Abstract
Splicing mutations account for approximately 10% of lesions causing genetic diseases, but few branchpoint sequence (BPS) lesions have been reported. In 3 families with autosomal recessive congenital erythropoietic porphyria (CEP) resulting from uroporphyrinogen III synthase (URO-synthase) deficiency, sequencing the promoter, all 10 exons and the intron/exon boundaries did not detect a mutation. Northern analyses of lymphoblast mRNAs from 2 patients and reverse-transcribed polymerase chain reaction (RT-PCR) of lymphoblast mRNAs from all 3 patients revealed multiple longer transcripts involving intron 9 and low levels of wild-type message. Sequencing intron 9 RT-PCR products and genomic DNA in each case revealed homozygosity for a novel BPS mutation (c.661-31T→G) and alternatively spliced transcripts containing 81, 246, 358, and 523 nucleotides from intron 9. RT-PCR revealed aberrant transcripts in both wild-type and CEP lymphoblasts, whereas BPS mutation reduced the wild-type transcript and enzyme activity in CEP lymphoblasts to approximately 10% and 15% of normal, respectively. Although the +81-nucleotide alternative transcript was in-frame, it only contributed approximately 0.2% of the lymphoblast URO-synthase activity. Thus, the BPS mutation markedly reduced the wild-type transcript and enzyme activity, thereby causing the disease. This is the first BPS mutation in the last intron, presumably accounting for the observed 100% intron retention without exon skipping.
Introduction
Congenital erythropoietic porphyria (CEP) is an autosomal recessive inborn error of heme biosynthesis resulting from markedly deficient, but not absent, activity of the fourth enzyme in the heme biosynthetic pathway, uroporphyrinogen III synthase (URO-synthase; EC 4.2.1.75).1 This enzyme catalyzes the conversion of the linear tetrapyrrole, hydroxymethylbilane to uroporphyrinogen (URO'gen) III, the only isomer of URO'gen that can ultimately be converted to heme.2,3 Deficient activity of URO-synthase in CEP patients results in the nonenzymatic cyclization of hydroxymethylbilane to URO'gen I. An excess of URO'gen I and COPRO'gen I and their oxidized forms, uroporphyrin (URO) I and coproporphyrin (COPRO) I, primarily in erythrocytes, leads to hemolysis. The released porphyrins accumulate in tissues and bones and are excreted in the urine and feces. Light activates the photocatalytic URO I and COPRO I isomers, resulting in tissue damage and the formation of bullous cutaneous lesions that rupture, often becoming infected, and lead to bone resorption and cutaneous deformity.4
The clinical manifestations of CEP are markedly heterogeneous, dependent primarily on the amount of residual URO-synthase activity, and range from nonimmune hydrops fetalis to milder, later-onset forms.1,4 Severely affected patients are transfusion-dependent throughout life and have splenomegaly and marked cutaneous involvement leading to severe scarring and/or deformities. Additional manifestations may include hypertrichosis, alopecia, and erythrodontia. Patients with a milder phenotype, who have sufficient residual URO-synthase activity to synthesize heme for hemoglobin and other essential heme-requiring proteins, are not transfusion-dependent and may only exhibit cutaneous involvement.
The approximately 34-kb human URO-synthase gene (UROS), located at chromosome 10q25.3-26.3, has 10 exons and 2 alternative promoters that generate housekeeping- and erythroid-specific transcripts5 that express the same enzyme polypeptide of 265 amino acids (molecular mass ∼ 29.5 kDa). To date, 38 mutations have been identified in unrelated CEP patients (Human Gene Mutation Database; http://www.hgmd.cfac.uk/). These include 24 missense mutations, 1 nonsense, 2 splice site (c.63 + 1G → A;c.245-2A → T), 2 deletions, 4 insertions, and 1 complex rearrangement. Only the C73R missense mutation is common, occurring in approximately one-third of the CEP alleles studied, whereas the other mutations have been detected in only 1 to 3 presumably unrelated families.4,6 Most of the missense mutations expressed in Escherichia coli had low activity (< 2% of wild-type), but several were found to have significant residual activity, which appeared to correlate with the milder, later-onset phenotypes.4
Disease-causing point mutations in the branchpoint consensus sequence (BPS) are rare. To date, 17 BPS mutations causing human disease have been described (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Of the 17, all but 2 are either mutations of the consensus branching adenosine or the −2 thymidine (consensus BPS = YNYTRAY). Exon skipping, intron retention, partial intron retention resulting from activation of cryptic 3′-splice sites, and combinations of these events have been observed, which presumably depends on the particular sequence context in each gene. Here, we report a novel branchpoint mutation in the last intron of the UROS gene in 3 CEP patients that resulted in at least a 90% reduction of the wild-type 1.5-kb message. Multiple alternatively spliced mRNAs were detected, ranging in size from 1.6 to 2.0 kb. Characterization and quantitation of the alternatively spliced mRNAs revealed that they were present in both normal and patient cells and resulted from retention of intronic sequences because of the use of cryptic splice sites in intron 9. There was no evidence of exon skipping. Of the alternatively spliced transcripts, only the +81 nucleotide (nt) in-frame insertion transcript encoded an active enzyme that, when pure, expressed approximately 4% of wild-type prokaryotically expressed enzymatic activity and when in crude extracts expressed 0.6% of wild-type activity. Thus, the branchpoint mutation caused a marked reduction of the normal transcript and encoded enzyme, thereby causing the disease in these patients.
Methods
Patients
Peripheral blood samples were collected at the Mount Sinai School of Medicine in New York with informed consent from 9 unrelated CEP patients and their family members. All studies involving patient samples were performed with approval from the Mount Sinai Institutional Review Board in accordance with the Declaration of Helsinki. Lymphoid cell lines were established as previously described.7 Patients 3 and 6 were 15- and 20-year-old white men of Ashkenazi descent. Their parents were not related to each other. Patient 3 was severely affected from birth with marked photosensitivity, hepatosplenomegaly, and anemia (hemoglobin 5 g/dL at 20 days of life8 ). He was treated with red cell transfusions until age 14, when worsening photosensitivity was managed with continued chronic hemotransfusions and marrow suppression with hydroxyurea.9 Patient 6 required red cell transfusions but had significant periods during adolescence without treatment. He had marked cutaneous involvement resulting from unprotected exposure to sunlight.
Patient 10, studied in Zurich, was a 44-year-old man of Lebanese descent, who was the product of consanguineous parents.10 He had had chronic, progressive skin ulcerations since adolescence that eventually disfigured his sun-exposed face and hands. CEP was recognized when he was in his 20s. After his initial evaluation, he remained moderately anemic with hemoglobins ranging from 10 to 12 g/dL over the next decade. At age 56, his hemoglobin was 8 g/dL and red cell transfusions were administered every 3 weeks.
The clinical diagnosis of CEP was confirmed in patients 3, 6, and 10 by finding markedly elevated levels of URO I in the urine. Erythrocyte URO-synthase activities could not be determined, as all patients were receiving transfusions of normal erythrocytes.
Initial molecular studies of patients 3 and 6 in New York included Northern, Southern, and allele-specific hybridization analyses, followed by the identification and characterization of the causative UROS BPS mutation. Subsequently, patient 10 was found to have the same BPS mutation in Zurich. Quantitative reverse-transcribed polymerase chain reaction (RT-PCR) studies were done on all 3 patients.
Northern hybridization analyses
Total RNA was isolated from cultured lymphoblasts by a guanidine isothiocyanate procedure, and poly(A)+ RNA was isolated by oligo (dT)-cellulose chromatography (type 3; Collaborative Research). Poly(A)+ RNA (5 μg) was analyzed by electrophoresis through 1% formaldehyde-agarose gels and transferred by capillary blotting to a nitrocellulose membrane (Schleicher and Schuell) and hybridized with the full-length UROS cDNA labeled with [α-32P]dCTP by random priming using the NEBlot kit according to the manufacturer's instructions (New England Biolabs).
cDNA synthesis amplification, cloning, and Southern hybridization analyses
First-strand cDNA was reverse-transcribed from 1 μg of poly(A)+ RNA using the AMV Reverse Transcriptase System (Invitrogen) and primer AS1, AS2, or AS3 (supplemental Table 2). The cDNA template (1 μg; 10% of the reaction) was PCR-amplified for 30 cycles. Primer set 1, located in exons 1 and 8, amplified the 5′ half of the cDNA; primer set 2, located in exon 7 and in the 3′ UTR of exon 10 amplified the 3′ half of the cDNA; and primer set 3, located in exons 9 and 10 amplified intron 9 (supplemental Table 2). The amplicons were analyzed by electrophoresis in 1.5% agarose gels.
The PCR products were digested with EcoRI, gel purified, and ligated into the EcoRI site of the pGEM4Z vector (Promega). Clones corresponding to the observed agarose gel bands were sequenced using an ABI 3730xl capillary array sequencer (Applied Biosystems) to determine the identity of the alternatively spliced transcripts.
Genomic DNA amplification and sequencing
Genomic DNAs for patients 1 to 10 were extracted from cultured lymphoblasts or isolated peripheral leukocytes, and each PCR-amplified UROS exon and flanking intronic region was sequenced. For detection of the mutation in intron 9, forward primer 5′-CAGTAACGTCCAACCGCAAAG-3′ and reverse primer 5′-CAGGTCAGGTCCCGATCCC-3′ were used to amplify a 407-bp segment of genomic DNA containing a portion of intron 9 adjacent to exon 10 and the entire exon 10. Primers for the amplification of a segment of the UROS cDNA spanning from exon 9 to exon 10 (nt 591-903) were: TCCCTCTGGCCTCACATACAG-3′ (forward) and 5′-GGAGTCTGACGGCAGC-3′ (reverse).
ASO hybridization analysis
UROS exon 10 with flanking intronic sequences was amplified with primer set 4 (supplemental Table 2) as described in “cDNA synthesis amplification, cloning, and Southern hybridization analyses.” The 624-bp amplicon was applied to duplicate nylon membranes (Zeta-Probe; Bio-Rad) and hybridized overnight at 55°C to 5′-[α-32P]dATP-radiolabeled allele-specific oligonucleotide wild-type (ASOwt) and mutant (ASOmut) allele-specific probes (supplemental Table 2, with the nucleotide corresponding to c.661-31 in bold type). Washing was performed at 55°C for the normal ASO and at 57°C for the mutant ASO.
Quantitative RT-PCR analyses
For quantitative real-time polymerase chain reaction (qRT-PCR) of the UROS wild-type and alternatively spliced transcripts from patients 3 and 6, total RNA was isolated from cultured lymphoblasts using TRI-Reagent (Ambion). Real-time PCR was performed using TaqMan Universal PCR 2X MasterMix and the SYBR Green PCR core reagents, both from Applied Biosystems. The primers used are specified in supplemental Table 2. Quantitation was performed on an ABI PRISM 7900HT (Applied Biosystems) normalized to the average of the glyceraldehyde-3-phosphate dehydrogenase, α-actin, β-tubulin, and rps11 transcript levels. The cycle threshold values were determined in triplicate for 2 or 3 RNA samples from each person. Relative quantitation and propagation of error calculations were carried out as described.11 The calibrator was the mean wild-type transcript level in normal controls, and the amounts of each transcript were expressed as a percentage of the total wild-type and alternatively spliced transcripts in normal persons. Similarly, for quantitation of wild-type and the abnormal transcripts of patient 10, total RNA was isolated from peripheral blood using the PAXgene Blood RNA Validation Kit (QIAGEN). Reverse transcription of 1-μg aliquots of RNA into cDNA was accomplished using the First Strand cDNA Synthesis Kit for reverse-transcribed polymerase chain reaction (RT-PCR; Roche Applied Sciences). The PCR primers and TaqMan probe sets (supplemental Table 2) were used to quantify the UROS cDNA by real-time PCR ABI PRISM 7000 (Applied Biosystems). Quantitation was normalized relative to the average of the 18S and β-actin RNAs.
Computer-assisted analyses
DNA sequence analyses were performed using the MacVector 9 DNA sequence analysis program (MacVector). Exon splice enhancers (ESEs) were identified using the ESEfinder3 program: http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home.12,13 The thresholds for significant SF2/ASF and SC35 scores were set at more than or equal to 4.0 and more than or equal to 4.5, respectively. Potential cryptic splice sites were identified using the NetGene2 program: http://www.cbs.dtu.dk/services/NetGene2/.14 The program's reported 5′-donor and 3-acceptor “Confidence Scores” were used as a measure of splicing potential and termed “Splice Scores” in the text.
In silico splice site analysis and variant enzyme structure prediction
Of note, the 81-bp intronic region common to all of the alternatively spliced transcripts was predicted to be an exon using the GENESCAN program,15 indicating that this region was suitable for splicing. Putative cryptic exons and their associated splice site scores, obtained using the NetGene2 program,16 the associated BPS and polypyrimidine sequences, and their 5′ and 3′ splice scores are shown in Table 1.
Potential cryptic exons in UROS intron 9
| Exon* . | BPS coordinate† . | BPS‡ . | Δ§ . | Polypyrimidine‖ . | 5′splice score¶ . | Exon size . | ESE/kb# . | 3′splice score¶ . |
|---|---|---|---|---|---|---|---|---|
| Exon 9 | 143 | tcttgat | 22 | gttttctgttccccacag | 0.56 | 99 | 0 | 0.83 |
| CE1 | 767 | ccttgaa* | 114 | tcacctgccttctcacag | 0.53 | 281 | 3.5 | 0.65 |
| CE2 | 2089 | ctctgag* | 35 | cctcgcctcttactacag | 0.25 | 208 | 9.6 | 0.54 |
| CE3 | 4868 | ttttgaa | 53 | actttattttttcctcag | 0.33 | 148 | 13.5 | 0.80 |
| CE4 | 5566 | *atttgat | 49 | atcttttcggtttgacag | 0.69 | 246 | 28.5 | 0.95 |
| CE5 | 5719 | ccttgat | 61 | catcctccccttgatcag | 0.65 | 81 | 37.0 | 0.95 |
| Exon 10 | 6110 | tgctgaa* | 28 | tctcttctgtctttatag | 0.94 | 423 | 26.0 | Term |
| Consensus | ynytray |
| Exon* . | BPS coordinate† . | BPS‡ . | Δ§ . | Polypyrimidine‖ . | 5′splice score¶ . | Exon size . | ESE/kb# . | 3′splice score¶ . |
|---|---|---|---|---|---|---|---|---|
| Exon 9 | 143 | tcttgat | 22 | gttttctgttccccacag | 0.56 | 99 | 0 | 0.83 |
| CE1 | 767 | ccttgaa* | 114 | tcacctgccttctcacag | 0.53 | 281 | 3.5 | 0.65 |
| CE2 | 2089 | ctctgag* | 35 | cctcgcctcttactacag | 0.25 | 208 | 9.6 | 0.54 |
| CE3 | 4868 | ttttgaa | 53 | actttattttttcctcag | 0.33 | 148 | 13.5 | 0.80 |
| CE4 | 5566 | *atttgat | 49 | atcttttcggtttgacag | 0.69 | 246 | 28.5 | 0.95 |
| CE5 | 5719 | ccttgat | 61 | catcctccccttgatcag | 0.65 | 81 | 37.0 | 0.95 |
| Exon 10 | 6110 | tgctgaa* | 28 | tctcttctgtctttatag | 0.94 | 423 | 26.0 | Term |
| Consensus | ynytray |
CE indicates cryptic exons. CE4 includes CE5, as there was no 5′ splice donor sequence in this 246-bp region.
Position of the BPS adenosine in the UROS genomic sequence where base 1 = 165 bases 5′ of exon 9.
The match to the BPS consensus YNYTRAY with one allowed mismatch in the first or last position as noted with an asterisk.
Δ indicates number of bases between the branchpoint adenosine and the first base of the putative or wild-type exon.
Match to the pyrimidine tract consensus (Y14)NCAG with 4 allowed mismatches in the 14 pyrimidine residues. An exception was made for exon 10 where the consensus (Y14)NYAG was permitted.
The 5′ splice donor and 3′ splice acceptor scores ranged from 0 to 1, and the cut-offs for acceptable scores were 0.5 and 0.2, respectively.
Density of high-scoring exonic splice enhancers SC-35 and SF2 per kilobase of sequence.
Prokaryotic expression, thermostability, and enzyme assay
The novel 27 in-frame residues encoded by the +81 nt alternatively spliced transcript were introduced by site-directed mutagenesis into the wild-type PKK-UROS prokaryotic expression construct, and the crude extracts were assayed for URO-synthase enzymatic activity as previously described.17 These constructs were also cloned into a high-level pET SUMO expression vector as a SUMO fusion protein, cleaved, and purified to homogeneity as previously described.18 The pure wild-type and mutant enzymes were incubated in 20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4, 0.1M NaCl, and 1mM dithiothreitol, for various times at 37°C, transferred to an ice bath, and then assayed to determine thermostability.17
Results
Detection and characterization of abnormal mRNAs
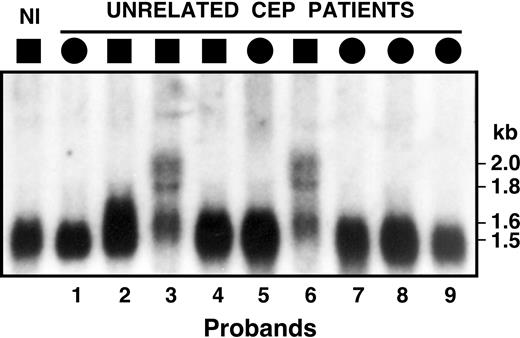
Among the 10 clinically diagnosed CEP patients with markedly elevated urinary URO I and COPRO I levels, including patients 3 and 6 in New York, and subsequently, patient 10 in Zurich, sequencing of the UROS exons, adjacent intron/exon boundaries, and promoter regions did not identify causative mutations. To screen for mRNA processing errors, lymphoblast poly(A)+ RNAs were analyzed from CEP patients 1 through 9 by Northern hybridization using the full-length UROS cDNA as a probe. As shown in Figure 1, patients 3 and 6 had markedly reduced amounts of the wild-type 1.5-kb transcript and readily observable amounts of several larger mRNA species with hybridization bands of approximately 1.6, 1.8, and 2.0 kb, whereas the other CEP patients and a normal control had a single UROS mRNA band of approximately 1.5 to 1.6 kb.
Northern hybridization analysis of UROS poly(A)+ RNA isolated from lymphoblasts of unrelated CEP patients. A single 1.5-kb transcript was observed for the normal person (Nl) and the CEP patients, with the exception of patients 3 and 6 who had longer transcripts of 1.6, 1.8, and 2.0 kb, suggesting that their CEP mutation(s) altered UROS pre-mRNA splicing.
Northern hybridization analysis of UROS poly(A)+ RNA isolated from lymphoblasts of unrelated CEP patients. A single 1.5-kb transcript was observed for the normal person (Nl) and the CEP patients, with the exception of patients 3 and 6 who had longer transcripts of 1.6, 1.8, and 2.0 kb, suggesting that their CEP mutation(s) altered UROS pre-mRNA splicing.
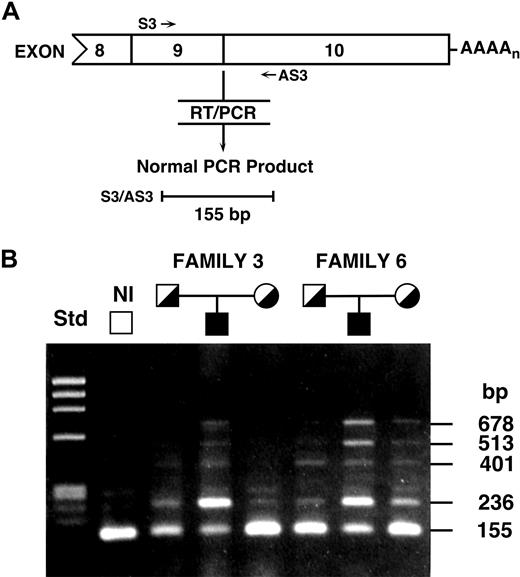
To determine the nature of the longer transcripts in patients 3 and 6, mRNAs isolated from cultured lymphoblasts were reverse-transcribed and exons 1 through 8 and 7 through 10 were amplified using primer sets 1 and 2, respectively (supplemental Table 2). In these CEP patients, PCR amplification from exons 1 through 8 generated only 1 amplicon of expected size, whereas amplification of exons 7 through 10 revealed longer amplicons in patients 3 and 6 and, to a lesser degree, in their parents (Figure 2). In the parents of patients 3 and 6, RT-PCR of UROS exons 9 through 10 (primer set 3; supplemental Table 2) generated predominantly the normal-sized 155-bp product as well as smaller quantities of some of the longer amplicons seen in patients 3 and 6 (Figure 2), whereas the normal control cDNA had primarily the expected 155-bp product with small amounts of longer amplicons. The cDNAs from patients 3 and 6 had variably reduced amounts of the 155-bp product from the normal transcript and additional longer amplicons of 236, approximately 282, 401, 513, and 678 bp. The variable degree of the normal mRNA amplicon in patients 3 and 6, and its greater amount compared with the normal transcript in the Northern analysis, was presumably the result of the more efficient amplification of shorter PCR products and probably equalization of band intensities at 30 PCR cycles. These results suggested the occurrence of the same or similar UROS hnRNA alternative splicing in patients 3 and 6.
RT-PCR of UROS mRNAs from lymphoblasts. RT-PCR analysis of the UROS mRNAs from lymphoblasts of patients 3 and 6, their parents, and a normal person demonstrated that the presence of additional sequences between exons 9 and 10 was responsible for the larger mRNAs. (A) Schematic overview of the RT-PCR analysis of alternative splicing in intron 9. The sequences of primers S3 and AS3 are provided in supplemental Table 2. (B) Agarose gel electrophoresis and ethidium bromide staining of UROS RT-PCR products revealed the 4 longer products. Sequencing the longer amplicons in patients 3 and 6 determined their location in intron 9 and their sizes of 236, 401, 513, and 678 bp in addition to the normal 155-bp product.
RT-PCR of UROS mRNAs from lymphoblasts. RT-PCR analysis of the UROS mRNAs from lymphoblasts of patients 3 and 6, their parents, and a normal person demonstrated that the presence of additional sequences between exons 9 and 10 was responsible for the larger mRNAs. (A) Schematic overview of the RT-PCR analysis of alternative splicing in intron 9. The sequences of primers S3 and AS3 are provided in supplemental Table 2. (B) Agarose gel electrophoresis and ethidium bromide staining of UROS RT-PCR products revealed the 4 longer products. Sequencing the longer amplicons in patients 3 and 6 determined their location in intron 9 and their sizes of 236, 401, 513, and 678 bp in addition to the normal 155-bp product.
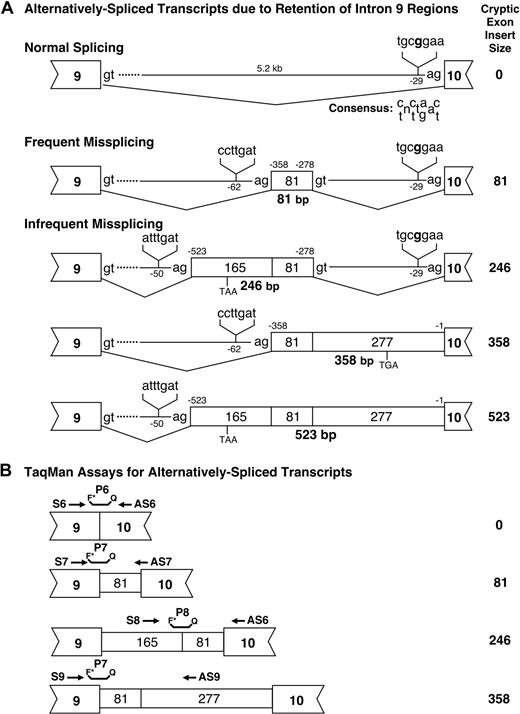
The individual abnormal RT-PCR products from patients 3 and 6 were subcloned and sequenced. Comparison of the 5.2-kb UROS intron 9 sequence with that of the abnormal RT-PCR products from these patients revealed that the alternatively spliced transcripts retained 1 or more intron 9 sequences of 81, 165, or 277 bp, respectively (Figure 3A). The 236-bp RT-PCR product (Figure 2) was the result of retention of an 81-bp intronic region in the UROS transcript (after codon 220 in the wild-type sequence) that predicted the in-frame addition of 27 amino acids. The +165 + 81-bp variant (401-bp RT-PCR product; Figures 2–3) encoded a 22-amino acid novel peptide after codon 220 with a premature termination TAA at codon 243. The +81 + 277-bp variant (513 bp, Figure 2) encoded a novel 91-amino acid extension terminating in TGA (codon 311). Finally, the +165 + 81 + 277-bp variant (678-bp RT-PCR product; Figures 2–3) encoded the same truncation product as did the 401-bp product (Figures 2–3). In all cases, except for the in-frame 27-amino acid insertion, the variant proteins lacked the 45-amino acid terminal region of UROS encoded by exon 10.
Analysis of alternative splicing. (A) Splicing in normal persons and CEP patients. Genomic positions of the alternative splice sites are indicated as base pairs before the first residue of exon 10 and the positions of the lariat “A” in their respective putative BPSs are indicated as base pairs before the first exonic residue of the nearest exon. The sizes of the cryptic exon inserts noted in Figure 2 are listed in the right column. (B) Design of TaqMan assays for quantitation of the normal and alternatively spliced transcripts. The primer sequences are listed in supplemental Table 2.
Analysis of alternative splicing. (A) Splicing in normal persons and CEP patients. Genomic positions of the alternative splice sites are indicated as base pairs before the first residue of exon 10 and the positions of the lariat “A” in their respective putative BPSs are indicated as base pairs before the first exonic residue of the nearest exon. The sizes of the cryptic exon inserts noted in Figure 2 are listed in the right column. (B) Design of TaqMan assays for quantitation of the normal and alternatively spliced transcripts. The primer sequences are listed in supplemental Table 2.
Expression, thermostability, and enzymatic activity of the variant enzyme
The additional 27-amino acid peptide of the URO-synthase variant protein encoded by the +81 nt in-frame insertion transcript was located far from the active site, so to determine whether it had any residual enzyme activity the +81-bp insertion transcript was prokaryotically expressed and compared with the wild-type construct, the pKK vector alone, and a previously reported CEP missense mutation (p.Ala69Thr) as a control. As shown in Table 2, in crude extracts, the +27-amino acid mutant protein had 0.6% of the expressed wild-type activity, whereas the p.A69T missense mutation had a residual activity similar to that previously reported, 1.4% of expressed wild-type activity.19 In addition, the +27-amino acid mutant protein was expressed from the pET SUMO construct, purified to homogeneity, and shown to have only 4.2% of wild-type expressed activity (Table 2). The pure wild-type and mutant enzymes were both stable at pH 7.4 and 37°C for 1 hour. In contrast, the other frame-shifted alternatively spliced transcripts all predicted the incorporation of different amino acids, with premature termination, which deleted the terminal 45 amino acids encoded by exon 10.
Prokaryotic expression of wild-type and alternatively spliced UROS transcripts
| . | Specific activity,* U/mg . | Net (-vector control), U/mg . | Percentage of wild-type . |
|---|---|---|---|
| pKK construct | |||
| Wild-type (n = 2) | 439.5 ± 3.1 | 431.6 | 100 |
| pKK vector (n = 3) | 7.9 ± 8.8 | 0 | NA |
| +81-bp insertion (n = 3) | 10.5 ± 7.3 | 2.6 | 0.6 |
| Mutation p.Ala69Thr† (n = 3) | 17.4 ± 0.8 | 9.5 | 2.2 |
| pET SUMO construct | |||
| Wild-type (n = 5) | 2.52 ± 0.13 × 106 | NA‡ | 100 |
| +81-bp insertion (n = 3) | 0.105 ± 0.008 × 106 | NA‡ | 4.2 ± 0.3 |
| . | Specific activity,* U/mg . | Net (-vector control), U/mg . | Percentage of wild-type . |
|---|---|---|---|
| pKK construct | |||
| Wild-type (n = 2) | 439.5 ± 3.1 | 431.6 | 100 |
| pKK vector (n = 3) | 7.9 ± 8.8 | 0 | NA |
| +81-bp insertion (n = 3) | 10.5 ± 7.3 | 2.6 | 0.6 |
| Mutation p.Ala69Thr† (n = 3) | 17.4 ± 0.8 | 9.5 | 2.2 |
| pET SUMO construct | |||
| Wild-type (n = 5) | 2.52 ± 0.13 × 106 | NA‡ | 100 |
| +81-bp insertion (n = 3) | 0.105 ± 0.008 × 106 | NA‡ | 4.2 ± 0.3 |
NA indicates not applicable.
Mean specific activity ± SD. Activities are for crude extracts for pKK constructs and for pure enzyme for pET SUMO constructs.
The previously published value for residual pKK-expressed activity for this mutation was 1.4% of wild-type.
Not applicable for purified enzyme.
Identification of the intron 9 branchpoint mutation, c.661-31T → G
Both exons 9 and 10 of UROS and their respective intron/exon junctions and flanking regions were amplified from genomic DNA from patients 3, 6, and 10, and sequenced. A homozygous T to G transversion 31 nucleotides upstream from exon 10 (c.661-31T → G) was identified in patients 3 and 6 in New York, and subsequently, in patient 10 in Zurich. Confirmation of the c.661-31T → G mutation was made in patients 3 and 6 and their parents by demonstrating the presence of the lesion in genomic DNAs from each by dot-blot hybridization with allele-specific oligonucleotides (data not shown). Amplified genomic DNAs from these patients hybridized strongly with the mutation-specific ASO, confirming that the patients were homoallelic for the base substitution. Genomic DNAs from each of their parents were heterozygous, hybridizing with both the normal and mutation-specific oligonucleotides.
To evaluate the possibility that the T to G substitution represented a polymorphism, genomic DNAs from 50 unrelated Ashkenazi Jewish normal persons were PCR-amplified and screened for the c.661-31T → G mutation by dot-blot analysis with the ASOs. None of the 100 wild-type alleles tested positive for this base substitution, suggesting that the base change was not a common polymorphism (data not shown).
In vivo abundance of the wild-type and alternatively spliced mRNAs
To assess the relative amounts of the normal and alternatively spliced transcripts in these 3 CEP patients, qRT-PCR primers and TaqMan probes were designed (supplemental Table 2) to specifically analyze each splice variant (Figure 3B). The results (Table 3) indicated that the normal 1.5-kb transcript accounted for approximately 81% of the total transcript mRNA, whereas the major alternatively spliced transcripts, corresponding to retention of intronic sequences of 81, 81 + 165, and 81 + 277 bp, represented 14%, 5%, and 0.3%, respectively, of the total transcript mRNA in normal persons. The mean percentage of wild-type 1.5-kb transcript in the lymphoblasts of all 3 patients was approximately 10% of that in normal lymphoblasts, whereas the mean levels of the alternatively spliced transcripts were similar or slightly higher than those in the normal controls. Note that, of the 5 independent replicate assays for the percentage of normal transcripts in the 3 CEP patients, 4 were 5% (all 3 patients) with 1 outlier of 33% (patient 6). The frequencies of occurrence of the normal and alternative transcripts in the latest UCSC Human Genome Browser assembly (ESTs, March 2006) were 90%, 8%, 2%, and 0%, respectively, consistent with our qRT-PCR data (81%, 14%, 5%, and 0.3%; Table 3). This supports our observation that similar amounts of alternative transcripts were found in normal and CEP patients.
Relative concentrations of alternatively spliced forms of UROS in normal and CEP lymphoblasts*
| Source . | 155-bp normal transcript . | 236-bp (+81 nt) variant . | 401-bp (+165/+81 nt) variant . | 513-bp (+81/+277 nt) variant . |
|---|---|---|---|---|
| Normal controls (n = 3) | 81 (58-111) | 14 (10-18) | 5 (4-8) | 0.3 (0.2-0.4) |
| Human UROS ESTs† | 90 | 8 | 2 | 0 |
| CEP patient 3 (n = 2) | 5 (5,5) | 12 (8,19) | 1 (1,1) | 1 (1,2) |
| CEP patient 6 (n = 2) | 14 (5,33) | 27 (12,60) | 4 (1,22) | 10 (3,33) |
| CEP patient 10 (n = 1) | 5 | 27 | 2 | 5 |
| CEP patients Average | 8 (5-19) | 22 (10-40) | 2 (1-11) | 5 (2-17) |
| Source . | 155-bp normal transcript . | 236-bp (+81 nt) variant . | 401-bp (+165/+81 nt) variant . | 513-bp (+81/+277 nt) variant . |
|---|---|---|---|---|
| Normal controls (n = 3) | 81 (58-111) | 14 (10-18) | 5 (4-8) | 0.3 (0.2-0.4) |
| Human UROS ESTs† | 90 | 8 | 2 | 0 |
| CEP patient 3 (n = 2) | 5 (5,5) | 12 (8,19) | 1 (1,1) | 1 (1,2) |
| CEP patient 6 (n = 2) | 14 (5,33) | 27 (12,60) | 4 (1,22) | 10 (3,33) |
| CEP patient 10 (n = 1) | 5 | 27 | 2 | 5 |
| CEP patients Average | 8 (5-19) | 22 (10-40) | 2 (1-11) | 5 (2-17) |
Values are percentage of all transcripts in normal controls (range).
The relative concentrations of the major UROS transcripts were calculated using the ΔΔCt procedure as described in “Quantitative RT-PCR analyses.” The abundance of the each transcript is expressed as a mean percentage of the total amount of normal and alternatively spliced transcripts in the normal controls.
Percentage of each EST found in the 90 ESTs containing human UROS exons 9 and 10 in the March 2006 Assembly of the Human Genome.
URO-synthase activities in normal and patient lymphoblasts
The mean URO-synthase activity in cytoplasmic extracts of normal lymphoblasts was 7.36 plus or minus 0.07 (SD) units/mg protein. For patients 3 and 6, their residual activities were 14.1% and 14.8%, respectively, an average residual activity of 14.4% plus or minus 0.5% of wild-type activity.
Characterization of the intron 9 cryptic splice sites
As shown in Figure 3, the alternatively spliced transcripts had intron 9 insertions of 81, 246 (165 + 81), 358 (81 + 277), and 523 (165 + 81 + 277) bp. The 81- and 358-bp insertions used the predicted ccttgat BPS located 62 bp upstream of the 81-bp insertion, whereas the 246- and 523-bp insertions used the predicted atttgat BPS 50 bp upstream of the 165-bp region. All 4 variant exonic sequences followed the GT-AG rule. The 81-bp insertion was in-frame and encoded an extra 27 amino acids (TQGPQHPKKNCLQLEPLRKDCTDTAVM). The other 3 variant insertions all resulted in premature stop codons and deleted the 45 residues encoded by exon 10. The 246- and 523-bp insertions encoded an additional 22 amino acids after exon 9 and terminated in a TAA stop codon 67 bp 3′ of the insertion, whereas the 513-bp insertion encoded an additional 91 amino acids after exon 9 and terminated in the stop codon, TGA, 274 bp 3′ of the insertion.
In silico prediction of splice site use
To evaluate the potential for the alternative splicing that resulted in intron 9 retention in normal persons and CEP patients, in silico analyses were performed to assess splice site favorability by identifying juxtaposed cryptic BPSs and pyrimidine tracts. The highest 5′ splice site score (0.94) was for the authentic exon 10 with the next highest scores associated with cryptic exons CE4 (0.69) and CE5 (0.64) that were the intronic sequences retained in the alternatively spliced transcripts that had the additional 165- and 81-bp intronic sequences, respectively (Figure 3; supplemental Figure 1). These 2 cryptic exons also had the highest 3′ splice scores (both were 0.95; Table 1). For comparison, the 5′ splice scores for normal exons 7, 8, 9, and 10 were 0.33, 0.43, 0.56, and 0.94, respectively. Thus, the presence of the alternatively spliced transcripts in normal persons and CEP patients was consistent with the favorable cryptic splice sites in intron 9.
The ESE densities for splicing factor 2/alternative splicing factor (SF2/ASF) and SC35 were calculated (Table 1) and plotted along with the NetGene2 results (supplemental Figure 1). ESE motifs SRp20, SRp40, and SRp55 were relatively uniform in distribution throughout intron 9 and thus were not informative. There were 3 to 10 times more SC35 and SF2/ASF motif ESE sites in exon 10 and in the retained intron 9 cryptic exons CE4 and CE5 than in the proximal 2.7 kb (CE1 and CE2) of intron 9 (Table 1; supplemental Figure 1). The frequency of SC35 and SF2/ASF motifs in CE4 and CE5 were 28.5 and 37 per kilobase, respectively, whereas in comparison, the frequency of the SC35 motif in 2626 exons and 2079 introns revealed an average of 5.9/kb in introns versus 9.0 /kb in exons, a 1.5-fold difference that was statistically significant.20 Thus, the cryptic exon use seen in both wild-type and the CEP c.661-31T → G transcripts was consistent with the presence in the intron 9 sequence of high-propensity cryptic BPS sites, polypyrimidine tracts, splice junctions, and ESE sites.
Discussion
Northern hybridization and RT-PCR analyses revealed molecular defects in the UROS transcripts causing CEP in 3 unrelated patients in whom no mutations were found in the promoter, exons, and intron/exon boundaries in the UROS gene. Subsequent sequence analysis identified the transversion c.661-31T → G in the intron 9 BPS in all 3 patients. That this BPS base change was the primary disease-causing mechanism was supported by: (1) its location at a highly conserved position in the BPS, −2 nt from the lariat-forming adenosine where base changes have previously altered interactions with the branchpoint-binding protein SF1 and with U2 snRNP (see below in this section), (2) the absence of any other UROS mutations that could account for the observed splicing defect, (3) the exclusion of the base change as a polymorphism in normal persons, (4) the fact that the in-frame +27-amino acid variant encoded by the +81 nt alternative transcript had little activity, and (5) the mutation's segregation with disease in 3 CEP families who claimed to be unrelated. However, the presence of the same mutation for a very rare disease in the 2 Ashkenazi Jewish families and a third Semitic family suggests that the Jewish families may be related, that they may share a possible common Middle Eastern ancestor, or, and much less probable, that independent mutation events occurred. Analysis of 100 Ashkenazi Jewish alleles did not identify a base substitution, indicating a frequency less than 1%.
With the exception of the in-frame +81 nt alternatively spliced transcript, each of the other splicing variants in these patients would result in frame-shifted, nonfunctional, and prematurely terminated URO-synthase mutant polypeptides lacking the terminal 45 amino acids encoded by exon 10. Exon 10 is the site of at least 6 known CEP mutations highlighting its functional importance. It is unlikely that an enzyme resulting from any of these aberrant transcripts that encoded novel peptides and/or deleted exon 10 would be active based on structural mapping studies.
Prokaryotic expression of the in-frame +81 nt insertion transcript resulted in 4.2% of expressed wild-type activity for the pure enzymes and 0.6% of wild-type URO-synthase enzymatic activity in crude extracts (Table 2). The lower yield in crude extracts was possibly the result of proteolytic instability because the mutant enzyme was not thermolabile. Because the average abundance of this transcript in lymphoblasts ranged from 12% to 27% of the normal transcript (Table 3), the low activity of the mutant expressed protein indicated that its contribution to residual activity would be only 0.2% to 1% (0.6% or 4.2% × 27%), a negligible contribution compared with the 14% residual enzyme activity observed in patient lymphoblasts because of the presence of normal transcript. Differences in tissue-specific expression could result in the residual activity being lower in erythroid cells because of message and/or enzyme instability. However, this level of residual activity is comparable with previous reports of 4% to 33% residual activity, mean = 16%, in early-onset CEP patients (eg, Freesemann et al21 ). Thus, these studies demonstrate that the disease-causing mechanism in these patients is the −2 BPS mutation, which markedly decreased the amount of the wild-type transcript, and hence, reduced the amount of normal URO-synthase.
The role of the BPS in the splicing of hnRNAs has been the subject of intense investigation.22-29 Nuclear magnetic resonance spectroscopy30 showed that the −2 uridine exhibits specific contacts with at least 6 residues in human SF1. The critical role of the −2 position also has been confirmed by functional studies and observed mutation frequencies; the most highly conserved positions are 0 and −2.31,32 The lariat adenosine and the −2 T are also the most critical bases in vivo because their alteration accounts for 16 of the 18 human disease-causing BPS mutations (supplemental Table 1). Thus, the observed decreased wild-type transcript in the CEP patients, along with the presence of larger transcripts using alternative BPSs and splicing sites might be expected, as inhibiting the binding of SF1 blocks the subsequent binding of the U2 snRNP.33
There was no evidence of exon skipping in association with this mutated BPS in the last intron of UROS. This contrasts with the previously published four −2 BPS mutations (for which in vivo data are available) that consistently caused partial or complete exon skipping (supplemental Table 1). It is interesting that no cryptic splice sites were available anywhere beyond the last intron. Exon skipping resulting from BPS mutations in the last intron may be selected against by elimination of potential exons in the 3′ untranslated region and beyond.
In conclusion, these studies demonstrated the occurrence of alternative splicing of the UROS transcript in normal persons and CEP patients, consistent with the presence of additional splice sites and ESE sites in intron 9 that generated these minor normally occurring alternative transcripts. Of these alternatively spliced transcripts, only the +81 nt in-frame insertion transcript encoded a viable enzyme protein, albeit contributing to only approximately 0.2% of wild-type activity. Thus, the BPS mutation markedly reduced the amount of wild-type UROS transcript, thereby causing CEP in these patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the late Raman Reddy and Weiming Xu for valued contributions of technical assistance and Ms Karin Walser and Mr Sulejman Ahmetovic for their technical contributions.
This work was supported in part by the National Institutes of Health (research grant 5 R01 DK26824), the Mount Sinai General Clinical Research Center from the National Center for Research Resources (grant 5 M01 RR00071), and the Mount Sinai Child Health Research Center (grant 5 P30 HD28822).
National Institutes of Health
Authorship
Contribution: D.F.B. designed the experiments, analyzed the data, and wrote the paper; X.S.-Y. designed the experiments, performed the experimental work, analyzed the data, and wrote the paper; S.C. designed the experiments, performed the experimental work, and analyzed the data; H.-W.Y. designed the experiments, performed the experimental work, and wrote the paper; E.I.M. designed the experiments and analyzed the data; R.J.D. obtained funding for the research, designed the experiments, analyzed the data, and wrote the paper; and all authors discussed results and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert J. Desnick, Department of Genetics and Genomic Sciences, Mount Sinai School of Medicine, Box 1498, Fifth Ave and 100th St, New York, NY 10029-6574; e-mail: Robert.Desnick@mssm.edu.
References
Author notes
D.F.B. and X.S.-Y. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal