Abstract
Human natural killer (NK) cells express Toll-like receptor 9 (TLR9) transcript and, upon exposure to microbial CpG oligodeoxynucleotide (ODN), release cytokines and kill target cells. Here we show that NK cell treatment with CpG ODN results in down-modulation of KIR3DL2 inhibitory receptor from the cell surface and in its cointernalization with CpG ODN. CpG ODN–induced interferon-γ (IFN-γ) release is mostly confined to KIR3DL2+ NK cells, thus suggesting a crucial role of KIR3DL2 in CpG ODN–mediated NK responses. Using soluble receptor molecules, we demonstrate the direct binding of KIR3DL2 to ODNs and we show that the D0 domain is involved primarily in this interaction. KIR3DL2 modulation is also induced in malignant cells of Sézary cutaneous T-cell lymphoma, a disease in which KIR3DL2 represents a typical marker of malignant T cells. Confocal microscopy analysis suggests that, in human NK cells, CpG ODN can encounter TLR9 in early endosomes after being shuttled to these sites by KIR3DL2, which functions as a CpG ODN receptor at the cell surface. This novel KIR-associated function emphasizes the antimicrobial role of NK cells in the course of infection.
Introduction
Innate immunity plays a crucial role in limiting or even in eradicating pathogens during the early phases of primary infections, before T and B cells can mount efficient adaptive responses. Natural killer (NK) cells, phagocytes, and other innate effector cells do not require clonal expansion to mediate their function and can enter and defend a tissue almost as soon as it becomes infected. These effector cells can eliminate pathogens by different mechanisms, including killing of infected cells and secretion of cytokines and chemokines, which promote inflammation and may further recruit/activate other cells of the innate immunity. In many instances, the prompt and different reactions of the innate immunity to pathogen invasion can lead to termination of infection with no further involvement of the adaptive immunity and no development of manifest disease. In addition, thanks to their interaction with dendritic cells, NK cells may influence the magnitude and the quality of subsequent adaptive immune responses.1-6 Recent advances in understanding the biologic role of different cell types of the innate immunity include mainly the discovery and the molecular characterization of an array of cell surface receptors. The structure and the evolution of genes coding for these receptors reflect mostly the need to adapt to mechanisms of pathogen evasion. A key example is represented by the human leukocyte antigen (HLA) class I–specific human killer immunoglobulin (Ig)–like receptor (KIR)–encoding genes. KIRs belong to the Ig superfamily and, in most instances, recognize determinants shared by groups of HLA-A, HLA-B, or HLA-C allotypes.7-11 From an evolutionary point of view, all KIRs derive from an ancestral molecule composed of 3 Ig-like domains, D0, D1, and D2 (KIR3D), and a long cytoplasmic tail. The predominant human KIRs are characterized by 2 Ig-like domains (KIR2Ds).12 There are 2 types of KIR2Ds: the first type (including KIR2DL1/L2/L3 and KIR2DS1/S2/S3/S4/S5) is composed of domains homologous to D1 and D2. The majority of these KIRs are specific for HLA-C molecules. KIRs belonging to the second type contain domains homologous to D0 and D2 (KIR2DL4/L5). KIRs composed of 3 Ig domains are specific either for the HLA-Bw4 group of alleles (KIR3DL1 and possibly KIR3DS1)13 or for some HLA-A alleles (KIR3DL2).14,15 All KIRs, with the exception of KIR2DL4, display a clonally distributed expression in human NK cells.7,16,17 KIRs were also detected on a small subset of cytolytic T lymphocytes18,19 and in the Sézary cutaneous T-cell lymphoma, in which both infiltrating and circulating malignant T cells are characterized by KIR3DL2 expression.20,21
Cells of the innate immune system of vertebrates recognize pathogen-associated molecular patterns and undergo activation mainly through specialized molecules called Toll-like receptors (TLRs). NK cells express different TLRs including TLR2,22 TLR5,23 TLR7,24 TLR3, and TLR9.25 Upon interaction with their specific ligands (poly I:C and CpG-ODNs, respectively), TLR3 and TLR9 promote cytokine release and increments of NK cytotoxicity. Responses to these stimuli were elicited by fresh NK cells as well as by interleukin-2 (IL2)–cultured NK cell populations.25 Among different ODNs, ODN C induced the highest release of cytokines and increments of cytotoxicity in NK cells.26
Leifer et al27 and Latz et al28 demonstrated that in B cells and plasmacytoid dendritic cells, respectively, prior to cell stimulation, TLR9 is localized in the endoplasmic reticulum while, upon cell exposure to CpG ODN, it translocates to endosomes where both binding of ODN and initiation of signal transduction take place.
In the present study, we show that exposure of NK cells or Sézary T cells to CpG ODNs results in sharp modulation of surface KIR3DL2 molecules that are rapidly internalized in intracellular compartments together with CpG ODNs. CpG ODN–induced interferon-γ (IFN-γ) production was confined mostly to NK cells expressing KIR3DL2. The use of KIR3D or KIR2D soluble receptors revealed specific binding of ODNs to KIRs bearing the D0 domain. These data suggest that certain KIRs not only may function as receptors for HLA class I molecules but also may mediate the uptake and internalization of CpG ODN, and the induction of rapid antimicrobial responses.
Methods
Sézary cell lines and analysis of TLR9 transcript
The Sézary cell lines used in this study were HUT78, PNO, and CS. Cells were cultured in RPMI1640 supplemented with 10% fetal calf serum, 10 ng/mL recombinant human IL7 (PeproTech), and 200 UI/mL recombinant human IL2 (Proleukin; Chiron Corp).
Total RNA was extracted from Sézary cell lines using RNeasy mini kit (QIAGEN) according to the manufacturer's instructions, and cDNA synthesis was performed on 500 ng of RNA using hexameric primers. To exclude that polymerase chain reaction amplifications were due to DNA contaminations, RNA was treated with RNase-free DNase (QIAGEN) and further retrotranscribed in the presence or absence of retrotranscripatse enzyme (Roche). The primers used for the TLR9 amplification (TLR9 up: 5′CAg CCA TAC CAA CAT CCT g and TLR9 down: 5′AAA ggA CAC CCT CTT TTg g) allow for amplification of a segment of 605 bp. The amplification conditions were 33 cycles: 30 seconds at 95°C, 30 seconds at 55°C, and 30 seconds at 72°C.
Stimulation of human NK cell populations, NK cell clones, and Sézary cell lines
Cells were cultured for the indicated time intervals in 24-well plates at a concentration of 106 in 1 mL of RPMI1640 supplemented with 10% fetal calf serum, 2 mM l-glutamine, 1% penicillin-streptomycin-neomycin (PSN) in the absence or presence of different stimuli.
The following modified CpG ODNs were synthesized and purified by TIB MOLBIOL Srl (Centro di Biotecnologie Avanzate) and used at the indicated final concentration: ODN 2216 (ODN A) 5′-ggG GGA CGA TCG TCg ggg gg, ODN 2006 (ODN B) 5′-tcg tcg ttt tgt cgt ttt gtc gtt, ODN 2395 (ODN C) 5′-tcg tcg ttt tcg gcg cgc gcc g, poly A 5′-aaa aaa aaa aaa aaa aaa aaa a, phosphodiester (PO) ODN B 5′-TCG TCG TTT TGT CGT TTT GTC GTT, and PO ODN C 5′-TCG TCG TTT TCG GCG CGC GCC G. Bases shown in capital letters are phosphodiester; those in lower case are phosphorothioate. ODN C was also synthesized by TIB MOLBIOL Srl with a 5′ fluorescein isothiocyanate (FITC) or Texas red (TxRed) label. Double-stranded ODN B (ds ODN B) was synthesized by Sigma-Aldrich.
After stimulation, cells were stained with the appropriate monoclonal antibodies (mAbs), followed by R-phycoerythrin–conjugated, isotype-specific, goat anti–mouse secondary antibodies (Southern Biotechnology). Samples were analyzed by 1-color cytofluorimetric analysis (FACScan; Becton Dickinson).
Results
CpG ODNs induce modulation of surface KIR3DL2 in NK cells and Sézary T cells
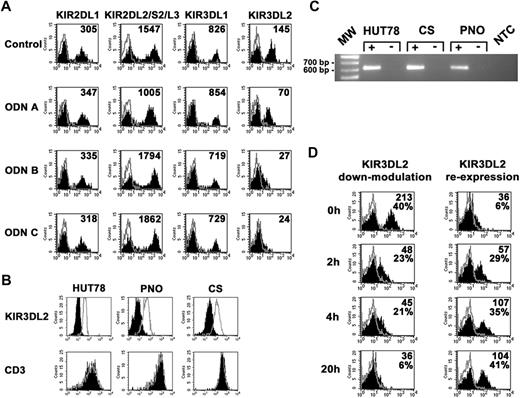
A panel of NK cell surface markers, including KIRs (Figure 1A), natural cytotoxicity receptors (NCRs), NKG2D, NKG2A, CD16, CD56, DNAM-1, CD2, LFA-1, 2B4, NTBA (not shown), was assessed by cytofluorimetric analysis in IL2-activated NK cell populations that had been cultured for 20 hours in the presence or absence of different CpG ODNs. NK cell markers did not reveal substantial differences in surface expression from unstimulated controls. A remarkable exception was represented by KIR3DL2. This KIR was sharply down-regulated, particularly upon cell treatment with ODN C (Figure 1A). Similar results were obtained in 10 different donors who were analyzed. Flagellin (TLR5 ligand), poly U, RNA40 or R848 (TLR7 and TLR8 ligands), and gardiquimod (TLR7 ligand) did not modify KIR3DL2 expression, whereas poly I:C (TLR3 ligand) induced some degree of modulation, although this was much less efficient compared with ODN C (not shown). Moreover, cross-linking of NKp46 or 2B4 using activating NK receptor–specific mAbs coated to plates did not modify KIR3DL2 expression (not shown).
CpG ODNs induce modulation of surface KIR3DL2 on IL2-activated NK cell populations and Sézary lymphoma T cells. (A) IL2-activated NK cell populations were cultured for 20 hours either in the absence or in the presence of ODN A (5 μg/mL), ODN B (5 μg/mL), or ODN C (5 μg/mL) and then assessed by cytofluorimetric analysis for the surface expression of various NK cell markers. Black profiles indicate the expression of different NK cell molecules. Gray profiles correspond to isotypic controls. The MFI of the positive peak is indicated in each histogram. Results are representative of 10 distinct experiments. (B) HUT78, PNO, and CS cell lines were cultured for 20 hours either in the absence (gray profiles) or in the presence (black profiles) of ODN C (5 μg/mL) and then analyzed for KIR3DL2 and CD3 surface expression. Results are representative of 4 distinct experiments. (C) Polymerase chain reaction analysis of TLR9 transcript in HUT 78, CS, and PNO cell lines. Plus signs (+) indicate samples obtained by retrotranscription in the presence of RT enzyme, minus signs (−) samples obtained in the absence of RT enzyme, used as control for DNA contaminations. Molecular weight marker (MW, given in bp) and no template control (NTC) are also reported. Results are representative of 3 independent experiments. (D) For KIR3DL2-modulation cytofluorimetric analysis of KIR3DL2, expression was assessed on IL2-activated NK cell populations stimulated with ODN C for different time intervals. For KIR3DL2 re-expression, after 20 hours of stimulation, cells were harvested, washed, and cultured without ODN for different time intervals. Surface re-expression of KIR3DL2 was analyzed at each time point. Black profiles indicate KIR3DL2 expression, whereas gray profiles correspond to isotypic controls. The percentage of positive cells and the MFI of the positive peaks are indicated in each histogram. Results are representative of 5 distinct experiments.
CpG ODNs induce modulation of surface KIR3DL2 on IL2-activated NK cell populations and Sézary lymphoma T cells. (A) IL2-activated NK cell populations were cultured for 20 hours either in the absence or in the presence of ODN A (5 μg/mL), ODN B (5 μg/mL), or ODN C (5 μg/mL) and then assessed by cytofluorimetric analysis for the surface expression of various NK cell markers. Black profiles indicate the expression of different NK cell molecules. Gray profiles correspond to isotypic controls. The MFI of the positive peak is indicated in each histogram. Results are representative of 10 distinct experiments. (B) HUT78, PNO, and CS cell lines were cultured for 20 hours either in the absence (gray profiles) or in the presence (black profiles) of ODN C (5 μg/mL) and then analyzed for KIR3DL2 and CD3 surface expression. Results are representative of 4 distinct experiments. (C) Polymerase chain reaction analysis of TLR9 transcript in HUT 78, CS, and PNO cell lines. Plus signs (+) indicate samples obtained by retrotranscription in the presence of RT enzyme, minus signs (−) samples obtained in the absence of RT enzyme, used as control for DNA contaminations. Molecular weight marker (MW, given in bp) and no template control (NTC) are also reported. Results are representative of 3 independent experiments. (D) For KIR3DL2-modulation cytofluorimetric analysis of KIR3DL2, expression was assessed on IL2-activated NK cell populations stimulated with ODN C for different time intervals. For KIR3DL2 re-expression, after 20 hours of stimulation, cells were harvested, washed, and cultured without ODN for different time intervals. Surface re-expression of KIR3DL2 was analyzed at each time point. Black profiles indicate KIR3DL2 expression, whereas gray profiles correspond to isotypic controls. The percentage of positive cells and the MFI of the positive peaks are indicated in each histogram. Results are representative of 5 distinct experiments.
As shown by previous studies, KIR3DL2 is also expressed in the Sézary cutaneous T-cell lymphoma.20,21 Therefore, we analyzed whether ODN stimulation could result in down-regulation of KIR3DL2 on different Sézary T-cell lines. All 3 analyzed cell lines (HUT78, PNO, and CS) were KIR3DL2+ (Figure 1B), KIR2DL4−, and KIR3DL1/S1− (not shown) and expressed the TLR9-encoding transcript (Figure 1C). Similarly to NK cells, a sharp modulation of KIR3DL2 was detected in Sézary T cells treated with ODN-C (Figure 1B).
The kinetics of KIR3DL2 modulation was analyzed in NK cell populations exposed for different time intervals to ODN C. As shown in Figure 1D, a marked modulation of KIR3DL2 occurred 2 hours after ODN C treatment, although the maximum effect was reached after 20 hours. Removal of CpG-ODN resulted in re-expression of KIR3DL2 molecules at the cell surface. Partial re-expression was detected after 2 to 4 hours, whereas it was fully restored after 20 hours (Figure 1D).
Uptake of ODN C by KIR3DL2+ cells
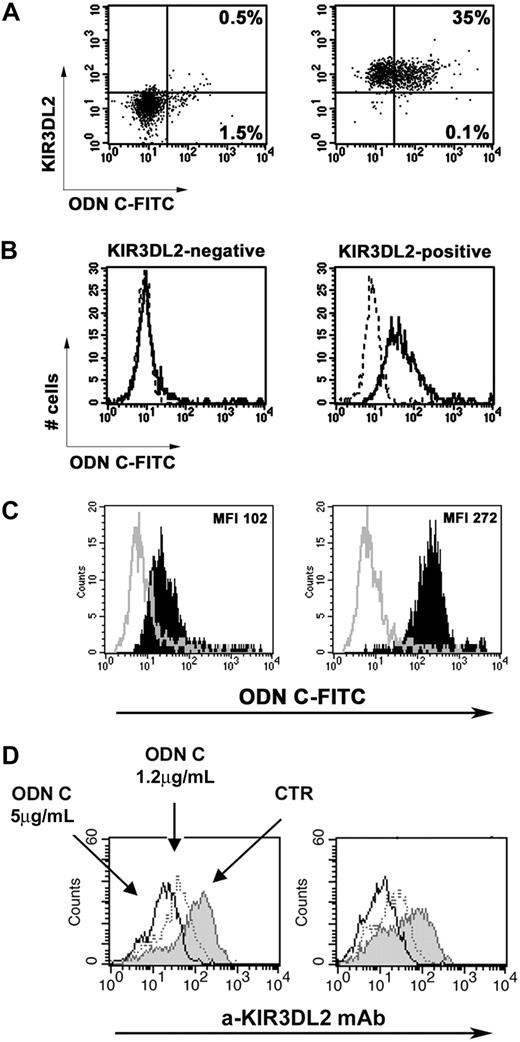
Because ODN C induced efficient cell surface modulation of KIR3DL2, we analyzed whether KIR3DL2 was involved in ODN C uptake. To this end, we first attempted to identify the NK cell subset capable of capturing FITC–ODN C. Fluorescence-activated cell sorting (FACS) analysis was performed 2 hours after NK cell stimulation with ODN C because, at this time point, KIR3DL2 was still partially detectable at the NK cell surface. Unfractionated NK cells pretreated with FITC-labeled ODN C were stained with phycoerythrin-labeled mAbs specific for informative NK cell markers, including NCRs, KIRs, and NKG2A. Double fluorescence analysis revealed that cells capturing FITC–ODN C expressed KIR3DL2. Because at 2 hours there was a partial down-modulation of KIR3DL2 (Figure 1D), further experiments were performed using KIR3DL2+ and KIR3DL2− NK cell fractions purified by FACS sorting. As shown in Figure 2A, KIR3DL2+ NK cells captured ODN C much more efficiently than KIR3DL2− NK cells. Thus, 35% of KIR3DL2+ and only 2% of KIR3DL2− NK cells were ODN C-FITC+. In further experiments, KIR3DL2+ and KIR3DL2− NK cell clones derived from the same donor were analyzed. As shown in Figure 2B, the representative KIR3DL2+ KIR3DL1/S1− KIR2DL1/S1− KIR2DL2/S2/L3− NKG2A+ NK cell clone did capture high amounts of FITC–ODN C, whereas the KIR3DL2− KIR3DL1/S1− KIR2DL1/S1− KIR2DL2/S2/L3− NKG2A+ clone did not. Treatment with dextran sulfate, a polyanion known to compete with ODN binding,29,30 strongly inhibited ODN C uptake by NK cells (Figure 2B).
Preferential uptake of ODN C by KIR3DL2+ cells. The uptake of FITC–ODN C by KIR3DL2+ or KIR3DL2− NK cell populations sorted from IL2-activated NK cell populations (A) or KIR3DL2+ or KIR3DL2− NK cell clones (B; derived from the same donor) was analyzed by flow cytometry after 2 hours of treatment with 5 μg/mL FITC–ODN C. In panel A, the percentage of ODN C–FITC+ cells is indicated in the right quadrants. In panel B, treatment with dextran sulfate was used as a negative control (dot profiles). (C) The uptake of FITC–ODN C (full black profiles) was analyzed on KIR3DL2-transfected HEK-293T cells (right) and untransfected control (left) after 1 hour of treatment with 1.2 μg/mL FITC–ODN C. Treatment with dextran sulfate was used as a negative control (empty profiles). Mean fluorescence intensities (MFIs) are reported. (D) HEK-293T cells transfected with plasmid coding for KIR3DL2 full-length receptor (left) or for its cytoplasmic truncated form (KIR3DL2Δ, right panel) were treated or not for 18 hours with the indicated doses of ODN C, and then KIR3DL2 expression was assessed by cytofluorimetric analysis. Results shown in each panel are representative of 3 distinct experiments.
Preferential uptake of ODN C by KIR3DL2+ cells. The uptake of FITC–ODN C by KIR3DL2+ or KIR3DL2− NK cell populations sorted from IL2-activated NK cell populations (A) or KIR3DL2+ or KIR3DL2− NK cell clones (B; derived from the same donor) was analyzed by flow cytometry after 2 hours of treatment with 5 μg/mL FITC–ODN C. In panel A, the percentage of ODN C–FITC+ cells is indicated in the right quadrants. In panel B, treatment with dextran sulfate was used as a negative control (dot profiles). (C) The uptake of FITC–ODN C (full black profiles) was analyzed on KIR3DL2-transfected HEK-293T cells (right) and untransfected control (left) after 1 hour of treatment with 1.2 μg/mL FITC–ODN C. Treatment with dextran sulfate was used as a negative control (empty profiles). Mean fluorescence intensities (MFIs) are reported. (D) HEK-293T cells transfected with plasmid coding for KIR3DL2 full-length receptor (left) or for its cytoplasmic truncated form (KIR3DL2Δ, right panel) were treated or not for 18 hours with the indicated doses of ODN C, and then KIR3DL2 expression was assessed by cytofluorimetric analysis. Results shown in each panel are representative of 3 distinct experiments.
The role of KIR3DL2 receptor in ODN C uptake was also evaluated using cell transfectants. Thus, as shown in Figure 2C, the internalization of ODN C was strongly increased in KIR3DL2-transfected HEK-293T cells compared with untransfected controls. Notably, also in this system, a sharp down-modulation of KIR3DL2 could be detected (Figure 2D left).
To analyze whether the cytoplasmic tail of KIR3DL2 could mediate the receptor internalization upon ODN stimulation, we transfected HEK-293T cells with a plasmid coding for a truncated KIR3DL2 molecule lacking the cytoplasmic region (KIR3DL2Δ). As shown in Figure 2D, the ODN C–induced internalization of KIR3DL2Δ was similar to that observed in cell transfectants expressing the KIR3DL2 full-length receptor. These data suggest that the KIR3DL2 cytoplasmic tail may not be strictly required for receptor internalization.
KIR3DL2+ NK cells release cytokines in response to ODN C stimulation
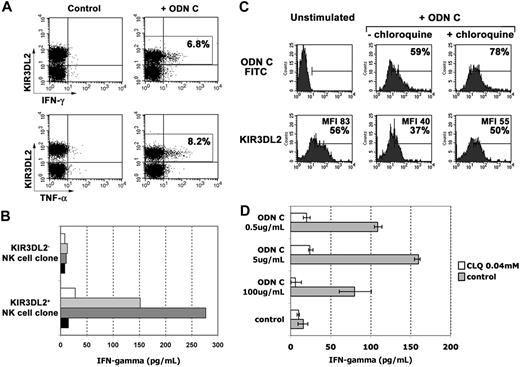
In the presence of IL12, human NK cells were shown to release abundant cytokines (such as IFN-γ and tumor necrosis factor-α [TNF-α]), in response to stimuli acting on TLRs.25 Thus, NK cell populations were analyzed for cytokine production in response to ODN C and IL12. IFN-γ and TNF-α production was assessed by double fluorescence and FACS analysis in combination with anti-KIR3DL2 mAb. Because KIR3DL2 was partially modulated 2 hours after stimulation with ODN C, to allow unequivocal identification of KIR3DL2+ NK cells, FACS analysis was performed after 1 hour. As shown in Figure 3A, NK cells releasing IFN-γ and TNF-α were confined mostly to KIR3DL2+ cells. This result was further confirmed on NK cell clones. Thus, as shown in Figure 3B, only KIR3DL2+ NK cell clones displayed dose-dependent responses (IFN-γ) to ODN C stimulation. Notably, both KIR3DL2+ and KIR3DL2− NK cell clones analyzed expressed high levels of TLR9 transcript and released similar levels of IFN-γ in response to PMA plus ionomycin (not shown).
KIR3DL2+ NK cells release cytokines in response to ODN C stimulation. (A) IL2-activated NK cell populations were cultured for 1 hour in medium supplemented with 1 ng/mL IL12 either in the absence or in the presence of ODN C (5 μg/mL) and then assessed by double fluorescence analysis for IFN-γ or TNF-α production and KIR3DL2 expression. In the upper-right quadrants is indicated the percentage of KIR3DL2+ cells that produce cytokines. Results are representative of 5 independent experiments. (B) IFN-γ production by KIR3DL2+ or KIR3DL2− NK cell clones was assessed by specific enzyme-linked immunosorbent assay after treatment for 2 hours with 1 ng/mL IL12 used alone ( ) or together with 0.5 μg/mL (▭), 5 μg/mL (
) or together with 0.5 μg/mL (▭), 5 μg/mL ( ), or 100 μg/mL (
), or 100 μg/mL ( ) ODN C. Results are representative of 3 independent experiments. (C) ODN C uptake by IL2-activated NK cell populations was analyzed in the absence or presence of chloroquine pretreatment. The upper histograms represent the FITC–ODN C fluorescence intensity, whereas the lower histograms represent the expression of KIR3DL2 under the same experimental conditions. The percentage of positive cells and the MFI of the positive peaks are indicated in each histogram. Results are representative of 4 distinct experiments. (D) ODN C–mediated IFN-γ production by IL2-activated NK cell populations was analyzed in the absence or presence of pretreatment with chloroquine 0.04 mM (CLQ). IFN-γ content was assessed by specific enzyme-linked immunosorbent assay. Data are represented as medians and interquartile ranges (IQRs) of 4 independent experiments.
) ODN C. Results are representative of 3 independent experiments. (C) ODN C uptake by IL2-activated NK cell populations was analyzed in the absence or presence of chloroquine pretreatment. The upper histograms represent the FITC–ODN C fluorescence intensity, whereas the lower histograms represent the expression of KIR3DL2 under the same experimental conditions. The percentage of positive cells and the MFI of the positive peaks are indicated in each histogram. Results are representative of 4 distinct experiments. (D) ODN C–mediated IFN-γ production by IL2-activated NK cell populations was analyzed in the absence or presence of pretreatment with chloroquine 0.04 mM (CLQ). IFN-γ content was assessed by specific enzyme-linked immunosorbent assay. Data are represented as medians and interquartile ranges (IQRs) of 4 independent experiments.
KIR3DL2+ NK cells release cytokines in response to ODN C stimulation. (A) IL2-activated NK cell populations were cultured for 1 hour in medium supplemented with 1 ng/mL IL12 either in the absence or in the presence of ODN C (5 μg/mL) and then assessed by double fluorescence analysis for IFN-γ or TNF-α production and KIR3DL2 expression. In the upper-right quadrants is indicated the percentage of KIR3DL2+ cells that produce cytokines. Results are representative of 5 independent experiments. (B) IFN-γ production by KIR3DL2+ or KIR3DL2− NK cell clones was assessed by specific enzyme-linked immunosorbent assay after treatment for 2 hours with 1 ng/mL IL12 used alone ( ) or together with 0.5 μg/mL (▭), 5 μg/mL (
) or together with 0.5 μg/mL (▭), 5 μg/mL ( ), or 100 μg/mL (
), or 100 μg/mL ( ) ODN C. Results are representative of 3 independent experiments. (C) ODN C uptake by IL2-activated NK cell populations was analyzed in the absence or presence of chloroquine pretreatment. The upper histograms represent the FITC–ODN C fluorescence intensity, whereas the lower histograms represent the expression of KIR3DL2 under the same experimental conditions. The percentage of positive cells and the MFI of the positive peaks are indicated in each histogram. Results are representative of 4 distinct experiments. (D) ODN C–mediated IFN-γ production by IL2-activated NK cell populations was analyzed in the absence or presence of pretreatment with chloroquine 0.04 mM (CLQ). IFN-γ content was assessed by specific enzyme-linked immunosorbent assay. Data are represented as medians and interquartile ranges (IQRs) of 4 independent experiments.
) ODN C. Results are representative of 3 independent experiments. (C) ODN C uptake by IL2-activated NK cell populations was analyzed in the absence or presence of chloroquine pretreatment. The upper histograms represent the FITC–ODN C fluorescence intensity, whereas the lower histograms represent the expression of KIR3DL2 under the same experimental conditions. The percentage of positive cells and the MFI of the positive peaks are indicated in each histogram. Results are representative of 4 distinct experiments. (D) ODN C–mediated IFN-γ production by IL2-activated NK cell populations was analyzed in the absence or presence of pretreatment with chloroquine 0.04 mM (CLQ). IFN-γ content was assessed by specific enzyme-linked immunosorbent assay. Data are represented as medians and interquartile ranges (IQRs) of 4 independent experiments.
Next, experiments of ODN C uptake were performed in NK cells pretreated with chloroquine. Because chloroquine blocks the acidification process occurring during endosome maturation, it can prevent ODN-C–mediated triggering of TLR9 localized in endosomes.31 Chloroquine treatment did not inhibit ODN C uptake and only marginally affected KIR3DL2 modulation (Figure 3C), although it strongly inhibited cytokine secretion (dependent on TLR9 signaling; Figure 3D).
Confocal microscopy analysis of KIR3DL2-ODN interactions in NK cells
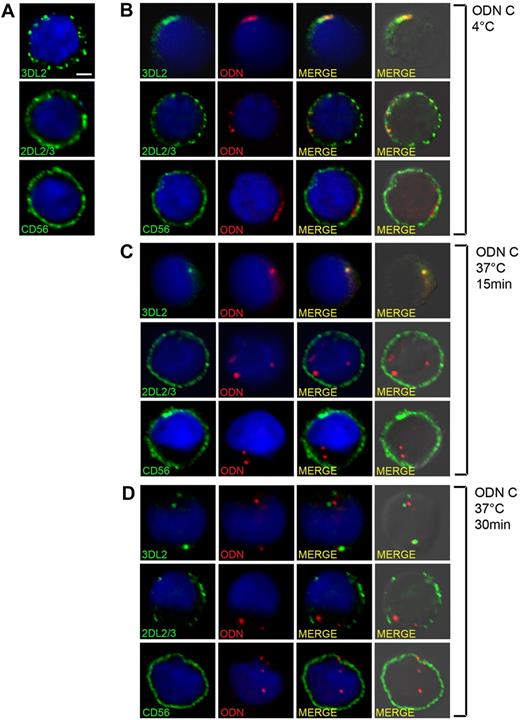
To directly visualize the interaction between KIR3DL2 and ODN, NK cells were incubated either at 4° or 37°C with TxRed-ODN and stained simultaneously with anti-KIR3DL2 mAbs. mAbs specific for KIR2DL2/S2/L3 or CD56 were used as control (Figure 4). In the absence of ODN, the analyzed surface molecules were evenly distributed on the plasma membrane (Figure 4A). In NK cells incubated with ODN at 4°C, marked surface clustering of KIR3DL2 occurred, accompanied by a clear colocalization with TxRed-ODN (Figure 4B). In contrast, KIR2DL2/S2/L3 and CD56 remained evenly distributed at the cell surface (Figure 4B). In NK cells incubated with ODN at 37°C for 15 minutes (Figure 4C), KIR3DL2 and ODN were clearly cointernalized and associated inside the cells (Mander coefficient [M] = 0.9 ± 0.04). After 30 minutes, association of KIR3DL2 with ODN was significantly reduced, as revealed by quantitative colocalization analysis (M = 0.56 ± 0.07; P < .001), suggesting that, after internalization, the 2 molecules segregate inside the cell (Figure 4D). Under the same conditions, no internalization of KIR2DL2/S2/L3 or CD56 was observed (Figure 4C-D).
Visualization of KIR3DL2-ODN interaction in NK cells. Confocal microscopy analysis of NK cells untreated (A) or incubated with 5 μg of TxRed-ODN (red) for 30 minutes at 4°C (B), or for 15 or 30 minutes at 37°C (C-D). Cells were then stained for KIR3DL2, KIR2DL2/S2/L3, or CD56 (green), as indicated. Images are single confocal sections taken with a 60× objective. Scale bar represents 2 μm. Quantitative colocalization analysis was performed to compare panels C and D, and it provided the following Mander coefficients of colocalization: 0.9 ± 0.04 (C), 0.56 ± 0.07 (D); P < .001.
Visualization of KIR3DL2-ODN interaction in NK cells. Confocal microscopy analysis of NK cells untreated (A) or incubated with 5 μg of TxRed-ODN (red) for 30 minutes at 4°C (B), or for 15 or 30 minutes at 37°C (C-D). Cells were then stained for KIR3DL2, KIR2DL2/S2/L3, or CD56 (green), as indicated. Images are single confocal sections taken with a 60× objective. Scale bar represents 2 μm. Quantitative colocalization analysis was performed to compare panels C and D, and it provided the following Mander coefficients of colocalization: 0.9 ± 0.04 (C), 0.56 ± 0.07 (D); P < .001.
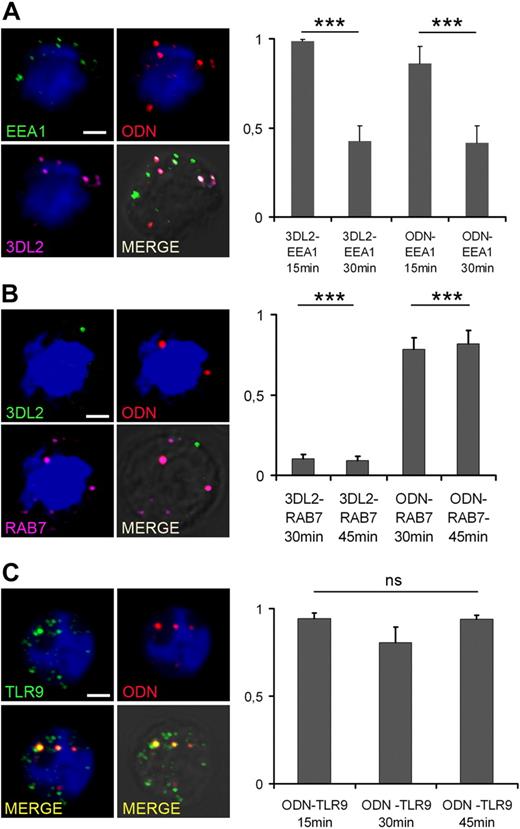
To verify whether early endosomes were the site of KIR3DL2/ODN colocalization, 3-color confocal analysis was performed using early endosome antigen-1 (EEA-1), a marker of early endosomes. As shown in Figure 5A, in NK cells incubated at 37°C for 15 minutes with TxRed-labeled ODN, there was a substantial overlap between KIR3DL2-ODN and EEA-1 staining. Not surprisingly, colocalization of KIR3DL2-ODN and EEA-1 was significantly reduced in NK cells incubated with ODN for 30 minutes (Figure 5A), suggesting the transfer of ODN to the late endocytic compartment. This hypothesis was confirmed by quantitative confocal analysis, using Rab7 as a marker for late endosomes (Figure 5B). Interestingly, in agreement with data shown in Figure 4, we found that ODN, but not KIR3DL2, colocalized with Rab7 in NK cells incubated with ODN for 30 or 45 minutes.
ODN traffics through early and late endosomes and associates with TLR9. Confocal microscopy analysis of NK cells incubated with 5 μg of TxRed-ODN (red) for 15, 30, or 45 minutes at 37°C. Cells were then stained for (A) EEA-1 (green) and KIR3DL2 (purple), (B) KIR3DL2 (green) and Rab7 (purple), or (C) TLR9 (green), as indicated. Images are single confocal sections, taken with a 60× objective, showing NK cells incubated with ODN for 15 minutes (A,C) or 30 minutes (B). Scale bar represents 2 μm. Quantitative colocalization analysis expressed as Mander coefficients of colocalization is shown. ***P < .001 was considered significant.
ODN traffics through early and late endosomes and associates with TLR9. Confocal microscopy analysis of NK cells incubated with 5 μg of TxRed-ODN (red) for 15, 30, or 45 minutes at 37°C. Cells were then stained for (A) EEA-1 (green) and KIR3DL2 (purple), (B) KIR3DL2 (green) and Rab7 (purple), or (C) TLR9 (green), as indicated. Images are single confocal sections, taken with a 60× objective, showing NK cells incubated with ODN for 15 minutes (A,C) or 30 minutes (B). Scale bar represents 2 μm. Quantitative colocalization analysis expressed as Mander coefficients of colocalization is shown. ***P < .001 was considered significant.
A previous, elegant study has demonstrated that a few minutes after CpG DNA stimulation TLR9 translocates to early endosomes.28 On the basis of our data showing the presence of ODN in early endosomes after 15 minutes of NK cell stimulation (Figure 5A), we hypothesized that from that time on TLR9 would colocalize with ODN. We addressed this hypothesis by performing quantitative colocalization analysis of ODN and TLR9 in NK cells incubated with ODN for 15, 30, or 45 minutes, and found that ODN and TLR9 were stably associated at all time points (Figure 5C).
Altogether, these data indicate that ODN and KIR3DL2 are cointernalized in the EEA-1+ compartment, where TLR9 translocates upon ODN stimulation.28 In EEA-1+ endosomes, KIR3DL2 and ODN dissociate, and ODN and TLR9 travel together toward Rab-7+ late endosomes.
Direct interaction between KIR3DL2 and ODNs
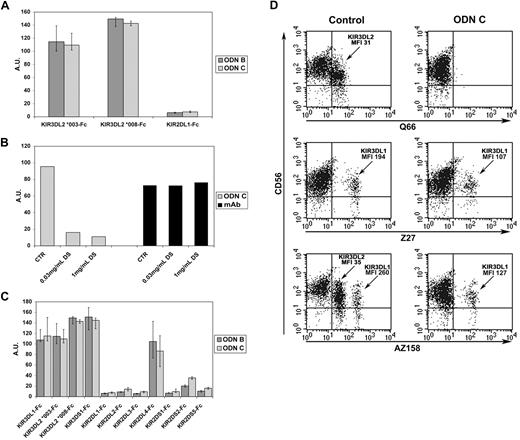
To further investigate whether KIR3DL2 can directly interact with ODNs, binding experiments were performed using KIR3DL2-Fc soluble receptors. In addition, because of the KIR3DL2 polymorphism (22 alleles have been identified), KIR3DL2*003 and KIR3DL2*008 (ie, the 2 alleles expressed by the donor shown in Figure 2) were used. Binding of biotinylated ODN B or ODN C to plate coated with KIR3DL2-Fc molecules was evaluated. KIR2DL1-Fc was used as control (KIR2DL1 was not modulated by ODNs, Figure 1A). As shown in Figure 6A, both KIR3DL2*003-Fc and KIR3DL2*008-Fc directly bound ODN B and ODN C, whereas no ODN/KIR2DL1-Fc interaction was detected. Addition of dextran sulfate virtually abrogated ODN/KIR3DL2-Fc interaction, whereas binding of specific anti-KIR3DL2 mAb to the same coated plates was not affected (Figure 6B). These data are in line with the finding that dextran sulfate inhibited FITC–ODN C uptake by KIR3DL2+ NK cells (Figure 2B). Taken together, these data indicate that KIR3DL2 can directly bind ODNs and strongly suggest that such interaction may occur at the NK cell surface as well.
ODNs directly bind KIR3DL2-Fc and down-regulate the expression of KIR3DL2 in freshly isolated NK cells. (A) Plates coated with KIR3DL2*003-, KIR3DL2*008-, or KIR2DL1-Fc soluble receptors were incubated with biotinylated sequence of ODN B or ODN C followed by streptavidin horseradish peroxidase (HRP)–conjugated second reagent or with anti–human IgG HRP-conjugated mAb. Arbitrary units (AU) on y-axis represent the ratio between optical density (OD) values obtained by analyzing ODN/KIR-Fc interaction and OD values obtained by testing binding of anti–human IgG HRP to the corresponding KIR-Fc molecules. Results are presented as medians of 3 to 8 independent experiments, each performed in duplicate. IQRs are reported. (B) Binding of ODN C to plates coated with KIR3DL2*003-Fc was tested in the absence or presence of dextran sulfate (DS) at the indicated concentrations. As control, we measured binding of an anti-KIR3DL2 mAb to KIR3DL2*003-Fc under the same experimental condition. AUs on y-axis represent the ratio between OD values obtained analyzing ODN/KIR3DL2-Fc interaction or mAb/KIR3DL2-Fc interaction and OD values obtained by testing the binding of anti–human IgG HRP to KIR3DL2-Fc–coated plates. A representative experiment of 3 independent ones is reported. (C) Binding of biotinylated sequences of ODN B or ODN C to plates coated with the indicated KIR-Fc molecules was determined and represented as described in panel A. Bars represent medians of 3 to 8 independent experiments performed in duplicate; IQRs are reported. (D) Freshly isolated NK cells were cultured for 20 hours either in the absence or in the presence of ODN C, and then assessed by double cytofluorimetric analysis for CD56 and KIR3DL2 (Q66 mAb) or KIR3DL1 (Z27 mAb) or KIR3DL2 and KIR3DL1 (AZ158 mAb) expression. AZ158dull and AZ158bright NK cells correspond to KIR3DL2+ and KIR3DL1+ NK cells, respectively.32 Mean fluorescence intensities (MFIs) are reported. Results are representative of 3 independent experiments.
ODNs directly bind KIR3DL2-Fc and down-regulate the expression of KIR3DL2 in freshly isolated NK cells. (A) Plates coated with KIR3DL2*003-, KIR3DL2*008-, or KIR2DL1-Fc soluble receptors were incubated with biotinylated sequence of ODN B or ODN C followed by streptavidin horseradish peroxidase (HRP)–conjugated second reagent or with anti–human IgG HRP-conjugated mAb. Arbitrary units (AU) on y-axis represent the ratio between optical density (OD) values obtained by analyzing ODN/KIR-Fc interaction and OD values obtained by testing binding of anti–human IgG HRP to the corresponding KIR-Fc molecules. Results are presented as medians of 3 to 8 independent experiments, each performed in duplicate. IQRs are reported. (B) Binding of ODN C to plates coated with KIR3DL2*003-Fc was tested in the absence or presence of dextran sulfate (DS) at the indicated concentrations. As control, we measured binding of an anti-KIR3DL2 mAb to KIR3DL2*003-Fc under the same experimental condition. AUs on y-axis represent the ratio between OD values obtained analyzing ODN/KIR3DL2-Fc interaction or mAb/KIR3DL2-Fc interaction and OD values obtained by testing the binding of anti–human IgG HRP to KIR3DL2-Fc–coated plates. A representative experiment of 3 independent ones is reported. (C) Binding of biotinylated sequences of ODN B or ODN C to plates coated with the indicated KIR-Fc molecules was determined and represented as described in panel A. Bars represent medians of 3 to 8 independent experiments performed in duplicate; IQRs are reported. (D) Freshly isolated NK cells were cultured for 20 hours either in the absence or in the presence of ODN C, and then assessed by double cytofluorimetric analysis for CD56 and KIR3DL2 (Q66 mAb) or KIR3DL1 (Z27 mAb) or KIR3DL2 and KIR3DL1 (AZ158 mAb) expression. AZ158dull and AZ158bright NK cells correspond to KIR3DL2+ and KIR3DL1+ NK cells, respectively.32 Mean fluorescence intensities (MFIs) are reported. Results are representative of 3 independent experiments.
Because confocal microscopy analysis revealed that ODN dissociates from KIR3DL2 in late endosomal compartments, we investigated whether the ODN/KIR3DL2 interaction is sensitive to acid pH. As shown in supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article), we could detect a sharp decrease of KIR3DL2 binding capability at pH 5.0. This acid pH, however, had no effect on KIR3DL2 expression at the NK cell surface as determined by the use of specific mAb (not shown).
ODN C binding by KIR3DS1, KIR3DL1, and KIR2DL4
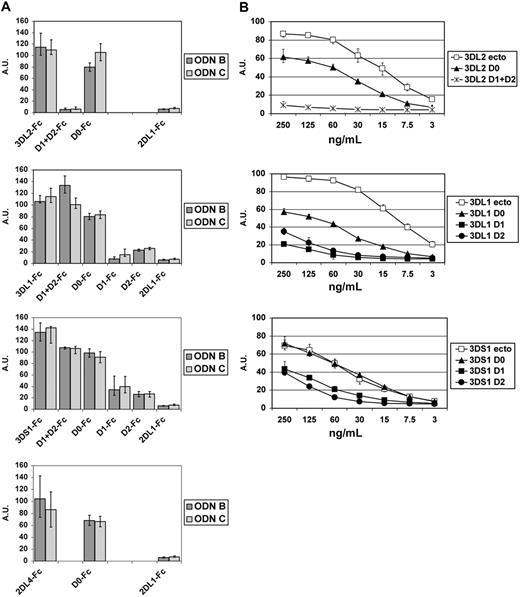
We next investigated whether other KIRs could bind ODNs. KIR-Fc chimeric molecules of KIR3DL1, KIR3DS1, KIR2DL2, KIR2DL3, KIR2DL4, KIR2DS1, KIR2DS2, and KIR2DS5 (supplemental Table 1) were analyzed. Also KIR3DL1-, KIR3DS1-, and KIR2DL4-Fc were found to bind biotinylated ODN B and ODN C (Figure 6C). Similar results were obtained using biotinylated ODN B and ODN C with a full phosphodiester backbone. As shown in supplemental Figure 2A, KIR3DL2 and KIR3DL1 bound phosphodiester (PO) and phosphotioate (PS) ODNs in a dose-dependent manner, whereas KIR2DL1 and KIR2DL3 did not. On the contrary, the interaction between KIR2DL4 and CpG ODNs could be detected with PS ODNs only. These data also suggest that the affinity of KIR3DL2 and KIR3DL1 is higher for PS ODNs than for PO ODNs.
We next analyzed whether ODN recognition could be sequence specific. To this end, we compared the capability of KIR3DL2 and KIR3DL1 to interact with ODN B, double-stranded (ds) ODN B, and poly A. As shown in supplemental Figure 2B, all of these short DNA molecules displayed a dose-dependent binding to KIRs. The same stimuli were then analyzed for their ability to induce KIR3DL2 surface modulation. As shown in supplemental Figure 3, a substantial effect could be observed with all stimuli when used at high concentration (100 μg/mL). Remarkably, only ODN C and ds ODN B could induce KIR3DL2 down-modulation when used at low concentration (0.5-5 μg/mL).
To investigate whether KIR3DL1, KIR3DS1, and KIR2DL4 also expressed at the cell surface interact with ODNs, we analyzed HEK-293T cells transfected with plasmids coding for the corresponding KIRs (or for KIR2DL1 or KIR3DL2 as negative and positive controls, respectively). Each cell transfectant was analyzed for KIR expression before and after ODN C treatment. As in the case of KIR3DL2+ NK cells, ODN C induced sharp modulation of KIR3DL2 also in KIR3DL2+ HEK-293T transfectants. Under these experimental conditions, KIR3DL1, KIR3DS1, and KIR2DL4 (but not KIR2DL1) were partially modulated upon cell exposure to ODN C (supplemental Figure 4A). These data suggest that surface KIR3DL1, KIR3DS1, and KIR2DL4, although to a lesser extent, also may be involved in the process of ODN uptake. We further investigated whether modulation of these KIRs also occurred in NK cells. Because only some persons express KIR3DL1, KIR3DS1, and the membrane-bound form of KIR2DL4,33 we analyzed selected donors expressing all the ODN-binding KIRs. As shown in supplemental Figure 4B, treatment of IL2-cultured NK cells with ODN C induced only a partial decrease of surface KIR3DL1/S1 and KIR2DL4 (much less than KIR3DL2). On the other hand, the surface expression of other members of the KIR family (ie, KIR2DL1/S1) was not modified (supplemental Figure 4B). Despite the binding of ODN to KIR3DL1-Fc, the partial ODN C–induced modulation of surface KIR3DL1 was detected only in some donors (Figure 1A and supplemental Figure 4B). KIR2DL4 is usually expressed at very low or undetectable levels by resting NK cells, whereas its expression is transiently up-regulated in IL2-cultured NK cells.34 Because NK cell populations analyzed in these experiments expressed low levels of KIR2DL4, only slight ODN C–induced modulation could be detected (supplemental Figure 4B). Similar to IL2-activated NK cells, ODN C treatment of freshly isolated peripheral blood NK cells induced strong KIR3DL2 but only partial KIR3DL1 modulation (Figure 6D).
Role of the D0 domain in KIR/ODN interaction
The analysis of the domain compositions of different KIRs suggested that the capability of a given KIR to bind ODN might correlate with the presence of the D0 domain. To assess whether the D0 domain is involved in CpG-ODN recognition, Fc chimeric soluble proteins composed of 1 or 2 domains were generated. In particular, we assessed the binding capability of single D0-Fc domains of KIR3DL1, KIR3DL2, KIR3DS1, and KIR2DL4; D1 + D2 of KIR3DL1, KIR3DL2, and KIR3DS1; single D1 of KIR3DL1 and KIR3DS1; and single D2 of KIR3DL1 and KIR3DS1 (supplemental Table 2). The results, summarized in Figure 7, strongly suggest a direct involvement of D0 domains in ODN binding. Moreover, KIR3DL1 and KIR3DS1 (alleles of the same locus) bound ODNs also through the D1 + D2 domains. Thus, the ability of single D1 or D2 of KIR3DL1 and KIR3DS1 to bind ODNs was also examined. As shown in Figure 7, the single D1 or D2 domains of both KIR3DL1 and KIR3DS1 receptors displayed only weak binding to ODNs (more evident at higher ODN concentration). This suggests that an appropriate spatial conformation, determined by D1 + D2 domains of KIR3DL1/S1, is required for binding ODN. On the contrary, D1 + D2-Fc of KIR3DL2 did not bind ODNs, indicating that, in KIR3DL2, the D0 domain is the only one responsible for the interaction with ODNs. Next, we comparatively analyzed the net charges of the ODN-binding KIRs with those of the nonbinding ones. This analysis, reported in supplemental Table 3, revealed that the extracellular portions of KIR3DL1, KIR3DL2, KIR3DS1, and KIR2DL4 display a positive charge, whereas those of KIR2DL1 and KIR2DS2 do not. Moreover, we found that the KIR domains showing the higher positive net charge were the ones that displayed the higher ODN-binding capability.
The D0 domains of KIR3DL2, KIR3DL1, KIR3DS1, and KIR2DL4 are involved in ODN recognition. (A) Each graph represents the ODN-binding capability of the full-length ectodomain of the reported KIRs and of their indicated extracellular domains. The starting and ending amino acids composing the indicated domains are listed in supplemental Table 2. Experimental conditions and data representations are described in Figure 6A. Results are reported as medians of 3 to 8 independent experiments, each performed in duplicate. IQRs are indicated. (B) KIR molecules and their domains were analyzed as titrations for the capability to bind ODN C. Experiments were performed and elaborated as described in Figure 6A. Data are represented as medians and IQRs of 4 independent experiments.
The D0 domains of KIR3DL2, KIR3DL1, KIR3DS1, and KIR2DL4 are involved in ODN recognition. (A) Each graph represents the ODN-binding capability of the full-length ectodomain of the reported KIRs and of their indicated extracellular domains. The starting and ending amino acids composing the indicated domains are listed in supplemental Table 2. Experimental conditions and data representations are described in Figure 6A. Results are reported as medians of 3 to 8 independent experiments, each performed in duplicate. IQRs are indicated. (B) KIR molecules and their domains were analyzed as titrations for the capability to bind ODN C. Experiments were performed and elaborated as described in Figure 6A. Data are represented as medians and IQRs of 4 independent experiments.
Discussion
KIRs were originally discovered and characterized as surface NK receptors specialized in HLA class I recognition.8-10 The present study provides evidence for a novel, relevant functional capability of these molecules, suggesting that they may function as sensors for microbial products. Primarily KIR3DL2, an inhibitory NK receptor specific for certain HLA-A alleles,14,15 can bind CpG ODNs at the cell surface and shuttle them to endosomes where TLR9s are localized. Interaction of KIR3DL2 with ODN resulted in sharp down-modulation of its surface expression and in induction of cytokine release. Remarkably, binding of ODN to KIR3DL2 did not result in the delivery of inhibitory signals, as it occurs when KIR3DL2 is engaged by its HLA-A–specific ligands. This may reflect simply the lack of appropriate KIR cross-linking. In this context, also KIR-specific mAbs do not deliver inhibitory signals in the absence of efficient cross-linking. It is also possible that the transient disappearance of KIR3DL2 from the cell surface (upon ODN uptake) may allow NK cells to kill target cells expressing “protective” HLA-A molecules.
It has been shown that KIR3DL2 represents a valuable marker for Sézary cutaneous T-cell lymphoma. Both cutaneous and circulating cells of this lymphoma express KIR3DL2.20,21 In agreement with a previous report,35 we show that Sézary cells express TLR9 transcripts. After exposure to ODN, a sharp down-regulation of KIR3DL2 occurred in T-cell lines established from different Sézary patients. However, in this case, we could not evaluate functional responses to ODN because Sézary cells do not secrete detectable levels of cytokines (despite TLR9 transcript expression). Notably, KIR3DL2, differently from other KIRs, has not been detected in T cells from most healthy persons. A role in the pathogenesis of Sézary T-cell lymphoma has been proposed for skin-associated microbes such as Staphylococcus aureus36 and Chlamydia.37 Other studies reported an association between this lymphoma and cytomegalovirus38 or Epstein-Barr virus infections.39 In this context, microbial ODN-mediated cell triggering may induce uncontrolled proliferation of KIR3DL2+ Sézary T cells, possibly representing an early event in the tumorigenic process leading to the malignant lymphoma.
One of the problems in TLR biology is represented by the delivery of ligands to endosomal/endoplasmic reticulum–resident TLRs. ODN binding, internalization, and traffic were analyzed by confocal microscopy. Notably, in these experiments, we used normal NK cells (and not cell lines or cell transfectants) to ensure a more physiologic experimental setting. KIR3DL2 molecules were found to be associated with ODN not only at the cell surface but also intracellularly. Our data indicate that KIR3DL2 and ODN are cointernalized through early (EEA-1+) endosomes, but then they separate and ODN proceeds in trafficking only through the late (Rab7+) compartment. Interestingly, we have found that, in contrast to KIR3DL2, TLR9 is stably associated with ODN during its residency in both early and late endosomes. Thus, although it cannot be excluded that a vanishing small amount of TLR9 may be expressed on the NK cell surface, our data suggest that, at least in NK cells, surface molecules such as KIR3DL2 may function as chaperons for TLR9 ligands. At the present, however, we cannot rule out the possibility that an additional yet-undefined molecule may be necessary for the process of KIR3DL2-mediated ODN internalization, as suggested by the experiments with KIR3DL2Δ. Importantly, the use of KIR3DL2-Fc soluble molecules allowed us to directly assess their binding to ODN. These experiments suggested that binding of ODN does not require additional molecules associated with KIR3DL2 at the NK (or T) cell surface. KIR-Fc specific for HLA-C alleles (KIR2DL1/S1 and KIR2DL2/L3/S2) did not bind ODN, whereas substantial binding was detected with other KIR-Fc soluble proteins including KIR3DL1, KIR3DS1, and KIR2DL4. Remarkably, all the ODN-binding KIRs are characterized by a D0 domain. Analysis of the ODN-binding capability of individual Ig domains revealed a predominant involvement of D0. Previous studies showed that KIR3DL1 recognizes HLA class I through D1 and D2 domains, whereas the D0 domain enhances the strength of KIR/HLA interaction.40 Our studies provide evidence for a novel major function of D0 (ie, to mediate direct pathogen recognition). Unfortunately, because none of the available anti-KIR3DL2 mAbs react with this domain, the binding of ODN could not be prevented by these antibodies. Although soluble KIR3DL1, KIR3DS1, and KIR2DL4 could bind ODNs, these did not induce, in normal NK cells, modulation of the corresponding membrane receptors comparable with that of KIR3DL2. Differences in KIR susceptibility to ODN-induced modulation could not be explained merely by differences in surface density. For example, modulation of KIR3DS1 was marginal, although its level of expression at the NK cell surface was similar to or weaker than that of KIR3DL2. Remarkably, IFN-γ production in response to ODN was confined mostly to KIR3DL2+ NK cell subsets, suggesting that other ODN-binding KIRs do not mediate efficient ODN-shuttling into NK cells.
The putative ancestral mammalian KIR was a 3-domain surface molecule carrying a D0 domain. All KIR2D-encoding genes are thought to have evolved from a KIR3D-encoding gene. Indeed, the majority of KIR2D genes contain a pseudoexon encoding a D0 domain.12,41 Among KIR2D, only KIR2DL4 and KIR2DL542 (this latter was not analyzed in the present study) carry D0 domain. They represent the most conserved KIRs in primates, being the only ones common to hominoids (KIR2DL4 and KIR2DL5) and monkeys (KIR2DL4).43 This might suggest that, in origin, these receptors had a function different from their ability to recognize major histocompatibility complex class I allele. In this context, it is worth noting that KIR3DL2 (together with KIR2DL4 and KIR3DL3) represents a framework gene (ie, it is present in all KIR haplotypes).41 As a consequence, NK cells of all persons can bind CpG ODN. This novel functional capability of KIR3DL2 may provide an important clue to understanding the driving forces that led to conservation of the KIR3DL2-encoding gene in all haplotypes, despite the low frequency, in the human population, of HLA-A3 or -A11 alleles (ie, the ligands of KIR3DL2). Indeed, the need of rapid NK-mediated responses to microbial products may represent an important factor of selective pressure. In this context, KIR3DL2 is characterized by a low inhibitory capability because of its low affinity for its HLA-A ligand. Moreover, such interaction is highly dependent on the peptide bound to the HLA pocket.44 This impaired inhibitory function may explain why KIR3DL2, in most NK cells, is coexpressed with other KIRs or NKG2A, which ensure a more efficient inhibitory effect upon interaction with their HLA class I–specific ligands. Thus, it is possible to speculate that the prevalent role of KIR3DL2 in humans may be its ability to promptly sense microbial CpG ODNs.
KIR3DL2 is expressed by NK cells and by Sézary T cells. However, because KIR3DL2-negative cell types (other than NK cells) are known respond to ODN, it is likely that other surface receptors may ensure ODN uptake and delivery to TLR-containing endosomes.45,46 For example, a 45-kDa protein expressed in kidney brush border membranes has been reported to act as a voltage-gated channel for the entry of ODN into these cells. However, no evidence exists that this protein is expressed in leukocytes as well.47 In macrophages, type I and II class A scavenger receptors (scavenger receptor AI/II, SR-A) and macrophage receptor with collagenous structure (MARCO) have been reported to bind CpG ODNs. However, SR-A– or MARCO-deficient macrophages could respond to ODN, suggesting a redundant function of these receptors in ODN uptake at least in macrophages.48-50 Although not shown, transcripts coding for SR-A and/or MARCO were not detected in IL2-activated NK cells.
Although KIR2DL4 was only partially down-regulated in NK cells exposed to CpG ODN, we speculate that it could also play a role in the uptake of microbial products. Differently from all the other KIRs, KIR2DL4 is present in all NK cells. Thus, in view of its broad expression profile and its evolutionary conservation, KIR2DL4 is likely to play a critical role in NK cell biology. It has been reported that HLA-G and some HLA class I alleles are ligands for membrane-bound KIR2DL4. In this regard, Rajagopalan et al showed that KIR2DL4 surface molecules, upon interaction with soluble HLA-G, undergo endocytosis and colocalize with Rab5, a marker of early endosomes where also TLR9s are located.16
In conclusion, our present study provides evidence of a novel, KIR-associated function. Notably, thanks to their ability to recognize HLA class I molecules, KIRs were known to play a fundamental role in viral infections, allowing discrimination between virus-infected and normal cells. This study underlines the important role of KIRs as sensors of CpG ODN. This would result in rapid NK responses, primarily characterized by cytokine production, also in the course of bacterial infections.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants awarded by Associazione Italiana Ricerca sul Cancro; Istituto Superiore di Sanità, no. 40G.41; Fondazione CARIPLO, no. 20060034085; Ministero dell'Istruzione, Università e Ricerca (MIUR-PRIN project 2006061378_003; MIUR-FIRB 2003 project RBLA039LSF-001); Ministero della Salute: Ricerca Finalizzata 2005/n.57; and Ricerca Oncologica-Project of Integrated Program 2006-08, agreements no. RO strategici 3/07.
Authorship
Contribution: S.S. and M.F. designed and performed research, interpreted data, and wrote the paper; S.C., E.R., and C.S. performed research and analyzed data; A.B. provided Sézary cell lines and analyzed data; A.V. supervised research and analyzed data on confocal microscopy; L.M. designed research and revised the paper; and A.M. designed research, interpreted data, and wrote the paper.
Conflict-of-interest disclosure: A.M. is founder and shareholder of Innate-Pharma. The remaining authors declare no competing financial interests.
Correspondence: Alessandro Moretta, Dipartimento di Medicina Sperimentale, Sezione di Istologia, Via G. B. Marsano 10, 16132 Genova, Italy; e-mail: alemoret@unige.it.
References
Author notes
S.S. and M.F. contributed equally to this study.