Abstract
Immunosuppression resulting in impaired Epstein-Barr virus (EBV)–specific T-cell immunity is involved in the pathogenesis of EBV-positive post-transplantation lymphoproliferative disorder (EBV+ PTLD). Restoration of EBV-specific T-cell immunity by adoptive immunotherapy can induce remission. EBV-nuclear antigen-1 (EBNA1) is unique in being expressed in all cases of EBV+ PTLD. Recent data demonstrate that EBNA1 is not immunologically silent and can be exploited as a T-cell target. There are no data on EBNA1-specific T cells in PTLD. EBNA1-specific T cells capable of proliferation, interferon-γ release, and CD107a/b degranulation were assayed in 14 EBV+ PTLD diagnostic blood samples and 19 healthy controls. EBNA1-specific CD4+ T cells predominated and were expanded in 10 of 14 patients and 19 of 19 controls. Although human leukocyte antigen class I alleles influenced the magnitude of the response, EBNA1-specific CD8+ effector T cells were successfully generated in 9 of 14 EBV+ PTLD patients and 16 of 19 controls. The majority of PTLD patients had a polymorphism in an EBNA1 epitope, and T-cell recognition was greatly enhanced when EBNA1 peptides derived from the polymorphic epitope were used. These results indicate that EBNA1-specific T cells should be included in adoptive immunotherapy for PTLD. Furthermore, expansion protocols should use antigenic sequences from relevant EBV strains.
Introduction
Post-transplantation lymphoproliferative disorder (PTLD) encompasses the spectrum of lymphoproliferative diseases that arise as a complication of organ transplantation. The incidence of PTLD varies depending on the type of organ transplanted (up to 30% for lung and small bowel transplantations vs 1%–5% for renal, cardiac, and liver transplantations) and the degree of immunosuppression.1 Management remains suboptimal, relapses are frequent, and treatment-related mortality is high.2 The rise in solid-organ transplantation, including high-risk solid-organ transplantation requiring more intense immunosuppression, coupled with improvements in long-term survival will probably result in a rise in the incidence of PTLD.
The majority of PTLD cases (50%-80%) are positive for Epstein-Barr virus (EBV+ PTLD). EBV is an oncogenic γ-herpes virus that infects more than 90% of adults worldwide. After primary infection, the virus persists for life within B cells. This asymptomatic state is, in part, maintained by T-cell-mediated immune control.3 EBV+ PTLD arises partly as an iatrogenic consequence of immunosuppression resulting in the loss of antiviral immunity.4 However, reduction of immunosuppression fails to induce a response in the majority of cases, suggesting that other mechanisms, including intratumoral immunosuppression, could operate in established PTLD. The expression of EBV-latent antigens in EBV+ PTLD provides an ideal target for in vitro-generated tumor antigen-specific T cells in an environment free from the constraints of ongoing iatrogenic and/or lymphoma-mediated immunosuppression. Adoptive immunotherapy has successfully been used in the prophylaxis and treatment of EBV+ PTLD.4-7 It can result in the reconstitution of EBV-specific T-cell immunity. Furthermore, transferred T cells are capable of homing to sites of disease, including traversing the blood-brain barrier.8
Improvements in transplantation surgery have resulted in increased numbers of transplantation survivors and increased numbers of late-onset PTLD (> 1 year after transplantation). Earlier studies defined PTLD as latency type III, a form of EBV latency characterized by expression of many latent antigens, including EBV nuclear antigens (EBNAs) 1, 2, 3, 4, and 6.9 However, more recent reports have established that the number of different EBV antigens expressed is variable between patients and within each tumor, with late-onset PTLD frequently showing restricted expression (type I/II EBV-latency).10 Importantly, EBNA1 is the only EBV-latent protein universally expressed in PTLD. EBNA1 is essential for maintenance of the EBV episome and is the only viral protein required for replication of EBV in its latent form. Earlier reports indicated that the EBNA1 protein evades T-cell recognition.11,12 However, recent studies indicate that the mechanisms of EBNA1 antigen processing require reevaluation and that it is an important target for CD4+ and CD8+ EBV-specific T-cell responses.13-17 If EBNA1-specific T-cell generation in vitro is viable, then EBV-specific T-cell expansion strategies should be modified to include T cells that target this protein. To our knowledge, there are no data on the feasibility of expanding EBNA1-specific T cells for use as adoptive immunotherapy in this setting.
Methods
Patient samples
Blood samples from 14 EBV+ PTLD patients were collected before therapy. Details of these 14 PTLD patients are outlined in Table 1. Blood samples from 19 EBV-seropositive healthy laboratory volunteers were also collected (mean age, 41 years; female:male 8:11). Peripheral blood mononuclear cells (PBMCs) were isolated from blood samples using a previously described technique.18 Written informed consent was provided by all donors in accordance with Queensland Institute of Medical Research and participating hospitals Human Research Ethics Committee guidelines and the Declaration of Helsinki. Approval for this study was granted by the Queensland Institute of Medical Research Human Research Ethics Committee and the Princess Alexandra Hospital Human Research Ethics Committee.
Patient data
| Patient no. . | Sex . | Type of transplantation . | Age at transplantation, y . | Time of PTLD onset* . | Histology . |
|---|---|---|---|---|---|
| 1 | Male | Renal | 30 | Late | Polymorphic |
| 2 | Male | Lung | 24 | Early | Polymorphic |
| 3 | Female | Heart | 38 | Late | Polymorphic |
| 4 | Male | Heart | 36 | Early | DLBCL |
| 5 | Male | Heart | 50 | Late | Polymorphic |
| 6 | Female | Heart | 34 | Late | DLBCL |
| 7 | Male | Heart | 0 | Early | B-cell unclassifiable |
| 8 | Male | Renal | 51 | Late | DLBCL |
| 9 | Male | Renal | 8 | Early | Plasmacytic hyperplasia |
| 10 | Female | Renal | 34 | Early | B-cell unclassifiable |
| 11 | Female | Lung | 24 | Early | B-cell unclassifiable |
| 12 | Male | Renal | 20 | Early | B-cell unclassifiable |
| 13 | Male | Renal | 40 | Late | DLBCL |
| 14 | Male | Renal | 33 | Early | DLBCL |
| Patient no. . | Sex . | Type of transplantation . | Age at transplantation, y . | Time of PTLD onset* . | Histology . |
|---|---|---|---|---|---|
| 1 | Male | Renal | 30 | Late | Polymorphic |
| 2 | Male | Lung | 24 | Early | Polymorphic |
| 3 | Female | Heart | 38 | Late | Polymorphic |
| 4 | Male | Heart | 36 | Early | DLBCL |
| 5 | Male | Heart | 50 | Late | Polymorphic |
| 6 | Female | Heart | 34 | Late | DLBCL |
| 7 | Male | Heart | 0 | Early | B-cell unclassifiable |
| 8 | Male | Renal | 51 | Late | DLBCL |
| 9 | Male | Renal | 8 | Early | Plasmacytic hyperplasia |
| 10 | Female | Renal | 34 | Early | B-cell unclassifiable |
| 11 | Female | Lung | 24 | Early | B-cell unclassifiable |
| 12 | Male | Renal | 20 | Early | B-cell unclassifiable |
| 13 | Male | Renal | 40 | Late | DLBCL |
| 14 | Male | Renal | 33 | Early | DLBCL |
PTLD indicates post-transplantation lymphoproliferative disorder; and DLBCL, diffuse large B-cell lymphoma.
Time of onset defined as late if greater than 1 year after transplantation.
Expansion of EBV-specific T cells
EBNA1 and BZLF1-specific T cells were expanded using EBNA1 and BZLF1 overlapping peptide pools. Pools consisted of 17-mer peptides overlapping by 10 amino acids (synthesized by Synbiosci). Peptides were dissolved in dimethyl sulfoxide and pooled at a stock concentration of 100 μg/mL and 150 μg/mL of each peptide for EBNA1 and BZLF1, respectively. The BZLF1 pool spanned the entire BZLF1 protein, and the EBNA1 pool spanned the immunogenic C-terminal region (51 peptides). The expansion protocol used is a modified version of a previously described method.19 Briefly, PBMCs were cultured for 14 days in RPMI 1640 supplemented with penicillin/streptomycin and 10% fetal bovine serum (Sigma-Aldrich). The relevant peptide pool was added on days 1 and 7 (at 2 μg/mL of each peptide). As a negative control, PBMCs were stimulated with medium alone. Interleukin-2 (IL-2) was added at 10 U/mL on days 4, 7, and 10.
Analysis of efficacy of polyclonal EBNA1-specific IFN-γ–secreting CD8+ T cells
Polyclonal EBNA1-specific T cells were generated from healthy PBMCs. On day 14, the cells were restimulated for 5 hours with the EBNA1 peptide pool (2 μg/mL of each peptide) and with both αCD28 (2 μg/mL) and αCD49d (1 μg/mL) as costimuli. The MACS interferon-γ (IFN-γ) Secretion Assay Detection kit (PE) was used for IFN-γ surface capture as per the manufacturer's instructions (Miltenyi Biotec). Cells were costained with peridinin chlorphyll protein (PerCP)–labeled anti–human CD8 monoclonal antibody and fluorescein isothiocyanate (FITC)–labeled anti–human CD14 monoclonal antibody (BD Biosciences). A FACSAria flow cytometer (BD Biosciences) was used to sort CD8+ IFN-γ+ cells and CD8+ IFN-γ− cells (gated on the CD14-negative population). The sorted cells were used as effectors in a standard 5-hour chromium (51Cr) release assay,20 using autologous lymphoblastoid B-cell lines transformed with B95-8 virus (lymphoblastoid cell line [LCL]) and K562 as targets. Samples were tested in duplicate with an effector/target ratio of 20:1.
Detection of IFN-γ and CD107a/b-producing EBV-specific T cells
The quantity of IFN-γ and CD107a/b-producing cells was determined on day 14 by intracellular cytokine staining (ICS).21 IFN-γ and CD107a/b were assayed separately. Briefly, 5 × 105 cytotoxic T lymphocytes (CTLs) were restimulated in fluorescence-activated cell sorter buffer (phosphate-buffered saline + 2% fetal bovine serum) for 5 hours with the relevant peptide pool and with both αCD28 (2 μg/mL) and αCD49d (1 μg/mL) as costimuli. For CD107a/b assays, FITC-labeled anti–human CD107a and CD107b monoclonal antibodies (BD Biosciences) were added before restimulation. The negative control was stimulated with only αCD28 and αCD49d. The positive control was stimulated with 10 ng/mL phorbol myristate acetate and 2 μg/mL ionomycin. After 1 hour of stimulation, 1:1000 brefeldin A (Golgiplug; BD Biosciences) was added for IFN-γ detection, and monensin (Golgistop; BD Biosciences) was added for CD107a/b detection. After stimulation, cells were washed in fluorescence-activated cell sorter buffer, permeabilized (Cytofix/Cytoperm; BD Biosciences), washed in permeabilization buffer (Permwash, BD Biosciences), and stained overnight with fluorophore-conjugated anti–human monoclonal antibodies for CD3, CD8, and IFN-γ (BD Biosciences). The fluorophores used were FITC, R-phycoerythrin (PE), PerCP, and allophycocyanin. Cells were run on a FACSCanto flow cytometer (BD Biosciences) and analyzed using FlowJo Version 7 software (TreeStar).
EBNA1 strain sequence analysis and effect of polymorphism on CTL recognition
DNA was extracted from patient PBMCs via the QIAamp DNA Blood Mini Kit (QIAGEN) and from primary tissue samples via RecoverAll Total Nucleic Acid Isolation kit (Ambion). The EBNA1 region was amplified by polymerase chain reaction using the primers: forward 5′-AAAAAGGAGGGTGGTTTGGA-3′ and reverse 5′-CATTCCAAAGGGGAGACGAC -3′. The polymerase chain reaction products were run on a gel, the appropriate band was removed, dissolved, and cleaned using QIAquick Gel Extraction Kit (QIAGEN). The products were then sequenced using BigDye Terminator, Version 3.1 (Applied Biosystems) and analyzed for previously described strain variation.22,23
YNLRRGIAL- HLA-B*0801 and YNLRRGTAL- HLA-B*0801-specific CTL bulk cultures were expanded using synthetic peptide stimulation (1 μg/mL; Mimotopes). Specificity of T-cell cultures was analyzed by peptide-major histocompatibility complex (MHC) pentamer staining. T-cell cultures were incubated for 20 minutes at 4°C with YNLRRGIAL-HLA-B*0801 or YNLRRGTAL-HLA-B*0801 allophycocyanin-labeled pentamer (ProImmune), washed, and then incubated for 20 minutes at 4°C with PerCP-labeled anti–human CD8 monoclonal antibody (BD Biosciences). Cells were washed again and run on a FACSCanto flow cytometer (BD Biosciences) and analyzed using FlowJo Version 7 software (TreeStar). The cytoxicity of the CTL cultures was assayed by a standard 5-hour chromium-release assay and by detection of IFN-γ and CD107a/b production via ICS. The 51Cr release assay was tested in duplicate with autologous phytohemagglutinin (PHA) blasts as the target cells. The autologous PHA blasts were either untreated or incubated for 1 hour with various concentrations of YNLRRGIAL or YNLRRGTAL peptide (10, 1, 0.1, and 0.01 μg/mL).
Statistical analysis
To compare EBV-specific T-cell values between groups, Mann-Whitney tests were performed using GraphPad Prism Version 5.01 for Windows (GraphPad Software).
Results
Polyclonal EBNA1-specific CD8+ T cells, expanded in vitro, are capable of recognizing endogenously expressed EBNA1
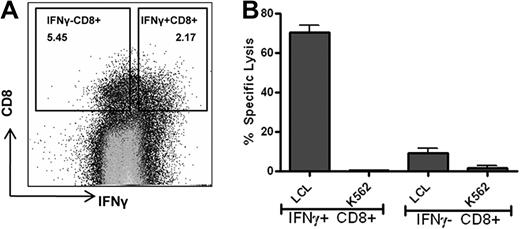
A 14-day expansion protocol has been previously used to quantify EBNA1-specific T cells in immunosuppressed patient populations.19 It measures the proliferation, as well as EBNA1-specific effector T-cell function, from an initial 106 cultured PBMCs. We wished to apply this protocol to ascertain the feasibility of EBNA1-specific T-cell generation for use in adoptive immunotherapy. To determine the applicability of this assay for this purpose, we initially tested the efficacy of this protocol in generating EBNA1-specific IFN-γ+ CD8+ T cells capable of lysing endogenously processed EBNA1-expressing B cells. We generated EBNA1-specific T cells from the PBMCs of a healthy person. EBNA1 specificity was confirmed by restimulation on day 14 with the EBNA1 peptide pool and live cell sorting of CD8+ T cells by IFN-γ surface capture using flow cytometry. Sorted CD8+ IFN-γ+ cells (and CD8+ IFN-γ− cells as controls) were used in a standard chromium release assay using autologous LCL and K562 cells as targets at an effector/target ratio of 20:1. As shown in Figure 1, EBNA1-specific CD8+ IFN-γ+ T cells demonstrated high levels of autologous LCL killing, whereas CD8+ IFN-γ− T-cell lysis was only marginally above background.
Polyclonal EBNA1-specific IFN-γ CD8+ T cells are capable of recognizing and killing endogenously expressed EBNA1-presenting B cells. Polyclonal EBNA1-specific T cells were generated from healthy PBMCs using the 14-day peptide pool expansion protocol. On day 14, the cells were restimulated with the EBNA1 peptide pool and live sorted for IFN-γ–producing CD8+ T cells using PerCP-labeled anti–human CD8 monoclonal antibody, FITC-labeled anti–human CD14 monoclonal antibody, and PE-labeled IFN-γ detection antibody/IFN-γ catch reagent. (A) Flow cytometry live sort of IFN-γ+ CD8+ T cells and IFN-γ− CD8+ T cells. The percentage of the cells within each quadrant is shown. Cells were gated on CD14-negative lymphocyte population. (B) Five-hour 51Cr release assay comparing the sorted IFN-γ+ CD8+ T cells and IFN-γ− CD8+ T cells using autologous LCL and K562 as targets. Samples were tested in duplicate with an effector/target ratio of 20:1. Error bars represent SEM.
Polyclonal EBNA1-specific IFN-γ CD8+ T cells are capable of recognizing and killing endogenously expressed EBNA1-presenting B cells. Polyclonal EBNA1-specific T cells were generated from healthy PBMCs using the 14-day peptide pool expansion protocol. On day 14, the cells were restimulated with the EBNA1 peptide pool and live sorted for IFN-γ–producing CD8+ T cells using PerCP-labeled anti–human CD8 monoclonal antibody, FITC-labeled anti–human CD14 monoclonal antibody, and PE-labeled IFN-γ detection antibody/IFN-γ catch reagent. (A) Flow cytometry live sort of IFN-γ+ CD8+ T cells and IFN-γ− CD8+ T cells. The percentage of the cells within each quadrant is shown. Cells were gated on CD14-negative lymphocyte population. (B) Five-hour 51Cr release assay comparing the sorted IFN-γ+ CD8+ T cells and IFN-γ− CD8+ T cells using autologous LCL and K562 as targets. Samples were tested in duplicate with an effector/target ratio of 20:1. Error bars represent SEM.
EBNA1-specific CD8+ and CD4+ T-cell expansion in PTLD patients is feasible
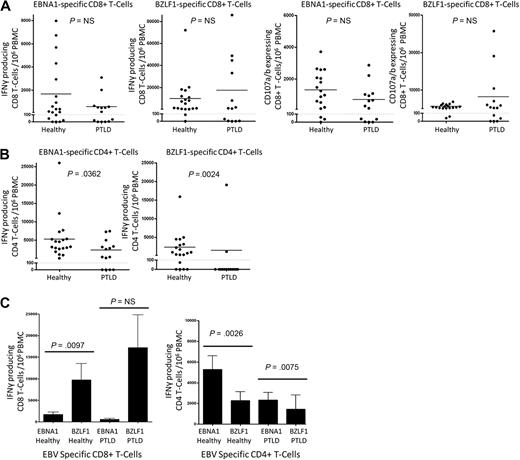
After the 14-day expansion protocol, EBV-specific T cells from PTLD patients and controls were assayed for IFN-γ secretion or CD107a/b degranulation by ICS assay after restimulation with peptide pools for 5 hours. Results from T cells expanded without peptide stimulation were used to obtain the background values. Results are presented as the number of IFN-γ and/or CD107a/b-producing cells from 106 cultured PBMCs, with the background values subtracted.19 As Figure 2 illustrates, values for both IFN-γ and CD107a/b were grouped to those with minimal response versus those with a response well above baseline. To delineate these 2 groups, we chose 100 cells grown out of 106 PBMCs as the cut-off for a positive response.
Expansion of EBV-specific effector T cells in PTLD. Polyclonal EBNA1-specific T cells and polyclonal BZLF1-specific T cells were generated from 19 healthy control and 14 EBV+ PTLD PBMCs using the 14-day peptide pool expansion protocol. On day 14, the cells were restimulated with the appropriate peptide pool and assayed for IFN-γ and CD107a/b production using flow cytometry. Results are as follows: (A) Number of IFN-γ or CD107a/b-producing CD8+ T cells expanded from 106 PBMCs. (B) Number of IFN-γ–producing CD4+ T cells (represented by the CD8− CD3+ population). (C) Comparison of EBNA1 and BZLF1-specific IFN-γ–producing CD8+ and CD4+ T cells
Expansion of EBV-specific effector T cells in PTLD. Polyclonal EBNA1-specific T cells and polyclonal BZLF1-specific T cells were generated from 19 healthy control and 14 EBV+ PTLD PBMCs using the 14-day peptide pool expansion protocol. On day 14, the cells were restimulated with the appropriate peptide pool and assayed for IFN-γ and CD107a/b production using flow cytometry. Results are as follows: (A) Number of IFN-γ or CD107a/b-producing CD8+ T cells expanded from 106 PBMCs. (B) Number of IFN-γ–producing CD4+ T cells (represented by the CD8− CD3+ population). (C) Comparison of EBNA1 and BZLF1-specific IFN-γ–producing CD8+ and CD4+ T cells
For both IFN-γ and CD107a/b, there was no significant difference between control and PTLD patients in terms of the magnitude of the EBNA1-specific CD8+ T-cell response (Figure 2A), and EBNA1-specific CD8+ T cells could be generated from the majority of the PTLD patients (9 of 14 for CD107a/b). Interestingly, and in contrast to the findings with CD8+ T cells, there were significant differences between healthy controls and PTLD IFN-γ+ EBNA1-specific CD4+ T cells (5280 ± 1319, 2336 ± 717.1, P = .036; Figure 2B). Nonetheless, we were able to expand IFN-γ+ EBNA1-specific CD4+ T cells above threshold from 10 of 14 of our PTLD patients.
BZLF1-specific CD4+ T cells were also impaired in PTLD patients (P > .002), whereas BZLF1-specific CD8+ T cells were not. The ex vivo CD8+ T-cell response to the lytic EBV-protein BZLF1 is strongly immunodominant relative to EBNA1.24 In accordance with this, we found that mean values of IFN-γ+ CD8+ T-cell were considerably higher in both the healthy EBV-seropositive volunteers (BZLF1: 9723 ± 3807; EBNA1: 1686 ± 588.1, P < .01) and the PTLD patients (BZLF1: 17 210 ± 7618; EBNA1: 581.6 ± 241.8, P < .166), although this did not reach significance for the PTLD patients (Figure 2C). EBNA1 is known to generate a relatively robust CD4+ T-cell response. However, the contribution of BZLF1-specific CD4+ T-cell immunity remains to be determined.24 In this regard, we found that, for both healthy subjects and PTLD patients, EBNA1-specific IFN-γ+ CD4+ T cells (as represented by gating on the CD8− CD3+ population) were more frequent than BZLF1-specific IFN-γ+ CD4+ T cells (P < .003 and P < .008; Figure 2C). Not surprisingly, CD107a/b CD4+ T-cell degranulation was negligible (data not shown).
HLA-B35-restricted EBNA1-specific T cells are dominant over EBNA1-specific T cells restricted by other HLA alleles
Because there was no difference in the EBNA1-specific IFN-γ+ CD8+ T-cell response between PTLD and controls, results were combined for the purposes of MHC class I analysis (Figure 3). Total EBNA1-specific IFN-γ+ CD8+ T-cell responses for each participant were tested for significance across 9 human leukocyte antigen (HLA) class I alleles (HLA-A1, A2, A3, B7, B8, B35, and B44). There was a significant association between the magnitude of HLA-B35-restricted EBNA1-specific IFN-γ+ CD8+ T cells compared with the other HLA alleles.
HLA-B35 EBNA1-specific T cells are dominant over EBNA1-specific T cells with other HLA class I alleles. Polyclonal EBNA1-specific IFN-γ CD8+ T-cell responses were analyzed for HLA class I associations. HLA class I alleles with a frequency in our control and patient samples of greater than 3 were analyzed, and EBNA1-specific IFN-γ+ CD8+ T-cell responses were compared using Mann-Whitney test. Although EBNA1-specific IFN-γ+ CD8+ T cells were expanded from all of the HLA class I alleles present in our samples, HLA-B35 EBNA1-specific T cells were statistically dominant. Error bars represent SEM.
HLA-B35 EBNA1-specific T cells are dominant over EBNA1-specific T cells with other HLA class I alleles. Polyclonal EBNA1-specific IFN-γ CD8+ T-cell responses were analyzed for HLA class I associations. HLA class I alleles with a frequency in our control and patient samples of greater than 3 were analyzed, and EBNA1-specific IFN-γ+ CD8+ T-cell responses were compared using Mann-Whitney test. Although EBNA1-specific IFN-γ+ CD8+ T cells were expanded from all of the HLA class I alleles present in our samples, HLA-B35 EBNA1-specific T cells were statistically dominant. Error bars represent SEM.
EBNA1 sequence variation influences the CD8+ T-cell response
We performed EBNA1 sequencing on PBMCs from all 14 EBV+ PTLD patients listed in Table 1. There was considerable genetic diversity with 4 different EBNA1 sequences present as outlined in Table 2. Three patients had 2 EBNA1 strain variations detectable within the peripheral blood (ie, 17 strains in 14 patients). In 4 cases, PTLD tissue was accessible for EBNA1 sequencing. In each case, the sequence was identical to that found in PBMCs. EBNA1 P-ala (the sequence observed in the EBV prototype EBV type 1 strain B95-8) was present in 43% (6 of 14) patients. Coinciding with previously published data, 71% (10 of 14) had the P-thr EBNA1 strain in which threonine is substituted with isoleucine at position 524.25 This substitution lies within a region that includes an endogenously processed HLA-B8-binding CD8+ epitope (P-ala: YNLRRGTAL; P-thr: YNLRRGIAL).25 To examine the impact of this substitution on generating effector CD8+ T cells for adoptive immunotherapy, YNLRRGIAL and YNLRRGTAL peptides were used to expand peptide-specific T cells from the PBMCs of an HLA-B8-positive healthy control and an HLA-B8-positive PTLD patient, each with the P-thr EBNA1 strain. In initial experiments, the EBNA1-specific CD8+ T cells were directly visualized by peptide-MHC pentamer staining (Figure 4A). Interestingly, significant staining was observed only with the YNLRRGIAL pentamer, demonstrating a predominance of CD8+ T cells specific for the YNLRRGIAL peptide but not for the YNLRRGTAL peptide, suggesting a dominance of YNLRRGIAL-specific CD8+ T cells within the memory T-cell repertoire in both the patient and healthy control. Functional assays confirmed this conclusion, with only the YNLRRGIAL peptide, and not the P-ala equivalent, stimulating IFN-γ release and CD107a/b degranulation (Figure 4B). Furthermore, peptide dose-response cytotoxicity assays revealed that EBNA1-specific T cells raised by in vitro stimulation with the YNLRRGIAL peptide could recognize 100-fold lower concentrations of the YNLRRGIAL peptide than the YNLRRGTAL peptide. In contrast, the T cells raised by in vitro stimulation with the YNLRRGTAL peptide were unable to recognize either the YNLRRGIAL or YNLRRGTAL peptide (Figure 4C).
EBNA1 subtypes in PTLD patients
| Sample . | EBNA1 subtypes . | |||
|---|---|---|---|---|
| P-ala . | P-ala-v2 . | P-thr . | V-val-v3 . | |
| PTLD1 | − | − | + | − |
| PTLD2 | − | − | + | − |
| PTLD3 | − | − | + | − |
| PTLD4 | +* | − | + | − |
| PTLD5 | − | − | + | − |
| PTLD6 | +* | − | + | − |
| PTLD7 | − | − | + | − |
| PTLD8 | +* | − | + | − |
| PTLD9 | +* | − | − | − |
| PTLD10 | − | + | − | − |
| PTLD11 | − | − | − | + |
| PTLD12 | − | − | + | − |
| PTLD13 | +* | − | − | − |
| PTLD14 | +* | − | − | − |
| Sample . | EBNA1 subtypes . | |||
|---|---|---|---|---|
| P-ala . | P-ala-v2 . | P-thr . | V-val-v3 . | |
| PTLD1 | − | − | + | − |
| PTLD2 | − | − | + | − |
| PTLD3 | − | − | + | − |
| PTLD4 | +* | − | + | − |
| PTLD5 | − | − | + | − |
| PTLD6 | +* | − | + | − |
| PTLD7 | − | − | + | − |
| PTLD8 | +* | − | + | − |
| PTLD9 | +* | − | − | − |
| PTLD10 | − | + | − | − |
| PTLD11 | − | − | − | + |
| PTLD12 | − | − | + | − |
| PTLD13 | +* | − | − | − |
| PTLD14 | +* | − | − | − |
PTLD indicates post-transplantation lymphoproliferative disorder.
EBNA1 subtype of the B95–8 laboratory strain.
EBNA1 sequence variation influences CD8+ effector T-cell responses. Peptide-specific T cells from an HLA-B8 EBV-seropositive control and an HLA-B8 PTLD patient were expanded from their PBMCs using EBNA1 peptides YNLRRGIAL and YNLRRGTAL. (A) Specificity of the expanded T cells was determined using YNLRRGIAL-HLA-B*0801 or YNLRRGTAL-HLA-B*0801 allophycocyanin-labeled pentamer, PerCP-labeled anti–human CD8 monoclonal antibody and FITC-labeled anti–human CD3 monoclonal antibody. The percentage of CD8+ T cells is shown. (B) The YNLRRGIAL and YNLRRGTAL expanded T cells were restimulated with YNLRRGIAL and YNLRRGTAL peptides at various concentrations and assayed for IFN-γ and CD107a/b production as previously described in Figure 2. The CD107a/b results are shown for YNLRRGIAL expanded CD8+ T cells at various concentrations of matched peptide and YNLRRGTAL expanded CD8+ T cells with the highest concentration of matched peptide. (C) The YNLRRGIAL and YNLRRGTAL expanded T cells were also used in a standard 5-hour 51Cr release assay against autologous PHA blast cells that were pretreated with various concentrations of YNLRRGIAL or YNLRRGTAL peptide. Samples were tested in duplicate with less than 3% SEM and an effector/target ratio of 20:1.
EBNA1 sequence variation influences CD8+ effector T-cell responses. Peptide-specific T cells from an HLA-B8 EBV-seropositive control and an HLA-B8 PTLD patient were expanded from their PBMCs using EBNA1 peptides YNLRRGIAL and YNLRRGTAL. (A) Specificity of the expanded T cells was determined using YNLRRGIAL-HLA-B*0801 or YNLRRGTAL-HLA-B*0801 allophycocyanin-labeled pentamer, PerCP-labeled anti–human CD8 monoclonal antibody and FITC-labeled anti–human CD3 monoclonal antibody. The percentage of CD8+ T cells is shown. (B) The YNLRRGIAL and YNLRRGTAL expanded T cells were restimulated with YNLRRGIAL and YNLRRGTAL peptides at various concentrations and assayed for IFN-γ and CD107a/b production as previously described in Figure 2. The CD107a/b results are shown for YNLRRGIAL expanded CD8+ T cells at various concentrations of matched peptide and YNLRRGTAL expanded CD8+ T cells with the highest concentration of matched peptide. (C) The YNLRRGIAL and YNLRRGTAL expanded T cells were also used in a standard 5-hour 51Cr release assay against autologous PHA blast cells that were pretreated with various concentrations of YNLRRGIAL or YNLRRGTAL peptide. Samples were tested in duplicate with less than 3% SEM and an effector/target ratio of 20:1.
Discussion
We present the first study of EBNA-1 effector T-cell immunity in EBV+ PTLD. In initial experiments, we demonstrate that EBNA1-specific CD8+ T cells grown in the 14-day expansion protocol are capable of recognizing endogenously expressed EBNA1. EBNA1-specific T cells assayed by ICS and CD107a/b degranulation were able to be generated in equivalent numbers in diagnostic blood samples from EBV+ PTLD patients and healthy EBV-seropositive controls. HLA class I influenced EBNA1-specific CD8+ effector T-cell recognition. Furthermore, we found genetic diversity in a portion of the EBNA1 genome from PTLD patients. The sequence polymorphism in an HLA-B8-binding EBNA1 epitope influenced peptide-specific T-cell responses.
The ex vivo CD8+ T-cell response to the lytic EBV-protein BZLF1 is known to be strongly immunodominant relative to EBNA1.24 Similarly, we observed that the 14-day in vitro proliferative potential of effector CD8+ T cells was significantly higher for BZLF1 compared with EBNA1 in healthy EBV-seropositive subjects. As would be expected, we found that EBNA1 generates a relatively robust CD4+ effector T-cell response. BZLF1-specific CD4+ T-cell epitopes have been identified.26 However, a systematic measure of the relative magnitude of BZLF1-specific CD4+ T-cell recall has not been established.24 In this regard, we saw that, for both healthy subjects and PTLD patients, the number of effector CD4+ T cells detected after 14-day expansion was higher for EBNA1 than BZLF1. Interestingly, there was no difference between healthy controls and PTLD patients in terms of the magnitude of the CD8+ effector T-cell response, whereas there were significant differences between healthy controls and PTLD for CD4+ T cells. Put together, these results indicate that the effect of in vivo iatrogenic immunosuppression on in vitro T-cell expansion is more difficult to reverse for CD4+ than CD8+ T cells. Nonetheless, we were able to expand effector EBNA1-specific CD4+ and CD8+ T cells from the majority of patients.
Despite some notable successes, a candid evaluation of adoptive cellular immunotherapy based on EBV-specific CD8+ T cells suggests limited success to date.3 EBV-specific adoptive immunotherapy can result in selective outgrowth of malignant cells harboring viral strains with mutations in targeted epitopes.27 Adoptive immunotherapies designed to target a broader range of epitopes covering multiple EBV antigens are probably efficacious. Protocols to date have focused on CD8+ T cells with EBNA3A, 3B, and 3C antigen specificities. This is because, in the hierarchy of EBV-specific CD8+ T-cell immunity, EBNA3A, 3B, and 3C proteins contain the most frequently recognized epitopes.24 Indeed, initial studies based on CD8+ T-cell expansion after LCL stimulation showed that EBNA1 was not recognized by CD8+ T cells.28 Subsequently, ex vivo analysis in healthy EBV-seropositive subjects, using EBNA1 peptide stimulation and cytokine release as a read-out, indicated that EBNA1-specific CD8+ T cells were detectable and in some persons were present in high numbers.15,29 Furthermore, EBNA1-specific CD8+ T-cell clones can efficiently recognize endogenously presented EBNA1 epitopes16 despite inefficient presentation of viral epitopes because of the long internal glycine-alanine repeat domain of EBNA1 adversely influencing translational efficiency.30 This results in a low but detectable level of MHC-EBNA1 peptide complexes that can be recognized by EBNA1-specific memory CD8+ T cells. A replication-deficient adenoviral construct encoding EBNA1 with the glycine-alanine repeat delete can successfully expand EBNA1-specific CD8+ effector T cells in EBV+ Hodgkin lymphoma.31 In addition, an nasopharyngeal carcinoma study using LCL stimulation to generate polyclonal EBV-specific CTL lines for autologous transfer found that 1 of 10 patients had a T-cell population specific for an EBNA1-derived, HLA class I-restricted epitope within the polyclonal CTL line. Interestingly, this was one of the 2 patients who established a complete response.32 Despite these recent findings, there are no prior data on the feasibility of expanding EBNA1-specific effector T cells in EBV+ PTLD.
A phase II trial of EBV-specific adoptive immunotherapy for EBV+ PTLD demonstrated a relationship between clinical outcome and the proportion of CD4+ T cells within the infused EBV-specific T-cell lines.6 Evidence from mouse models implies that cognate CD4+ T-cell help is essential for effective CD8+ T-cell surveillance.33 IFN-γ secretion (but not CD107a/b degranulation) of proliferating CD4+ T cells was observed in our experiments, and EBNA1-specific CD4+ T cells are known to be predominantly Th1 polarized.34 Approximately two-thirds of all healthy white donors have a CD4+ T-cell response to the C-terminal half of EBNA1.14,35 CD4+ T-cell clones to some EBNA1 epitopes can recognize and kill HLA-matched LCL, and HLA class II is expressed by PTLD lymphoma cells. The importance of CD4+ effector T-cell function directly inhibiting tumor cells (relative to indirect inhibition via T-cell help) in PTLD is not known.
The CD8+ T-cell repertoire is in part influenced by the peptide avidities for its MHC alleles. Although a number of HLA class I-restricted EBNA1 epitopes have been identified,31 to our knowledge no formal evaluation of the influence of HLA class I alleles on the magnitude of in vitro expanded EBNA1-specific effector T cells has been performed. We demonstrate that the magnitude of the EBNA1-specific IFN-γ+ CD8+ T-cell response was significantly higher for HLA-B35+ persons compared with persons expressing other HLA alleles. Interestingly, HLA-B35 shows preferential binding of atypically long CD8+ T-cell target peptides.36 The 11-mer B35 binding HPVGEADYFEY EBNA1 peptide is known to be strongly immunogenic. By contrast, although the shorter sequences HPVGEADY and HPVGEADYF conform to the HLA-B35 binding motif, they induce markedly reduced T-cell responses.37,38 Our use of 17-mer containing peptide pools that overlapped by 10 amino acids prevented bias toward CD8+ T-cell reactivities against only 8- to 10-mer peptides.
EBNA1 has several strain variations, which can impact T-cell recognition in healthy persons. The ability of T cells to recognize and respond to EBNA1 is probably crucial in maintaining immune control over EBV. However, outside of EBV+ Burkitt lymphoma,39 EBNA1 sequence analysis in EBV+ lymphomas is limited. We report the most extensive EBNA1 sequence analysis conducted to date on viral strains from EBV+ PTLD and demonstrate considerable genetic diversity, with the dominant strains not well represented by the prototype EBV laboratory strain B95-8. In line with previous data in EBV-seropositive subjects,25 we demonstrate that a sequence polymorphism within EBNA1 can impact on immune recognition of this antigen. Our data suggest that most HLA-B8 EBV-seropositive PTLD patients will recognize the variant sequence but not the EBV B95-8 YNLRRGTAL encoding epitope. Thus, EBV-specific adoptive immunotherapy expansion systems that deliver the P-ala EBNA1 sequence probably stimulates YNLRRGTAL specific T cells, which have a limited capacity in recognizing the YNLRRGIAL sequence typically encoded by EBV strains present within the lymphoma. Our results illustrate that T-cell expansion is influenced by the genetic diversity of EBV, and can be enhanced by the inclusion of antigenic determinants encoded by common polymorphisms.
We demonstrate that overlapping EBNA1 peptides can generate EBNA1-specific CD4+ and CD8+ T cells for adoptive immunotherapy in the majority of PTLD patients. Furthermore, we demonstrate that these T-cell responses can be enhanced by the inclusion of peptides encoded by common polymorphisms. A peptide-based strategy would permit generation of T cells for adoptive immunotherapy without recourse to methods using genetic manipulation. From a regulatory perspective, this is attractive. However, the cost of Good Manufacturing Practice grade EBNA1 peptides remains too high for this to be a viable option for most investigators. A number of alternative approaches might be used to expand EBNA1-specific T cells. One strategy is to use vectors encoding glycine alanine repeat-deleted EBNA1. These have been shown to efficiently stimulate EBNA1-specific CD8+ T cells in healthy virus carriers and patients with EBV+ Hodgkin lymphoma.31 A Good Manufacturing Practice grade replication-deficient adenoviral vector encoding a truncated EBNA1 is currently in clinical trials for patients with relapsed/refractory EBV+ lymphomas (M.K.G., NCT00779337). The use of novel adjuvants may further enhance EBNA1-specific CTL recognition.40 An alternative approach would be to use vectors encoding known immunogenic EBNA1 epitopes encompassing a spectrum of HLA class I and II alleles.3 To date, only a relatively limited number of CD4+ and CD8+ EBNA1-specific T-cell epitopes have been described.24 This is the result of their low frequency ex vivo. One strategy to facilitate their recognition might be to use the 14-day in vitro expansion protocol before epitope screening. Notably, this protocol resulted in preferential expansion of EBNA1-specific CD4+ T cells. Endogenous recognition of memory CD4+ T cells is thought to be dependent on lysosomal processing. More recently, autophagy has been shown to contribute to endogenous EBNA1 presentation by MHC class II.41 To manipulate the EBNA1 CD4+ T-cell pathway, further understanding of EBNA1 antigen processing is required.
In conclusion, we demonstrate, for the first time, that EBNA1-specific effector T cells should be included to broaden the antigen specificity of EBV-specific adoptive immunotherapy for PTLD. To maximize efficacy, expansion protocols should use EBNA1 antigenic sequences from relevant EBV strains.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Paul Fahey (Queensland Institute of Medical Research) for providing statistical advice.
The Clinical Immunohaematology Laboratory is funded by the NHMRC (Australia), Cancer Council of Queensland and the Queensland Smart State. K.J. is supported by the Leukemia Foundation of Queensland, and M.K.G. by the NHMRC (Australia).
Authorship
Contribution: M.K.G. and K.J. designed this study, interpreted the data, and wrote the manuscript; K.J. conducted the experimental studies, analyzed the data, and performed statistical analysis; L.M. collected the majority of the samples and assisted with some experimental studies; D.N.-V. designed the EBNA1 sequencing assay; and J.P.N., D.J.M., and S.R.B. contributed to study design and the writing of this manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Maher K. Gandhi, Clinical Immunohaematology Laboratory, Queensland Institute of Medical Research, 300 Herston Rd, Brisbane, Australia, QLD, 4006; e-mail: Maher.Gandhi@qimr.edu.au.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal