutes of Health (grant DK50107; E.H.B.), Grants-in-Aid for Creative Scientific Research (M.Y.), Scientific Research on Priority Areas (H.M., M.Y.), Scientific Research (H.M., M.Y.), and the Cell Science Research Foundation (H.M.).
National Institutes of Health
Authorship
Contribution: H.M. designed and performed research, collected and analyzed data, and wrote the paper; M.K., R.F., A.I., M.T., and X.P. performed research and collected data; F.K. designed research, interpreted data, and wrote the paper; H.A. contributed vital analytical tools; E.H.B. contributed vital analytical reagents and wrote the paper; and M.Y. interpreted data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hozumi Motohashi, Center for Radioisotope Sciences, Tohoku University Graduate School of Medicine, 2-1 Seiryo-cho, Aoba-ku, Sendai 980-8575, Japan; e-mail: hozumim@m.tains.tohoku.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal