Abstract
Regulatory T cells (Tregs) are a specific subset of lymphocytes that are critical for the maintenance of self-tolerance. Expression levels of the transcription factor Foxp3 have been causally associated with Treg differentiation and function. Recent studies show that Foxp3 can also be transiently expressed in effector T cells; however, stable Foxp3 expression is required for development of a functional Treg suppressor phenotype. Here, we demonstrate that Foxp3 is acetylated, and this can be reciprocally regulated by the histone acetyltransferase p300 and the histone deacetylase SIRT1. Hyperacetylation of Foxp3 prevented polyubiquitination and proteasomal degradation, therefore dramatically increasing stable Foxp3 protein levels. Moreover, using mouse splenocytes, human peripheral blood mononuclear cells, T cell clones, and skin-derived T cells, we demonstrate that treatment with histone deacetylase inhibitors resulted in significantly increased numbers of functional Treg cells. Taken together, our data demonstrate that modulation of the acetylation state of Foxp3 provides a novel molecular mechanism for assuring rapid temporal control of Foxp3 levels in T cells, thereby regulating Treg numbers and functionality. Manipulating Foxp3 acetylation levels could therefore provide a new therapeutic strategy to control inappropriate (auto)immune responses.
Introduction
Regulatory T cells (Tregs) are a specific subset of lymphocytes that play a crucial role in the maintenance of self-tolerance.1,2 These CD4+CD25+ cells can be distinguished from conventional T cells by the expression of a distinct subset of molecules, both on their cell surface as well as intracellularly.3,4 The transcription factor Foxp3 is crucial for Treg differentiation and function, and various Foxp3 mutations, both in scurfy mice and IPEX (immune dysregulation polyendocrinopathy, enteropathy, X chromosome-linked syndrome) patients, result in the development of complex autoimmune disease, resulting from Treg deficiency.5,6 Ectopic expression of Foxp3 in CD4+CD25− T cells has also been shown to induce a suppressive phenotype, suggesting that stable Foxp3 expression is sufficient for development of functional Tregs.2,3,7
Foxp3 expression is not unique to lymphocytes; recent reports demonstrate that respiratory, thymic, prostate, and mammary epithelium cells also express Foxp3, although expression levels were low compared with Tregs.8 Furthermore, it has been reported that in vitro T-cell receptor (TCR) stimulation of CD4+CD25− effector T (Teff) cells can result in transient Foxp3 expression, which does not however generate T cells with a suppressive phenotype.9,10 In contrast, TCR-stimulated CD4+CD25− cells expressing high and stable Foxp3 levels develop suppressive capacity, illustrating that persistent Foxp3 expression is probably an essential step in the conversion of Teff cells into Tregs.11,12
Because Foxp3 is an essential transcription factor for maintenance of immune homeostasis, its activity must be tightly and specifically regulated. However, surprisingly little is currently understood concerning Foxp3 posttranslational regulation of this transcription factor. A recent study has demonstrated that Foxp3 can interact with histone deacetylase (HDAC) 7 and 9, and with the histone acetyltransferase (HAT) TIP60.13 Although these data suggest that Foxp3 is able to form a multiprotein complex containing both HAT/HDAC molecules, the functional relevance of these observations remains to be further clarified. Furthermore, mice treated with HDAC inhibitor trichostatin A (TSA) have increased numbers of functionally improved Tregs correlating with reduced disease severity in an induced colitis model as well as an increased donor-specific allograft tolerance in a cardiac and islet transplantation model.14 However, the molecular mechanism underlying improved Treg function by TSA treatment remains unclear, and it is not evident whether these are direct or indirect effects.
Here, we demonstrate that Foxp3 acetylation can be reciprocally regulated by the HAT p300 and the HDAC SIRT1. We show that Foxp3 protein has a short half-life and that acetylation prevents proteasomal degradation, dramatically increasing Foxp3 levels. Furthermore, modulating SIRT activity in mouse and human primary T cells regulates Foxp3 protein levels as well as the number and suppressive capacity of Tregs. Taken together, directly modulating the acetylation state of Foxp3 provides a novel molecular mechanism for assuring rapid temporal control of Foxp3 levels in T cells. Increasing Foxp3 acetylation levels may thus be a critical switch in the generation of induced Tregs from activated peripheral T cells.
Methods
Antibodies, DNA constructs, and reagents
The following antibodies were used: mouse anti-Foxp3 clone PCH101 for fluorescence-activated cell sorter (FACS) analysis (eBioscience), rabbit anti-p300 (Santa Cruz Biotechnology), rabbit anti–acetyl-lysine (Cell Signaling Technology), mouse anti-Flag M2 from Sigma-Aldrich (Zwijndrecht), mouse antihemaglutinin (HA) clone 12CA5 from Santa Cruz Biotechnology, mouse antitubulin (Sigma-Aldrich), and anti-Myc monoclonal mice antibody were made using a hybridoma cell line. Foxp3 was cloned from MIGR1-Foxp3 (kindly provided by Dr S. Sakaguchi7 ) into pMT2 that already contained a Flag tag resulting in pMT2-Flag-Foxp3. Using side-directed mutagenesis, the pMT2-Flag-Foxp3ΔE250 mutant was constructed. pcDNA3 (Invitrogen), pcDNA3-HA-p300, pcDNA3-HA-TIP60 (kindly provided by Dr D. Trouche15 ), 6xHis-p300 (kindly provided by Dr W. L. Kraus16 ), pRSV-NFATc/A,17 and Myc-SIRT118 (both kindly provided by Dr B. M. T. Burgering, University Medical Center, Utrecht, The Netherlands) have been described earlier. pCDNA3-p300-HA was generated by cloning a Not I-HindIII fragment from CMVβ-p300-HA (a gift from Dr R. Eckner, ESBATech AG, Zurich, Switzerland) into the respective cloning sites of pCDNA3. PEI (#23966) was purchased from Polysciences. TSA, nicotinamide (NAD), cycloheximide (CHX), epoxomycin, lactacystin, and MG132 were from Sigma-Aldrich.
Transfection of cells and luciferase assays
HEK293 cells were maintained in Dulbecco modified Eagle medium (Invitrogen) supplemented with 8% heat-inactivated fetal calf serum (FCS), penicillin, and streptomycin (Invitrogen) at 37°C and 5% CO2. Cells were grown to 50% confluence in 6-well plates (Nunc) and transfected with a mixture of 1.5 μg DNA and 7.5 μL PEI overnight; the next day, cells were washed twice with phosphate-buffered saline (PBS) and cultured for 24 hours in medium. Cell lysates were prepared for Western blot analysis. For the luciferase assay, cells were transfected. Calcium-phosphate was used with 1 μg interleukin-2 (IL-2) promoter luciferase reporter from Panomics, 0.5 μg of pMT2-Foxp3, 0.5 μg pcDNA3-HA-p300, pcDNA3-NFATc/A, or 0.5 μg pcDNA3 empty vector and 7 μg pMT2 empty vector and 0.05 μg pRLTK Renilla (Promega) to normalize for transfection efficiency. Cells were transfected in a 6-well plate; 3 days after transfection, the cells were washed twice with PBS and lysed in 50 μL passive lysis buffer for 15 minutes, insoluble cell debris were spun down, and the supernatant fraction was assayed for luciferase activity using Dual-Luciferase Reporter Assay System (Promega).19
In vitro acetylation assay
Glutathione S-transferase (GST) fusion proteins were induced and purified as described previously.20 The wild-type and catalytic AT2 mutant p300 proteins were synthesized in Sf9 cells using a baculovirus expression system and purified as previously described. A total of 1 μg GST-Foxp3, 0.5 μL (10 nCi) [14C]-acetylCoA (PerkinElmer), and 2 μg His(6x)-p300 or His(6x)-p300mutAT2 were incubated in AIPH buffer (20mM Tris-HCI pH 8.0, 60mM NaCI, 2mM EDTA, 0.2% NP-40, 40μM PMSF) for 40 minutes at 30°C. The reaction was stopped by 10 μL 5 × sample buffer. Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Kodak XB films.
Confocal studies
HEK 293 cells were cultured on poly-L-lysine–coated (Sigma-Aldrich) microscope glasses, fixed in PBS containing 3% paraformaldehyde (Merck) for 15 minutes at 15°C, and subsequently 100% methanol (Merck) for 30 minutes at −20°C. Cells were preincubated with 10% normal goat serum (Jackson ImmunoResearch Laboratories) before mouse anti-Foxp3 (5 μg/mL; eBioscience) and of rabbit anti-p300 (5 μg/mL) was added for 1 hour, followed by PBS washes and incubation with 2 μg/mL goat anti–mouse Cy3 (Jackson ImmunoResearch Laboratories) or goat anti–rabbit Cy5 (Jackson ImmunoResearch Laboratories) conjugates (all antibody stainings were in PBS with 10% goat serum). Slides were then washed extensively, and cells were mounted in mowiol containing 3% 1,4-diazabiclo-(2,2,2)-octan followed by a glass cover as described.21 Cells were examined with a Zeiss LSM 710 microscope (Carl Zeiss).
Western blots
Cells were lysed in Laemmli buffer (0.12M Tris-HCL, pH 6.8, 4% SDS, 20% glycerol, 0.05 μg/μL bromophenol blue, 35mM β-mercaptoethanol) and boiled for 5 minutes, and the protein concentration was determined. Equal amounts of sample were analyzed by SDS-PAGE, electrophoretically transferred to polyvinylidene difluoride membrane (Millipore), and probed with the respective antibodies. Immunocomplexes were detected using enhanced chemiluminescence (GE Healthcare).
Isolated primary T cells
T cells from human PBMCs.
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density-gradient centrifugation (Pharmacia) and CD4+ cells were isolated by magnetic-activated cell sorting (MACS). Cells were cultured in for 7 days with RPMI 1640 supplemented with 2mM l-glutamine, 100 units/mL of penicillin/streptomycin (Invitrogen), and 10% (vol/vol) AB heat-inactivated (60 minutes at 56°C) human serum (Sanquin Blood Bank), and stimulated with plate-bound anti-CD3 (1.5 μg/μL), 300 IU/mL human IL-2, and 2 μg/mL anti–human CD28 in combination with 10mM NAM or 5μM resveratrol. Cells were cultured in triplicate in round-bottom 96-well plates (Nunc) at 37°C in an atmosphere of 5% CO2 with 100% relative humidity.
T cells from human skin.
Normal human skin was obtained from patients undergoing cosmetic surgery procedures. Three-dimensional matrices (Statamatrix) were obtained from Cell Sciences. Explant cultures were established as described.22 Skin was cut into very small fragments and placed on the surface of a matrix. Each matrix was placed into 1 well of a 24-well plate in 2 mL/well of Iscove modified medium (Mediatech) with 20% heat-inactivated fetal bovine serum (Sigma-Aldrich), penicillin, and streptomycin, and 3.5 mL/L β-mercaptoethanol. Cultures were fed 3 times a week by careful aspiration of 1 ml of culture medium and replacement with fresh medium. Cells were harvested at 12 days. IL-15 (10 ng/mL) and IL-2 (5 IU/mL; R&D Systems) were added and refreshed with each feeding, in combination with 10mM NAM or 5μM resveratrol.
T cells from mouse splenocytes.
Spleens were removed from healthy C57BL/6 mice after they were killed, and CD4+ T cells were isolated from splenocytes using MACS separation. Cells were cultured for 5 days in flat-bottom plates coated with mu-aCD3 (1 μg/mL in pbs) in Iscove modified Dulbecco medium supplemented with 2 mM l-glutamine, 100 units/mL of penicillin/streptomycin (Invitrogen), and 10% (vol/vol) heat-inactivated (60 minutes at 56°C) FCS, 0.2 ng/mL murine IL-2, and 0.5 μg/mL anti–murine CD28 and 2 ng/mL transforming growth factor-β in combination with 10mM NAM or 5μM resveratrol. Cells were cultured in triplicate in round-bottom 96-well plates (Nunc) at 37°C in an atmosphere of 5% CO2 with 100% relative humidity. The University Medical Center Utrecht approved this study.
T cell clone.
The CD4+ T-cell clone N3CA8 was generated as described elsewhere23 and cultured for 6 days with RPMI 1640 supplemented with 2mM l-glutamine, 100 units/mL of penicillin/streptomycin (Invitrogen), and 10% (vol/vol) heat-inactivated pooled human serum (Sanquin Blood Banks) in 48-well tissue culture plates (BD Biosciences). The T-cell clone was stimulated with 40 IU/mL human IL-2 and anti-CD3/anti-CD28 Dynabeads (Invitrogen) at a bead/T-cell ratio of 1:10 in combination with 5mM NAM or 2.5μM resveratrol. Staining of surface markers and Foxp3 for flow cytometry was done according to the manufacturer's protocol (eBioscience).
Suppression assay
Mice CD4 T cells were cultured as described in “Methods” and stained by anti–mouse CD4 peridinin chlorophyll protein and anti–mouse CD25 antigen-presenting cells (APCs) (clone RM4-5 and PC61, respectively; both from BD Biosciences PharMingen). CD4+CD25− cells Teff (10 000) were sorted by FACSAria directly into a 96-well plate, and CD4+CD25+ sorted cells were added at a ratio of 1:5. A CD4-depleted fraction of spleen cells (using anti–mouse CD4 microbeads and a single run on an LS column, MACS; Miltenyi Biotec) was irradiated at 35 Gy and served as a source for APCs. CD4+CD25+ and CD4+CD25− cells were cultured in Iscove modified Dulbecco medium supplemented with 10% FCS, 2mM l-glutamine, 100 units/mL penicillin, 100 μg/mL streptomycin, and 5 × 10−5M 2-mercaptoethanol in the presence of 50 000 APCs and 1 μg/mL soluble anti-CD3 (clone 145-2C11; BD Biosciences PharMingen) without NAM for 120 hours. During the last 18 hours, 1 μCi (0.037 MBq) 3H-thymidine (GE Healthcare) was added per well, and 3H uptake was measured using a liquid scintillation β counter. Proliferative responses were calculated as the mean 3H incorporation (cpm) of triplicate wells. To determine suppression of proliferation by measuring carboxyfluorescein succinimidyl ester (CFSE) dilution within the CD4 T effector cell population, we labeled freshly isolated splenocytes with 2μM CFSE for 7 minutes at 37°C. A total of 10 000 labeled splenocytes per well in a 96-well plate were stimulated with 1 μg/mL soluble anti-CD3, and 2000 CD4+CD25+ sorted cells were added. On day 4, cells were harvested, stained with anti-CD4, and CFSE dilution was measured on FACSCanto (BD Biosciences).
Quantitative PCR
mRNA was isolated using the Trizol according to the manufacturer's protocol (Invitrogen), and cDNA synthesis was performed using IScript cDNA synthesis kit (Bio-Rad). cDNA samples were amplified using SYBR Green supermix (Bio-Rad), in a MyiQ single-color real-time PCR detection system (Bio-Rad) according to the manufacturer's protocol. The following primers were used: Foxp3 forward: TCAAGCACTGCCAGGCG, Foxp3 reverse: CAGGAGCCCTTGTCGGAT, β2-microglobulin (β2M) forward: CCAGCAGAGAATGGAAAGTC, β2M reverse: GATGCTGCTTACATGTCTCG. To quantify the data, the comparative threshold cycle method was used. Relative quantity was defined as 2−ΔΔCt. β2M was used as reference gene.
Statistical analysis
Statistical analysis was performed using the Mann-Whitney test (Prism GraphPad Software). P less than .05 was considered statistically significant.
Results
Foxp3 interacts with and is acetylated by p300
To assess whether Foxp3 can be acetylated, cells were transfected with a Flag-tagged Foxp3 construct and incubated with the HDAC inhibitors (HDACi) TSA and NAM. Together these HDACi can inhibit a majority of HDACs. Foxp3 acetylation was analyzed by Flag immunoprecipitation and immunoblotting using an anti–acetyl lysine antibody. We observed a basal level of Foxp3 acetylation in the absence of HDACi, which was increased by addition of TSA/NAM in a dose-dependent manner (Figure 1A). To determine whether the ubiquitously expressed HAT p300 could acetylate Foxp3, cells were transfected with Foxp3 and increasing amounts of p300, and Foxp3 acetylation was again analyzed. Indeed, addition of p300 dose-dependently increased Foxp3 acetylation (Figure 1B). Because a recent study demonstrated interaction between Foxp3 and the HAT TIP60,13 we wished to determine whether TIP60 could also acetylate Foxp3 in living cells. Lysates were prepared from HEK 293 cells cotransfected with Flag-Foxp3 and HA-TIP60 or HA-p300. Immunoblots reveal that, in contrast to p300, TIP60 cotransfection did not increase Foxp3 acetylation (Figure 1C). We next determined whether p300 and TIP60 could associate with Foxp3 by coimmunoprecipitation after cotransfection of cells with Flag-Foxp3 and HA-p300 or HA-TIP60. Both p300 and TIP60 were found to associate with immunoprecipitated Foxp3 (Figure 1D-E). Finally, we tested p300 for its ability to directly acetylate Foxp3 using an in vitro acetylation assay. GST-Foxp3 fusion protein coupled to Sepharose beads was incubated with [14C]-labeled acetyl-CoA and p300, or an acetylase dead p300mutAT2, and samples were separated on SDS-PAGE and analyzed by autoradiography (Figure 1F). Incubation of Foxp3 with p300, but not the catalytically inactive variant, resulted in Foxp3 acetylation. These data demonstrate that, although both TIP60 and p300 can associate with Foxp3, Foxp3 acetylation is selectively mediated by p300.
p300 promotes Foxp3 acetylation. (A) Flag immunoprecipitation from lysates from Flag-Foxp3–transfected cells. HEK 293 cells were treated with HDACi: + indicates 50 nm TSA and 1mM NAM; or ++, 250 nm TSA and 5mM NAM for 16 hours. Equal amounts of immunoprecipitated Foxp3 were separated by SDS-PAGE, and Western blots were probed for acetylated lysines (Ac-Lys) or Flag. (B) Cells were transfected with Flag-Foxp3 and/or HA-p300. Flag-Foxp3 was immunoprecipitated with anti-Flag beads and analyzed using acetyl lysines (Ac-Lys), Flag, or HA antibodies. (C) Cells were cotransfected with Flag-Foxp3 and HA-p300 or HA-TIP60 and immunoprecipitated using anti-Flag beads. Immunoblots were analyzed with acetyl-lysine (Ac-Lys) antibody, anti-Flag, or anti-HA. Cells were cotransfected with Flag-Foxp3, HA-Tip60 (D), or HA-p300 (E). Cell lysates were coimmunoprecipitated using anti-HA beads and analyzed using anti-Flag or anti-HA. (F) GST-Foxp3 fusion protein coupled to Sepharose beads was incubated with [14C]-labeled acetyl-CoA and p300 or the acetylase dead p300mutAT2; samples were separated on SDS-PAGE and analyzed using films sensitive for radioactivity. Results are representative of at least 3 independent experiments. *Aspecific band. IP indicates immunoprecipitation; and WB, Western blot.
p300 promotes Foxp3 acetylation. (A) Flag immunoprecipitation from lysates from Flag-Foxp3–transfected cells. HEK 293 cells were treated with HDACi: + indicates 50 nm TSA and 1mM NAM; or ++, 250 nm TSA and 5mM NAM for 16 hours. Equal amounts of immunoprecipitated Foxp3 were separated by SDS-PAGE, and Western blots were probed for acetylated lysines (Ac-Lys) or Flag. (B) Cells were transfected with Flag-Foxp3 and/or HA-p300. Flag-Foxp3 was immunoprecipitated with anti-Flag beads and analyzed using acetyl lysines (Ac-Lys), Flag, or HA antibodies. (C) Cells were cotransfected with Flag-Foxp3 and HA-p300 or HA-TIP60 and immunoprecipitated using anti-Flag beads. Immunoblots were analyzed with acetyl-lysine (Ac-Lys) antibody, anti-Flag, or anti-HA. Cells were cotransfected with Flag-Foxp3, HA-Tip60 (D), or HA-p300 (E). Cell lysates were coimmunoprecipitated using anti-HA beads and analyzed using anti-Flag or anti-HA. (F) GST-Foxp3 fusion protein coupled to Sepharose beads was incubated with [14C]-labeled acetyl-CoA and p300 or the acetylase dead p300mutAT2; samples were separated on SDS-PAGE and analyzed using films sensitive for radioactivity. Results are representative of at least 3 independent experiments. *Aspecific band. IP indicates immunoprecipitation; and WB, Western blot.
Acetylation regulates Foxp3 protein levels
Because acetylation of the related Foxo Forkhead transcription factors has been reported to mediate their subcellular localization,18,24 we analyzed Foxp3 subcellular localization under conditions of hyperacetylation. Using confocal microscopy, we found Foxp3 to be exclusively localized in the nucleus (Figure 2A top panel). In contrast to previous reports analyzing Foxo transcription factor localization, cotransfecting cells with p300 did not influence Foxp3 localization (Figure 2A bottom panel). However, ectopically expressed as well as endogenous p300 colocalized with Foxp3 in the nucleus, underscoring a role for p300 in nuclear Foxp3 acetylation.
Acetylation modulates Foxp3 protein levels. (A) Representative examples of cells that were transfected with only Foxp3 (green; top panel). Subcellular distribution of cells that were cotransfected with Foxp3 (green) and p300 (red) are shown in the bottom panel. p300 was localized using an anti-p300 antibody that recognizes both endogenous and ectopically expressed p300. Colocalization of Foxp3 and p300 is indicated in yellow. (B) Foxp3 function was assessed by evaluating IL-2 promoter reporter activity. IL-2 promoter luciferase activity was analyzed in HEK 293 cells by cotransfecting NFAT with Foxp3 del E250 (▭), Foxp3 ( ), or Foxp3 with p300 (
), or Foxp3 with p300 ( ). Repression of IL-2 luciferase activity by wild-type Foxp3 was set as 100%. Values were all normalized for cotransfected Renilla. **P < .01. (C) HEK 293 cells were transfected with Flag-Foxp3 and treated with HDACi: + indicates 50 nm TSA and 1mM NAM; ++, 250 nm TSA and 5mM NAM. Immunoblots were probed for Flag expression or tubulin as loading control. (D) Cells were transfected with Flag-Foxp3 and increasing amounts of HA-p300. Western blots were incubated with antibodies against Flag, HA, or tubulin as indicated. (E) HEK 293 cells were transfected with Flag-Foxp3 or Flag-Foxo3 with or without HA-p300. Cells were treated with 100nM TSA and 2.5mM NAM for 16 hours (HDACi). Data are representative of at least 3 independent experiments.
). Repression of IL-2 luciferase activity by wild-type Foxp3 was set as 100%. Values were all normalized for cotransfected Renilla. **P < .01. (C) HEK 293 cells were transfected with Flag-Foxp3 and treated with HDACi: + indicates 50 nm TSA and 1mM NAM; ++, 250 nm TSA and 5mM NAM. Immunoblots were probed for Flag expression or tubulin as loading control. (D) Cells were transfected with Flag-Foxp3 and increasing amounts of HA-p300. Western blots were incubated with antibodies against Flag, HA, or tubulin as indicated. (E) HEK 293 cells were transfected with Flag-Foxp3 or Flag-Foxo3 with or without HA-p300. Cells were treated with 100nM TSA and 2.5mM NAM for 16 hours (HDACi). Data are representative of at least 3 independent experiments.
Acetylation modulates Foxp3 protein levels. (A) Representative examples of cells that were transfected with only Foxp3 (green; top panel). Subcellular distribution of cells that were cotransfected with Foxp3 (green) and p300 (red) are shown in the bottom panel. p300 was localized using an anti-p300 antibody that recognizes both endogenous and ectopically expressed p300. Colocalization of Foxp3 and p300 is indicated in yellow. (B) Foxp3 function was assessed by evaluating IL-2 promoter reporter activity. IL-2 promoter luciferase activity was analyzed in HEK 293 cells by cotransfecting NFAT with Foxp3 del E250 (▭), Foxp3 ( ), or Foxp3 with p300 (
), or Foxp3 with p300 ( ). Repression of IL-2 luciferase activity by wild-type Foxp3 was set as 100%. Values were all normalized for cotransfected Renilla. **P < .01. (C) HEK 293 cells were transfected with Flag-Foxp3 and treated with HDACi: + indicates 50 nm TSA and 1mM NAM; ++, 250 nm TSA and 5mM NAM. Immunoblots were probed for Flag expression or tubulin as loading control. (D) Cells were transfected with Flag-Foxp3 and increasing amounts of HA-p300. Western blots were incubated with antibodies against Flag, HA, or tubulin as indicated. (E) HEK 293 cells were transfected with Flag-Foxp3 or Flag-Foxo3 with or without HA-p300. Cells were treated with 100nM TSA and 2.5mM NAM for 16 hours (HDACi). Data are representative of at least 3 independent experiments.
). Repression of IL-2 luciferase activity by wild-type Foxp3 was set as 100%. Values were all normalized for cotransfected Renilla. **P < .01. (C) HEK 293 cells were transfected with Flag-Foxp3 and treated with HDACi: + indicates 50 nm TSA and 1mM NAM; ++, 250 nm TSA and 5mM NAM. Immunoblots were probed for Flag expression or tubulin as loading control. (D) Cells were transfected with Flag-Foxp3 and increasing amounts of HA-p300. Western blots were incubated with antibodies against Flag, HA, or tubulin as indicated. (E) HEK 293 cells were transfected with Flag-Foxp3 or Flag-Foxo3 with or without HA-p300. Cells were treated with 100nM TSA and 2.5mM NAM for 16 hours (HDACi). Data are representative of at least 3 independent experiments.
To examine a possible role for p300 in Foxp3-mediated transcriptional activity, we performed transcription reporter assays in which cells were transfected with NFAT and an IL-2 promoter luciferase reporter together with Foxp3 in the absence or presence of p30025 (Figure 2B). Foxp3 expression resulted in clear repression of IL-2 promoter activity, whereas the IPEX mutant Foxp3 ΔE250, which does not dimerize, was not transcriptionally functional as expected. Cotransfection of p300 significantly increased Foxp3 transcriptional repression (P = .008), indicating an increased functionality of acetylated Foxp3.
Because p300-mediated acetylation resulted in increased Foxp3 activity, Foxp3 protein expression levels were analyzed. Cells were transfected with Foxp3, incubated with HDACi, and Foxp3 levels were determined by immunoblot analysis. We observed a dramatic, dose-dependent increase in Foxp3 protein levels in HDACi-treated cells (Figure 2C). Similarly, p300 cotransfection increased Foxp3 protein levels in a dose-dependent manner (Figure 2D). Expression levels of the related Foxo3 transcription factor, which has previously been described to be acetylated, was unaffected by HDACi treatment or p300 cotransfection18 (Figure 2E). These results demonstrate that p300-dependent Foxp3 acetylation can regulate its protein expression level.
Because it has been described that Foxp3 function depends on homodimerization,26 we also investigated whether dimerization is also necessary for p300-mediated acetylation. Flag-Foxp3 or Flag-Foxp3ΔE250 was immunoprecipitated, and acetylation levels were analyzed. Foxp3ΔE250 acetylation was comparable with that of wild-type Foxp3 in the presence or absence of p300 (supplemental Figure 1A, available on the Blood website; see the Supplemental Materials link at the top of the online article). To determine whether acetylation of Flag-Foxp3ΔE250 also resulted in increased protein expression levels, cells transfected with Flag-Foxp3 or Flag-Foxp3ΔE250 were treated with HDACi or cotransfected with p300. Both Foxp3 and Foxp3ΔE250 protein levels were increased by HDACi and p300 to a similar extent (supplemental Figure 1B), indicating that acetylation and modulation of protein expression are not dependent on dimerization per se.
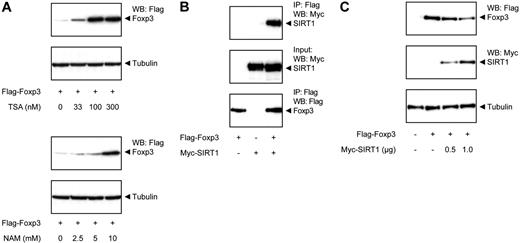
Because we observed that Foxp3 protein levels are modulated by p300-mediated acetylation, we also wished to examine the molecular mechanism regulating deacetylation. To further pinpoint which HDAC is responsible for the effects observed, cells were treated with either TSA or NAM, and Foxp3 expression levels were analyzed. TSA is an inhibitor of multiple HDAC families, whereas NAM selectively inhibits the SIRT HDAC family. Both TSA and NAM treatment increased Foxp3 protein levels in a dose-dependent manner (Figure 3A), indicating that Foxp3 can be deacetylated by members of at least 2 independent HDAC families.
SIRT1 deacetylates Foxp3 and decreases its expression. (A) Flag-Foxp3–transfected HEK 293 cells were treated with increasing amounts of TSA (top panel) or NAM (bottom panel) for 16 hours. Foxp3 levels were determined using antibodies against Flag or tubulin as loading control. (B) Cells were cotransfected with Flag-Foxp3 and Myc-SIRT1. Cell lysates were immunoprecipitated using anti-Flag beads and analyzed using anti-Flag or anti-Myc antibodies. (C) Cells were cotransfected with Flag-Foxp3 and Myc-SIRT. Cell lysates were quantified and analyzed by Western blotting using anti-Flag antibody and tubulin as loading control.
SIRT1 deacetylates Foxp3 and decreases its expression. (A) Flag-Foxp3–transfected HEK 293 cells were treated with increasing amounts of TSA (top panel) or NAM (bottom panel) for 16 hours. Foxp3 levels were determined using antibodies against Flag or tubulin as loading control. (B) Cells were cotransfected with Flag-Foxp3 and Myc-SIRT1. Cell lysates were immunoprecipitated using anti-Flag beads and analyzed using anti-Flag or anti-Myc antibodies. (C) Cells were cotransfected with Flag-Foxp3 and Myc-SIRT. Cell lysates were quantified and analyzed by Western blotting using anti-Flag antibody and tubulin as loading control.
To verify that SIRT is involved in modulation of Foxp3 expression, interaction between Foxp3 and SIRT was confirmed by coimmunoprecipitation after cotransfection of cells with Flag-Foxp3 and myc-tagged SIRT1 (Figure 3B). We next assessed whether SIRT1 could also modulate Foxp3 expression by cotransfecting cells with Flag-Foxp3 and myc-SIRT1. Immunoblots revealed that Foxp3 protein levels were clearly reduced in a dose-dependent manner in cells cotransfected with SIRT1 (Figure 3C). Taken together, these data support a role for acetylation/deacetylation in regulating Foxp3 protein expression levels, a process that can be reciprocally regulated by p300 and SIRT.
Acetylation impairs proteasome-mediated Foxp3 degradation.
To further evaluate the molecular mechanism underlying acetylation-dependent modulation of Foxp3 expression, proteasomal degradation was abrogated using the inhibitor MG132. Cells transfected with equal amounts of Flag-Foxp3 were incubated with HDACi with or without MG132, and Foxp3 protein levels were analyzed. Treatment with MG132 alone dramatically increased Foxp3 levels, indicating that Foxp3 is rapidly degraded by the proteasome. Importantly, treatment with HDACi did not further increase Foxp3 protein levels when cells were simultaneously incubated with MG132 (Figure 4A). To validate these observations, we repeated the experiments using additional specific proteasome inhibitors epoxomycin and lactacystin (Figure 4B). Similar results were obtained compared with MG132, indicating that Foxp3expression level is regulated in a proteasome-dependent manner.
Foxp3 acetylation prevents proteasomal degradation. Flag-Foxp3–transfected HEK 293 cells were treated with 2μM MG132 (A), 100nM epoxomycin, or 10μM lactacystin (B) for 16 hours to inhibit proteasome function and/or HDACi TSA 100nM and NAM 2.5mM also for 16 hours. Foxp3 expression was analyzed using a Foxp3 antibody, and equal loading was verified by analyzing tubulin expression. (C) Cells were transfected with equal amounts of Flag-Foxp3. Half of the cells were treated with or without TSA 100nM and NAD 2.5mM for 16 hours (right panel) and 5 μg/mL CHX for the indicated time points. Foxp3 expression was analyzed using a Flag antibody, and tubulin expression was used as a loading control. Results are representative of at least 3 independent experiments. (D) Flag-Foxp3 or a Flag-tagged Foxp3 mutant in which all the lysines are mutated to arginines (Flag-Foxp3 K22xR) was transfected into HEK 293 cells. The cells were treated with the HDACi TSA (100nM) and NAM (2.5mM) for 16 hours. Cell lysates were made and immunoblotted for Flag and tubulin as control. (E) HEK 293 cells were transfected with Flag-Foxp3, treated with or without TSA 100nM and NAD 2.5mM for 16 hours, and cell lysates were immunoprecipitated using anti-Flag beads and analyzed using anti-ubiquitin and anti-Flag as transfection control.
Foxp3 acetylation prevents proteasomal degradation. Flag-Foxp3–transfected HEK 293 cells were treated with 2μM MG132 (A), 100nM epoxomycin, or 10μM lactacystin (B) for 16 hours to inhibit proteasome function and/or HDACi TSA 100nM and NAM 2.5mM also for 16 hours. Foxp3 expression was analyzed using a Foxp3 antibody, and equal loading was verified by analyzing tubulin expression. (C) Cells were transfected with equal amounts of Flag-Foxp3. Half of the cells were treated with or without TSA 100nM and NAD 2.5mM for 16 hours (right panel) and 5 μg/mL CHX for the indicated time points. Foxp3 expression was analyzed using a Flag antibody, and tubulin expression was used as a loading control. Results are representative of at least 3 independent experiments. (D) Flag-Foxp3 or a Flag-tagged Foxp3 mutant in which all the lysines are mutated to arginines (Flag-Foxp3 K22xR) was transfected into HEK 293 cells. The cells were treated with the HDACi TSA (100nM) and NAM (2.5mM) for 16 hours. Cell lysates were made and immunoblotted for Flag and tubulin as control. (E) HEK 293 cells were transfected with Flag-Foxp3, treated with or without TSA 100nM and NAD 2.5mM for 16 hours, and cell lysates were immunoprecipitated using anti-Flag beads and analyzed using anti-ubiquitin and anti-Flag as transfection control.
These data suggest that Foxp3 proteasomal turnover is abrogated by HDACi. To confirm this, we made use of CHX, a widely used inhibitor of protein translation (Figure 4C). Cells were transfected with Flag-Foxp3 and then treated with CHX before analyzing Foxp3 expression levels. CHX treatment led to a decrease in Foxp3 protein levels within 6 hours. However, when the cells were also treated with HDACi, Foxp3 protein levels stabilized, confirming that Foxp3 protein stability is indeed increased through acetylation.
Proteasomal degradation is mediated by polyubiquitination, which, similar to acetylation, occurs at lysine residues. This suggests that acetylation of lysine residues may prevent ubiquitination and thereby inhibit Foxp3 degradation. To test this hypothesis, we generated a Foxp3 mutant in which all 22 lysines were mutated into arginines (Foxp3 K22xR). If direct Foxp3 ubiquitination is indeed responsible for increased protein turnover, then this mutant should exhibit enhanced stability; importantly, Foxp3 protein levels should no longer be modulated by HDACi. Cells were transfected with equal amounts of Flag-Foxp3 or Flag-Foxp3 K22xR and incubated with HDACi (Figure 4D). As previously demonstrated, hyperacetylation increased the levels of wild-type Foxp3. In contrast, Foxp3 K22xR protein levels were not altered by treatment with HDACi. Importantly, we observed that the levels of Foxp3 K22xR were considerably increased compared with wild-type Foxp3. To confirm our hypothesis that there is competition for the same lysines by acetylation and ubiquitination, we analyzed the effect of HDACi on the level of Foxp3 ubiquitination. Cells were transfected with Foxp3 and treated with HDACi. Subsequently, Foxp3 was immunoprecipitated, blotted, and evaluated using an antiubiquitin antibody. As shown in Figure 4E, there is a basal level of polyubiquitinated Foxp3. Treatment with HDACi, however, considerably reduced the amount of polyubiquitinated Foxp3.
Taken together, these data demonstrate that Foxp3 protein levels are rapidly turned over in a proteasome-dependent manner and that acetylation probably prevents entry of Foxp3 into this pathway by competing with polyubiquitination.
Acetylation increases Foxp3 levels in primary T cells
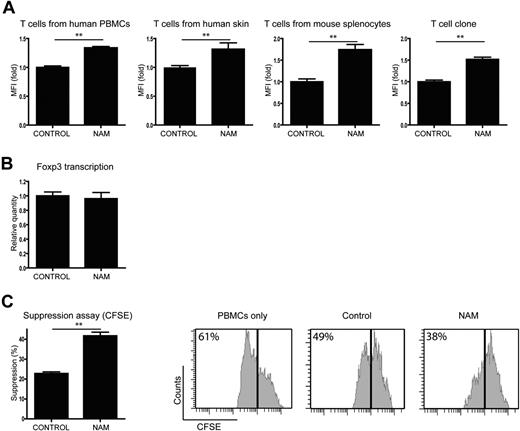
We next assessed the effect of acetylation on Foxp3 levels using primary T cells in 4 different models. T cells from 4 different sources were stimulated with IL-2, anti-CD3, and anti-CD28 to induce Foxp3, and Foxp3+ cells were analyzed by flow cytometry. First, CD4+ cells from human PBMCs were treated with either the SIRT activator resveratrol,27 or the SIRT inhibitor NAM or control (Figure 5A; supplemental Figure 2). We observed significantly decreased numbers of Foxp3+ cells in the resveratrol-treated cells, whereas the cells treated with NAM showed a significantly higher percentage of Foxp3+ cells compared with control. Second, because it has recently been reported that Tregs play a crucial role in immune homeostasis of the skin,28 T cells were isolated from human skin samples and treated with resveratrol or NAM for 14 days (Figure 5B). Analysis of Foxp3+ cells again revealed that treatment with resveratrol decreased the percentage of Foxp3+ cells, whereas NAM increased Foxp3+ cell numbers. T cells were also isolated from healthy C57BL/6 mice spleens and cultured in the presence of resveratrol or NAM (Figure 5C). Activation of SIRT resulted in a diminished percentage of Foxp3+ cells. Treatment with NAM, however, increased the number of Foxp3+ cells by up to 10-fold. We also generated human T-cell clones, which were treated with resveratrol or NAM (Figure 5D). Again, resveratrol treatment decreased the numbers of Foxp3+ cells, whereas inhibition of SIRT in these cells resulted in a significantly increased percentage of Foxp3+ cells. Taken together, these data demonstrate that SIRT-mediated regulation of acetylation influences Foxp3 expression levels in both mouse and human primary T cells from a variety of origins.
Treatment of primary T cells with a SIRT inhibitor results in increased Foxp3+ cell numbers. Isolated primary T cells were cultured in the presence of IL-2, anti-CD3, and anti-CD28 (mouse splenocytes were also cultured in the presence of transforming growth factor-β). Cells were treated with the SIRT activator resveratrol, the SIRT inhibitor NAM, or carrier as control. The percentage of Foxp3+ cells was determined using FACS technology. T cells originated from human PBMCs (A), human skin (B), mouse spleen (C), and a human T-cell clone (D). Results are the means of at least 4 independent experiments. **P < .01.
Treatment of primary T cells with a SIRT inhibitor results in increased Foxp3+ cell numbers. Isolated primary T cells were cultured in the presence of IL-2, anti-CD3, and anti-CD28 (mouse splenocytes were also cultured in the presence of transforming growth factor-β). Cells were treated with the SIRT activator resveratrol, the SIRT inhibitor NAM, or carrier as control. The percentage of Foxp3+ cells was determined using FACS technology. T cells originated from human PBMCs (A), human skin (B), mouse spleen (C), and a human T-cell clone (D). Results are the means of at least 4 independent experiments. **P < .01.
Hyperacetylation of Foxp3 increases Treg function
Because we observed that acetylation impairs proteasomal degradation (Figure 4), we analyzed whether Foxp3 protein levels per cell were regulated by SIRT, as are the Foxp3+ cell numbers. T cells from all 4 sources were treated with NAM or control and the mean fluorescence intensity of Foxp3+ cells was analyzed by FACS (Figure 6A). The mean fluorescence intensity of the NAM-treated cells was significantly higher compared with the control-treated cells for all T-cell types, indicating that SIRT inhibition not only increased the number of Foxp3+ cells but also the Foxp3 levels per cell compared with control. To elucidate whether the changes in Foxp3 expression are the result of increased protein stability, and not increased transcription, Foxp3 mRNA levels were analyzed. mRNA was isolated from control and NAM-treated T cells from the human T-cell clone that was previously used (Figure 5D). cDNA was prepared and analyzed using quantitative PCR for Foxp3 and β2M as reference. The ratio of Foxp3 versus β2M per Foxp3+ cell is shown in Figure 6B. No significant difference in Foxp3 mRNA levels per Foxp3+ cell was observed, demonstrating that Foxp3 transcription is not influenced by SIRT.
SIRT inhibition increases Treg functionality. Isolated primary T cells were cultured in the presence of IL-2, anti-CD3, and anti-CD28. Cells were treated with the SIRT inhibitor NAM or carrier as control. (A) Foxp3 levels per cells of different T-cell sources were analyzed by FACS, and data represent the mean fluorescence intensity of the Foxp3+ population. (B) Cells from a human T-cell clone were stimulated and treated with NAM or control. Foxp3 transcription was determined with quantitative PCR, and results were corrected for the housekeeping gene β2M and Foxp3+ cell numbers. (C) CD4 T cells isolated from mouse splenocytes were stimulated using IL-2, anti-CD3, and anti-CD28 beads and treated with NAM or control. CD25+ cells were sorted, and the function of these induced Tregs was analyzed using a standard suppression assay. A total of 10 000 effector T cells were cocultured with 2000 sorted CD25+ cells for 4 days (without NAM). Proliferation of effector T cells on day 4 was measured by CFSE dilution within the CD4+ effector T-cell population. Data shown are representative of at least 3 independent experiments. **P < .01.
SIRT inhibition increases Treg functionality. Isolated primary T cells were cultured in the presence of IL-2, anti-CD3, and anti-CD28. Cells were treated with the SIRT inhibitor NAM or carrier as control. (A) Foxp3 levels per cells of different T-cell sources were analyzed by FACS, and data represent the mean fluorescence intensity of the Foxp3+ population. (B) Cells from a human T-cell clone were stimulated and treated with NAM or control. Foxp3 transcription was determined with quantitative PCR, and results were corrected for the housekeeping gene β2M and Foxp3+ cell numbers. (C) CD4 T cells isolated from mouse splenocytes were stimulated using IL-2, anti-CD3, and anti-CD28 beads and treated with NAM or control. CD25+ cells were sorted, and the function of these induced Tregs was analyzed using a standard suppression assay. A total of 10 000 effector T cells were cocultured with 2000 sorted CD25+ cells for 4 days (without NAM). Proliferation of effector T cells on day 4 was measured by CFSE dilution within the CD4+ effector T-cell population. Data shown are representative of at least 3 independent experiments. **P < .01.
To confirm that acetylation not only regulates Foxp3+ cell numbers and Foxp3 protein levels, but also Treg suppressor capacity, a suppression assay was performed. Primary T cells were stimulated and cultured with NAM for 5 days. Subsequently, the functionality of Foxp3+ cells was analyzed in CFSE-based suppression assay (Figure 6C). NAM or control-treated CD4+CD25+ T cells were cocultured with effector T cells in a 1:5 ratio, and proliferation of Teff cells was measured. As expected, Tregs from the control-treated sample were able to suppress the proliferation of effector T cells. Tregs from the NAM-treated cultures suppressed T-cell proliferation compared with the control significantly better. These data clearly demonstrate that SIRT-mediated regulation of acetylation not only modulates Foxp3+ cell numbers but also Foxp3 protein levels independently of increased Foxp3 transcription. Importantly, inhibition of SIRT also results in functionally improved Treg cells.
Discussion
Foxp3 is a key transcription factor controlling immune homeostasis through regulating both the numbers and functionality of immunosuppressive Tregs. Although recent studies have focused on identifying Foxp3 transcriptional targets or the molecular mechanisms underlying transcriptional regulation of Foxp3 expression, analysis of specific posttranslational modifications regulating Foxp3 function have thus far not been critically addressed. Here we report, for the first time, that Foxp3 protein levels are directly controlled by acetylation, a process mediated by inhibition of proteasomal degradation. Moreover, we identified novel interaction partners that are key mediators of this of this process: the HAT p300 and the HDAC SIRT1. Furthermore, hyperacetylating Foxp3 in primary T cells resulted in more Foxp3+ cells, higher Foxp3 protein levels per cell, and better suppressive capabilities. Our data provide novel insights into posttranslational mechanisms regulating Foxp3 function and provide a model by which Treg numbers and functionality can be rapidly modulated by the extracellular milieu.
A recent study by Tao et al has demonstrated that in vivo HDAC inhibition results in enhanced Treg-mediated suppression of homeostatic proliferation, decreased inflammatory bowel disease through Treg-dependent effects, and induction of permanent tolerance against islet and cardiac allografts.14 Because we have demonstrated that HDACi increase Foxp3 expression through protein stabilization, this suggests a molecular mechanism explaining these in vivo observations. Importantly, we demonstrate that p300 increases Foxp3 expression levels through inhibition of proteasomal degradation. This is a novel and unique observation for members of the forkhead transcription factor family.3 Degradation of other transcription factors is mediated by polyubiquitination of these proteins. Because lysine acetylation and ubiquitination are mutually exclusive, acetylation may thus prevent polyubiquitination through a competition-based mechanism. Indeed, in Figure 4, we demonstrate that, by mutating all Foxp3 lysines into arginines, we significantly increased Foxp3 protein levels, a process most probably mediated by inhibition of proteasomal degradation because Foxp3 K22xR cannot be polyubiquitinated. Inhibition of the proteasome-mediated degradation dramatically increased and stabilized Foxp3 protein levels, which could not be further increased by treatment with HDACi. Furthermore, treatment with HDACi did not further increase Foxp3 K22xR protein levels as observed in wild-type. In addition, we also show that treatment with HDACi dramatically decreased polyubiquitination of Foxp3. Taken together, we propose that polyubiquitination-mediated Foxp3 degradation can be impaired by acetylation of lysines in a competition-based mechanism. A similar mechanism has been previously described for RUNX3, Smad7, and p53 where polyubiquitination of these transcription factors was significantly impaired by hyperacetylation.29-32
Although p300 and TIP60 were both found to associate with Foxp3, we found that only p300 was able to acetylate Foxp3 (Figures 1, 2). Li et al recently demonstrated that Foxp3 is part of a transcriptional complex containing both HDAC and HAT.13 This study reported that TIP60 overexpression promotes Foxp3-mediated transcriptional repression. However, as we did not observe TIP60-mediated Foxp3 acetylation, our data suggest that TIP60 predominantly acts indirectly on other (non) histone proteins in the transcription/repression complex, rather than directly modulating Foxp3 transcriptional function.
We also examined which HDAC is responsible for regulating Foxp3 deacetylation. Treatment with either TSA or NAM alone was sufficient to result in increased Foxp3 expression (Figure 3). TSA has a broad specificity, inhibiting HDAC families I, II, and IV. It has also recently been reported that both HDAC7 and HDAC9 associate with Foxp3 in a multimeric protein complex,13 and it is possible that Foxp3 deacetylation can also be mediated by one of these HDACs. Because NAM is a specific inhibitor of the SIRT HDAC III family, we conclude that there is also a role for SIRT in regulating Foxp3 activity, which we could subsequently further substantiate by coimmunoprecipitation experiments (Figure 3). Furthermore, ex vivo treatment of CD4+ T cells from 4 different sources with the SIRT activator resveratrol decreased the number of Foxp3+ cells. Interestingly, multiple studies have reported that resveratrol has antitumor activity, although through an as yet undefined mechanism.33,34 Because Tregs repress immune function,35,36 it is interesting to speculate that resveratrol may act by reducing Foxp3 protein levels, thereby relieving Treg-mediated immune suppression and ultimately resulting in enhanced immune activity toward tumors.
Although there has been a general paradigm that Foxp3 is exclusively expressed by regulatory T cells, it was recently shown that respiratory, thymic, prostate, and mammary epithelial cells also express this transcription factor.8 Foxp3 function has also been directly linked with tumor suppressor activity; it was demonstrated that Foxp3 binds and represses the promoter of the HER-2/erbB2 and SKP2 oncogenes.37,38 In addition, somatic mutations and down-regulation of Foxp3 were found in human breast cancer samples and correlated significantly with HER-2/erbB2 and SKP2 expression. Because we have demonstrated that acetylation increases Foxp3 expression, it is plausible that treatment with HDACi may enhance the tumor suppressor activity of Foxp3 and therefore have beneficial effects on tumor formation in the breast epithelium.
It has now become clear that CD4+ effector T cells can also express Foxp3 after TCR stimulation in vitro, and Foxp3 expression has been strongly correlated with hyporesponsiveness of activated T cells.9-11 However, not all TCR-stimulated Foxp3+ cells have suppressive capabilities. In nonsuppressive Foxp3+ T-cell populations, Foxp3 expression was found to be transient, whereas stably expressing Foxp3 cells had true suppressive capabilities with a phenotype similar to that of naturally occurring Tregs.9,10 We speculate that this critical difference in transient versus stable Foxp3 expression is a key switch in the generation of “true” suppressor Tregs and that this is directly regulated by acetylation. It is possible that normally antigen exposure activates T cells, resulting in transient Foxp3 expression levels, which may act as an “immune brake” preventing T-cell hyperactivation through repression of IL-2 and interferon-γ transcription.39 However, when T cells are chronically stimulated, for example, by self-antigen, Foxp3 is acetylated stabilizing protein levels and allowing the initiation of a unique transcriptional program defining the Treg phenotype. Further work will be required to determine whether this is indeed the case.
Taken together, we have established that Foxp3 protein levels can be tightly regulated by acetylation. p300-mediated acetylation of Foxp3 plays a critical role in stabilizing Foxp3 protein levels. In contrast, we found that SIRT1-mediated deacetylation of Foxp3 results in reduced protein expression levels in cell lines as well as primary T cells. In addition, we demonstrate that the mechanism by which Foxp3 protein levels are stabilized by acetylation is by inhibition of proteasomal degradation. Our findings have important consequences for the development of novel molecular therapies regulating Treg numbers through pharmacologic stabilization of Foxp3 protein levels.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr S. Sakaguchi (Institute for Physical and Chemical Research, Yokohama, Japan) for providing the MIGR1-Foxp3 construct, Dr L. W. Kraus for providing His(6x)-p300 and His(6x)-p300mut-AT2, Dr D. Trouche (Université Paul Sabatier, Toulouse, France) for providing pCDNA3-HA-TIP60, Dr B. M. T. Burgering (University Medical Center Utrecht, The Netherlands) for providing pcDNA-MYC-SIRT1 and pcDNA3-NFATc/A, and K. Dreijerink for technical assistance with the bacalovirus system.
J.v.L. and Y.V. were supported by a grant from the Dutch Rheumatology Foundation. J.M.B. was supported by a grant from the Dutch Scientific Organization (NWO 917.36.316).
Authorship
Contribution: J.v.L. wrote the paper, designed and performed the research, and analyzed data; Y.V., T.G., J.M.B., and O.v.B. designed and performed the research and analyzed data; Y.Y.J.G performed research and analyzed data; A.B.B., D.-J.H., T.M., E.K., and B.J.P. designed the research and analyzed data; and P.J.C. designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paul J. Coffer, Molecular Immunology Lab, Department of Immunology, University Medical Centre Utrecht, Lundlaan 6, 3584 EA Utrecht, The Netherlands; e-mail: p.j.coffer@umcutrecht.nl.

![Figure 1. p300 promotes Foxp3 acetylation. (A) Flag immunoprecipitation from lysates from Flag-Foxp3–transfected cells. HEK 293 cells were treated with HDACi: + indicates 50 nm TSA and 1mM NAM; or ++, 250 nm TSA and 5mM NAM for 16 hours. Equal amounts of immunoprecipitated Foxp3 were separated by SDS-PAGE, and Western blots were probed for acetylated lysines (Ac-Lys) or Flag. (B) Cells were transfected with Flag-Foxp3 and/or HA-p300. Flag-Foxp3 was immunoprecipitated with anti-Flag beads and analyzed using acetyl lysines (Ac-Lys), Flag, or HA antibodies. (C) Cells were cotransfected with Flag-Foxp3 and HA-p300 or HA-TIP60 and immunoprecipitated using anti-Flag beads. Immunoblots were analyzed with acetyl-lysine (Ac-Lys) antibody, anti-Flag, or anti-HA. Cells were cotransfected with Flag-Foxp3, HA-Tip60 (D), or HA-p300 (E). Cell lysates were coimmunoprecipitated using anti-HA beads and analyzed using anti-Flag or anti-HA. (F) GST-Foxp3 fusion protein coupled to Sepharose beads was incubated with [14C]-labeled acetyl-CoA and p300 or the acetylase dead p300mutAT2; samples were separated on SDS-PAGE and analyzed using films sensitive for radioactivity. Results are representative of at least 3 independent experiments. *Aspecific band. IP indicates immunoprecipitation; and WB, Western blot.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/5/10.1182_blood-2009-02-207118/4/m_zh89990948230001.jpeg?Expires=1767704095&Signature=XJikg2MeB6~2RVN8WY33jHLSQ7SWE4FycrM3kwmU9-n0IETQggixm1m-Xw~iSH50-clDRdJ1ymdNZMzVDSKpykv34yW~BOyKIYqwlAlgwr0rK6h6SfWA1ore6aSmBVkcXc~e0tqdRHuEfk81QhfwGQ4B-G80Vqrmh~fDcgJNzXYoz~KYXEhmuAxy98ykKbHcUnFDOsEwJ2Sgfm4afFpQmvx2Nyk2kGmOLEWt20pd0G3mBDHshiwC5idu4Izcc1n~LyU2mqGdPP3~S8Mtp2NEuljbr3V3S7dVSrPgfyrqYPIaadrbPcvfKWKGlSa97r3N4fxJTfrwKIqMms8baZ0zKw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal