Abstract
Notch signaling is an evolutionary conserved pathway that is mediated by cell-cell contact. It is involved in a variety of developmental processes and has an essential role in vascular development and angiogenesis. Delta-like 4 (Dll4) is a Notch ligand that is up-regulated during angiogenesis. It is expressed in endothelial cells and regulates the differentiation between tip cells and stalk cells of neovasculature. Here, we present evidence that Dll4 is incorporated into endothelial exosomes. It can also be incorporated into the exosomes of tumor cells that overexpress Dll4. These exosomes can transfer the Dll4 protein to other endothelial cells and incorporate it into their cell membrane, which results in an inhibition of Notch signaling and a loss of Notch receptor. Transfer of Dll4 was also shown in vivo from tumor cells to host endothelium. Addition of Dll4 exosomes confers a tip cell phenotype on the endothelial cell, which results in a high Dll4/Notch-receptor ratio, low Notch signaling, and filopodia formation. This was further evidenced by increased branching in a tube-formation assay and in vivo. This reversal in phenotype appears to enhance vessel formation and is a new form of signaling for Notch ligands that expands their signaling potential beyond cell-cell contact.
Introduction
Notch signaling is a cell-cell signaling pathway mediated by membrane-tethered receptor-ligand interactions. It is highly conserved during evolution and has a role in several biologic processes, such as cell fate determination and differentiation.1 Recently, Notch signaling has been implicated in vascular development and angiogenesis.2 Delta-like 4 (Dll4) expression is particularly critical for angiogenesis, because haploinsufficiency results in vascular abnormalities and embryonic lethality.3 Its expression is largely restricted to the endothelium of developing vessels, and it is found at the tip cell of developing arteries, where it regulates the number of tip cells to control vessel sprouting and branching.2
Although much is known about Notch signaling after cleavage, the mechanism by which Delta-like ligands (DLLs) trigger this cleavage is less understood. Recent evidence suggests that DLL endocytosis is crucial in regulating its function, and several models have been proposed to explain the requirement for endocytosis (reviewed in Le Borgne4 ). One hypothesis suggests that endocytosis of DLL allows the ligand to undergo posttranslational modification or an association with a cofactor in the recycling endosome to make it a more effective ligand.5 Trafficking of DLL on recycling endosomes could also serve to cluster the protein and increase its affinity for Notch, because clustering of a recombinant form of DLL has been shown to be more effective at binding and activating Notch.6 Accumulation of DLL in intracellular vesicles has been described in Drosophila and zebrafish with localization to endocytic vesicles, which correlates well with Notch signaling.7 This has led to speculation that DLL may be incorporated into exosomes.8
Exosomes are produced in multivesicular bodies that are produced via endocytosis. Invagination of the limiting membrane of multivesicular bodies generates intraluminal vesicles that contain endocytic and transmembrane proteins for recycling. Multivesicular bodies can fuse with lysosomes to degrade their cargo or fuse with the plasma membrane to release these vesicles into the extracellular space. These vesicles, now called exosomes, display the endocytosed transmembrane proteins in the same orientation as at the plasma membrane.9 They can be detected in the circulation and have roles in antigen presentation and coagulation.10 It has been postulated that exosomes have signaling potential11 ; however, there is no direct evidence that DLL is incorporated into exosomes, although it has been noted that conditioned media from Delta-expressing S2 cells contains soluble full-length Delta that can activate Notch signaling.12
In this study, we investigated the possibility that Dll4 is incorporated into exosomes. We demonstrated that human umbilical vein endothelial cells (HUVECs) naturally produce exosomes that contain Dll4. Dll4 could also be incorporated into the exosomes of tumor cells that overexpressed Dll4. These exosomes can transfer the Dll4 protein from one cell type to another and incorporate it into the plasma membrane in vitro and in vivo. Surprisingly, the Dll4-containing exosomes can inhibit Notch signaling in vitro and appear to switch the endothelial cell phenotype toward tip cells. This results in an increase in vessel density in vitro and an increase in branching in vivo. These results suggest a new aspect to DLL/Notch signaling that does not require direct cell-cell contact.
Methods
Cell culture and reagents
HUVECs (Lonza) were cultured in M199 media supplemented with 10% fetal calf serum (Sigma-Aldrich), 50 mg/L ECGS (endothelial cell growth supplement; BD Biosciences), and 10 U/mL heparin (Sigma-Aldrich). The cells between passages 3 and 7 were used. U87GM cells (including Dll4-overexpressing cells) were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. Production of the U87 cell line overexpressing Dll4 has been described previously.13 Recombinant human Dll4 extracellular domain was purchased from R&D Systems. It was coated onto tissue culture dishes overnight at a concentration of 1 μg/mL in 0.2% gelatin. The γ-secretase inhibitor dibenzazepine (Calbiochem) was dissolved in dimethyl sulfoxide and used at a concentration of 10nM.
Quantitative polymerase chain reaction protocol
RNA extraction, the quantitative polymerase chain reaction protocol, and primers have been described previously.14
Western blotting
Proteins were separated with sodium dodecyl sulfate–polyacrylamide gel electrophoresis by use of standard techniques. Antibodies were purchased from the following companies: Notch1 cleaved intracellular domain (NICD) and Dll4 from Cell Signaling; β-tubulin, β-actin, and green fluorescent protein from Sigma-Aldrich; Notch1 extracellular domain, TSG101, and mouse Dll4 from Abcam; Rab5 from Santa Cruz; and Lamp1 from BD Biosciences. Anti–human Dll4 (Regeneron 242) was a gift from Regeneron. Secondary antibodies were purchased from DAKO.
Protein identification by mass spectrometry
Isolated U87 control and U87 Dll4 exosome material was precipitated and desalted with chloroform/methanol.15 Samples were resuspended in buffer that contained 6M urea and 100mM Tris pH 7.8, followed by reduction, alkylation, and in-solution trypsin digestion as described previously.16 Trypsin digested protein material was analysed by liquid chromatography–mass spectrometry/mass spectrometry with an UltiMate (LC Packings, Dionex) high-performance liquid chromatography system coupled online to a 3-dimensional high-capacity ion trap (HCTplus, Bruker Daltonics) mass spectrometer as described previously.16
Protein identification and label-free quantitative analysis by nano-UPLC-MSE tandem mass spectrometry
The digested material prepared above was subjected to nano-ultraperformance liquid chromatography tandem mass spectrometry analysis (nano-UPLC-MSE or nano-MS/MS) as described previously.17
NanoSight analysis of exosome size
U87 control and HUVEC exosome preparations were assessed with the NanoSight LM10 (NanoSight, Ltd) system. Exosome preparations were stored at −80°C, thawed at room temperature, and then diluted in phosphate-buffered saline (PBS) that had been shown to be free of any contaminant particles. U87 control and HUVEC exosome samples were diluted accordingly so that approximately 1000 particles were analyzed per sample. The culture media from both cell types were analyzed at the same dilution, independent of the sample, and no contaminating particles were detected. Particles in the range of 80 to 300 nm have been detected previously in the medium of live cells by a related technique, dynamic light scattering. Larger apoptotic vesicles are detected between 400 and 1200 nm.18
Exosome isolation and labeling
U87 cells were incubated in Opti-Mem (Gibco) for 48 hours to obtain exosomes. At the point of harvest, the cells were stained with trypan blue to check viability. HUVECs were cultured in endothelial cell growth medium-2 (EGM2) for 48 hours before being harvested. HUVECs were plated on Dll4-coated plates to induce the production of Dll4 and subsequently the production of exosomes that contained Dll4. The EGM2 had been centrifuged previously at 110 000g for 16 hours to remove contaminating exosomes. Exosomes were harvested from cell media by ultracentrifugation as described previously.19 The resulting exosomes were resuspended in PBS or lysed in radioimmunoprecipitation assay buffer. The exosomes were labeled with the fluorescent membrane dye PKH67 (Sigma-Aldrich) according to the manufacturer's instructions.
Biotinylation of cell-surface proteins
U87 and HUVEC cell-surface proteins were labeled with biotin and purified with the Pierce cell-surface protein isolation kit according to the manufacturer's instructions.
Tube-formation assay
Normal dermal fibroblasts were seeded into the wells of a 24-well plate at a density of 2 × 104/well. The cells were allowed to adhere for 4 hours before HUVECs were seeded on top at a density of 4 × 103/well. Cells were cultured in EGM2, and the medium was changed every 2 days (+50 μg/mL exosomes) for 10 days before fixation in methanol. Endothelial cells were stained with anti-CD31 and goat anti–mouse antibodies (both from Serotec). The vessels were then visualized with Sigma-Fast BCIP (Sigma-Aldrich).
Proliferation assay
HUVEC proliferation was assessed with the CellTiter 96 AQueous One Solution cell proliferation assay from Promega according to the manufacturer's instructions.
Xenograft mouse models
All protocols were conducted with approval from the United Kingdom Home Office and have been described previously.13 Exosomes (100 μL at 50 μg/mL) were mixed with U87 cells and Matrigel before injection with a 1-mL syringe and a 25-gauge needle. Exosomes (100 μL at 50 μg/mL) were then injected into the edge of the growing tumor with a single injection with a 1-mL syringe and a 27-gauge needle. This was performed 3 times weekly for 5 weeks.
Immunohistochemistry and immunocytochemistry
The mouse CD31 staining protocol has been described previously.13 For immunocytochemistry, cells were fixed in acetone and stained with anti–human Dll4 (a gift from Regeneron) according to standard procedures. Filopodia were stained with tetramethylrhodamine B isothiocyanate–phalloidin (Sigma-Aldrich) according to the manufacturer's procedures.
Results
Dll4 is incorporated into exosomes
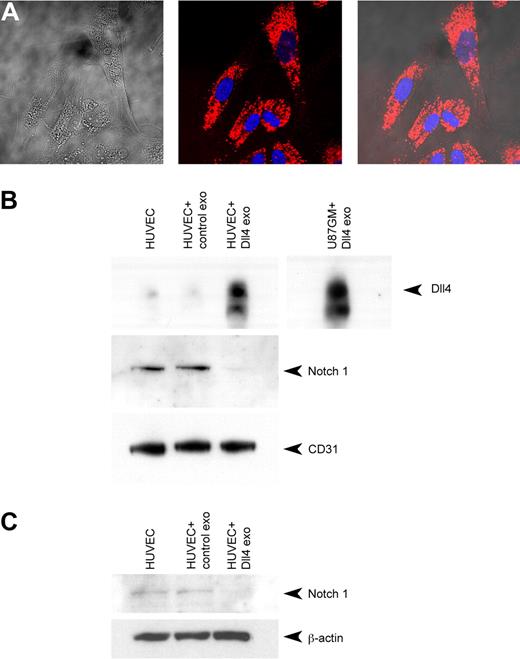
HUVECs contain low levels of Dll4 under normal culture conditions. They can be stimulated to produce Dll4 by plating the cells onto Dll4-coated plates.14 This results in the activation of Notch signaling and the transcription and production of Dll4, which can be inhibited by the addition of the γ-secretase inhibitor dibenzazepine (Figure 1A). Under these conditions, Dll4 can easily be detected in HUVEC lysates and in the exosomes that the cells produce (Figure 1B). The Dll4 in the exosomes appears to have the same molecular weight as the lysate, which suggests that no substantial protein cleavage has occurred.
Dll4 is incorporated into exosomes. (A) HUVECs were seeded onto 6-well plates at 3 × 105/well with or without the presence of 10nM dibenzazepine (DBZ). The plates had been coated previously with either bovine serum albumin (BSA; 1 μg/mL) or recombinant hDll4 (1 μg/mL) to induce Notch signaling. The cells were lysed after 24 hours and subjected to Western blotting to detect Dll4 or β-actin as a loading control. (B) HUVECs were grown on recombinant hDll4–coated plates for 48 hours in exosome-depleted EGM2. The medium was harvested, and the cells were lysed in radioimmunoprecipitation assay buffer. The medium was ultracentrifuged to isolate the exosomes (exo), then the fractions were analyzed by Western blotting to detect Dll4. (C) U87 Dll4 and U87 control cells were grown for 48 hours in Opti-Mem before harvesting of the media and cell lysis. The exosomes were harvested by ultracentrifugation and subjected to Western blotting to detect Dll4 and exosomal markers. (D) Exosome size was analyzed with a NanoSight LM10; vertical axis units 1 × 107 particles/mL per nanometer.
Dll4 is incorporated into exosomes. (A) HUVECs were seeded onto 6-well plates at 3 × 105/well with or without the presence of 10nM dibenzazepine (DBZ). The plates had been coated previously with either bovine serum albumin (BSA; 1 μg/mL) or recombinant hDll4 (1 μg/mL) to induce Notch signaling. The cells were lysed after 24 hours and subjected to Western blotting to detect Dll4 or β-actin as a loading control. (B) HUVECs were grown on recombinant hDll4–coated plates for 48 hours in exosome-depleted EGM2. The medium was harvested, and the cells were lysed in radioimmunoprecipitation assay buffer. The medium was ultracentrifuged to isolate the exosomes (exo), then the fractions were analyzed by Western blotting to detect Dll4. (C) U87 Dll4 and U87 control cells were grown for 48 hours in Opti-Mem before harvesting of the media and cell lysis. The exosomes were harvested by ultracentrifugation and subjected to Western blotting to detect Dll4 and exosomal markers. (D) Exosome size was analyzed with a NanoSight LM10; vertical axis units 1 × 107 particles/mL per nanometer.
For subsequent experiments, a U87 cell line (which normally does not express Dll4) was used that overexpresses human Dll4 (U87 Dll4). This allowed us to produce larger quantities of exosomes with and without Dll4. Western blotting confirmed that Dll4 was present in the exosomes derived from the cell line that overexpressed Dll4 (Figure 1C). Both sets of exosomes contained known exosomal markers: LAMP1 (lysosomal-associated membrane protein 1, a marker of late exosomes), TSG101 (tumor-susceptibility gene 101, a member of the ESCRT [endosomal sorting complex required for transport] machinery), and Rab5 (a small GTPase involved in clathrin-mediated endocytosis).20
The level of exosome production was compared between the cell lines (supplemental Table 2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). U87 control cells produce approximately 1.48 times more exosomes than U87 cells that overexpress Dll4. U87-Dll4 cells also grow more slowly because of increased Notch signaling.13 Control HUVECs produce 2.87 times more exosomes than HUVECs plated onto Dll4. Activation of Notch signaling in these cells also reduces their proliferation rate.2 Control U87 cells produce 8 times more exosomes than control HUVECs. These data suggest that exosome production is related to growth rate.
Characterization of the exosomes
Exosomes are observed to be between 40 and 100 nm in diameter by electron microscopy.11 To assess particle size and size distribution, we used the NanoSight LM10 system for real-time visualization and assessment of particles in solution. The modal sizes of the U87 control exosomes and HUVEC exosomes were similar and were estimated to be 120 and 114 nm, respectively (Figure 1D).
To further characterize the properties of our exosome preparations, we performed an analysis by tandem mass spectrometry on exosomes derived from U87 control cells. The analysis revealed 40 proteins, 95% of which have been described previously as exosomal proteins (supplemental Table 1). These included extracellular matrix and cell-surface proteins such as collagens, integrins, galectin, and CD44/63/151, as well as intracellular components such as metabolic enzymes, G proteins, and cytoskeletal components consistent with previous analyses.21 The remaining proteins are other family members of previously known exosomal proteins such as β-enolase and may be more specific to U87-derived exosomes. The high degree of correlation between the composition of proteins identified and the previously known content of these particles confirms the validity of the procedure we used to isolate exosomes from these cells. Further analysis of control and Dll4-containing U87 exosomes by quantitative, comparative mass spectroscopy revealed an additional 5 proteins that were only present in the Dll4-containing exosomes, which have been described as having a role in angiogenesis (supplemental Table 3) and have been reported to have roles in endothelial cell migration, adhesion, proliferation, and vessel stabilization.22 It is unclear whether their presence is a result of Notch signaling or a direct interaction with the Dll4 itself, because a known Dll4-interacting protein, DLG1, was also identified in the Dll4-containing exosomes.23
Dll4-containing exosomes inhibit Notch signaling
To assess the effects of Dll4-containing exosomes on Notch signaling, we measured the effects on downstream Notch targets with a quantitative polymerase chain reaction protocol. As an internal control, we activated Notch signaling by plating HUVECs on Dll4-coated plates for 24 hours before RNA extraction (compared with a bovine serum albumin control). This treatment resulted in increased transcription of the Notch target genes Hey1 and Hey2 (Figure 2A). During plating on Dll4-coated plates, U87 exosomes (50 μg/mL) were added to assess their effects. Control exosomes had no significant effect on Notch signaling, but Dll4-containing exosomes produced down-regulation of downstream Notch target expression (Figure 2A). HUVEC exosomes isolated from cells plated on Dll4-coated plates were also capable of inhibiting Notch signaling (Figure 2B). To confirm these results, we investigated the production of NICD in response to Notch activation by Western blotting. After a 24-hour incubation of HUVECs on Dll4-coated plates, there was an accumulation of NICD compared with the bovine serum albumin control (Figure 2C). This level was unaffected when HUVECs were plated onto Dll4-coated plates in the presence of 50 μg/mL control exosomes; however, incubation with Dll4-containing exosomes resulted in an inhibition of NICD accumulation (Figure 2C).
Dll4-containing exosomes inhibit Notch signaling. (A) HUVECs were plated onto bovine serum albumin (BSA)– or Dll4-coated 6-well plates at a density of 3 × 105/well in the presence of either control exosomes (50 μg/mL) or Dll4-containing exosomes harvested from U87 (50 μg/mL). After 24 hours, the cells were extracted in TRI reagent to isolate the RNA. Quantitative polymerase chain reaction protocol was conducted to examine the Notch target genes Hey1 and Hey2. (B) Identical experiment with exosomes isolated from HUVEC control cells or HUVECs plated on Dll4. Samples were analyzed in triplicate. One-way analysis of variance with a Tukey multiple-comparison post hoc test was performed with GraphPad Prism 4.0b software. (C) The same experiment as shown in panel A, except after 24 hours, the cells were lysed in radioimmunoprecipitation assay buffer and subjected to Western blotting to detect the level of NICD or β-actin as a loading control. GAPDH indicates glyceraldehyde-3-phosphate dehydrogenase; and exo, exosomes.
Dll4-containing exosomes inhibit Notch signaling. (A) HUVECs were plated onto bovine serum albumin (BSA)– or Dll4-coated 6-well plates at a density of 3 × 105/well in the presence of either control exosomes (50 μg/mL) or Dll4-containing exosomes harvested from U87 (50 μg/mL). After 24 hours, the cells were extracted in TRI reagent to isolate the RNA. Quantitative polymerase chain reaction protocol was conducted to examine the Notch target genes Hey1 and Hey2. (B) Identical experiment with exosomes isolated from HUVEC control cells or HUVECs plated on Dll4. Samples were analyzed in triplicate. One-way analysis of variance with a Tukey multiple-comparison post hoc test was performed with GraphPad Prism 4.0b software. (C) The same experiment as shown in panel A, except after 24 hours, the cells were lysed in radioimmunoprecipitation assay buffer and subjected to Western blotting to detect the level of NICD or β-actin as a loading control. GAPDH indicates glyceraldehyde-3-phosphate dehydrogenase; and exo, exosomes.
Notch signaling in HUVECs reduces cell proliferation2 ; therefore, the rate of proliferation was assessed in exosome-treated HUVECs with an MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. The addition of Dll4-containing exosomes to HUVECs increased the rate of proliferation, which is consistent with an inhibition of Notch signaling; however, control exosomes were also capable of increasing proliferation (supplemental Figure 1). Tumor exosomes have been reported to induce endothelial cell proliferation.24 Mass spectroscopy revealed that U87 exosomes contained several proteins that are known to be involved in proliferation (eg, G proteins), and their transfer to HUVECs may contribute to their proliferative effects (supplemental Table 1).
Dll4-containing exosomes increase vessel branching and vessel density in vitro
To investigate the function of Dll4-containing exosomes in angiogenesis, HUVECs were mixed with fibroblasts to induce tube formation in vitro. After plating of the cells, the medium was changed to contain either 50 μg/mL control exosomes or 50 μg/mL Dll4-containing exosomes. The medium was changed every 2 days, with new exosomes added for a period of 10 days. The vessels were stained with an anti-CD31 antibody, and the tube length, size, density, and degree of branching were assessed with ImageJ Version 1.41 software (National Institutes of Health). A typical example of tube formation after 10 days is shown in Figure 3B. Statistical analysis revealed that there was a significant increase in the number of nodes (branching) and number of tips formed when the cells were incubated with Dll4-containing exosomes compared with controls. This also resulted in a significant increase in vessel density (Figure 3A). This phenotype is consistent with an inhibition of Notch signaling.25
Dll4-containing exosomes increase vessel branching. (A) Normal dermal fibroblasts were seeded onto 24-well plates at a density of 2 × 104/well, and HUVECs were seeded onto the fibroblasts at a density of 4 × 103/well. The cells were cultured for 10 days in EGM2 with either control exosomes (50 μg/mL) or Dll4-containing exosomes (50 μg/mL), with the media changed every 2 days. Cells were fixed and stained as described in “Methods,” and pictures were taken with a Zeiss Axiovert 135 microscope at 5× magnification. (A) Three batches of control exosomes and 3 batches of Dll4-containing exosomes were tested, each in triplicate. Ten images were captured from each well for analysis (a total of 90 pictures for control and 90 for Dll4-containing exosome–treated cells). (B) Representative images were analyzed with ImageJ software for vessel length, size, and density. The number of tips and nodes was also counted. (C) U87 xenografts were grown and injected 3 times per week for 5 weeks with either PBS, control exosomes (Control exo), or Dll4 exosomes (Dll4 exo, isolated from U87 cells) at a concentration of 50 μg/mL. The animals were then killed and the tumors sectioned for vessel staining. A Chalkley vessel count (CVC) was performed for vessel density, and the vessel length and lumen size were estimated with ImageJ. The number of branches was counted manually and normalized to the CVC. Data were analyzed by t test or 1-way analysis of variance with a Tukey multiple-comparison post hoc test with GraphPad Prism 4.0b software and shown as mean with standard deviation. (D) Representative images taken on a Nikon Eclipse E800 microscope at 10× magnification.
Dll4-containing exosomes increase vessel branching. (A) Normal dermal fibroblasts were seeded onto 24-well plates at a density of 2 × 104/well, and HUVECs were seeded onto the fibroblasts at a density of 4 × 103/well. The cells were cultured for 10 days in EGM2 with either control exosomes (50 μg/mL) or Dll4-containing exosomes (50 μg/mL), with the media changed every 2 days. Cells were fixed and stained as described in “Methods,” and pictures were taken with a Zeiss Axiovert 135 microscope at 5× magnification. (A) Three batches of control exosomes and 3 batches of Dll4-containing exosomes were tested, each in triplicate. Ten images were captured from each well for analysis (a total of 90 pictures for control and 90 for Dll4-containing exosome–treated cells). (B) Representative images were analyzed with ImageJ software for vessel length, size, and density. The number of tips and nodes was also counted. (C) U87 xenografts were grown and injected 3 times per week for 5 weeks with either PBS, control exosomes (Control exo), or Dll4 exosomes (Dll4 exo, isolated from U87 cells) at a concentration of 50 μg/mL. The animals were then killed and the tumors sectioned for vessel staining. A Chalkley vessel count (CVC) was performed for vessel density, and the vessel length and lumen size were estimated with ImageJ. The number of branches was counted manually and normalized to the CVC. Data were analyzed by t test or 1-way analysis of variance with a Tukey multiple-comparison post hoc test with GraphPad Prism 4.0b software and shown as mean with standard deviation. (D) Representative images taken on a Nikon Eclipse E800 microscope at 10× magnification.
Dll4-containing exosomes increase vessel branching, vessel length, and vessel size in vivo
Control and Dll4-containing exosomes were injected into U87 xenografts 3 times per week for 5 weeks. At the end of the experiment, the tumors were measured, and the Dll4-containing exosome–treated tumors appeared to be slightly larger, although this was not significant (supplemental Figure 2). The tumors were sectioned and stained to visualize the vessels. Vessel density was high in all of the tumors, and there was no significant difference in density between the groups (supplemental Figure 2). There was, however, a significant increase in vessel branching observed in the Dll4-containing exosome–treated tumors compared with the controls, which is consistent with an inhibition in Notch signaling (Figure 3C). There was also a significant increase in vessel length and size in the Dll4-containing exosome–treated tumors (Figure 3C). Representative images of vessels are shown in Figure 3D.
Dll4 can be transferred to another cell via exosomes
To determine the mechanism of Dll4-containing exosome–mediated inhibition of Notch signaling, we looked at the subcellular location of the Dll4 after incubation with the exosomes. U87GM cells, which contain no Dll4, were incubated for 24 hours with 50 μg/mL control or Dll4-containing exosomes and then washed thoroughly with PBS before lysis in radioimmunoprecipitation assay buffer. Western blotting revealed that the Dll4 from the exosomes had been transferred to the U87GM cells (Figure 4A). The association of Dll4-containing exosomes with the U87GM cells was confirmed by immunocytochemistry (Figure 4B). To resolve whether the exosomes were being internalized or were strongly associated with the plasma membrane, U87 cells were pelleted, after a 24-hour incubation with exosomes, then embedded and sectioned to obtain slices through the cell. The sections were then stained for human Dll4 (hDll4). Dll4 was only detected in the cells incubated with Dll4-containing exosomes (50 μg/mL), and it appeared to be at the membrane and intracellular level (Figure 4C). The exosomes were also labeled with the fluorescent membrane dye PKH67 and incubated with HUVECs for 16 hours. The cells were then analyzed by confocal microscopy to assess the position of the exosomes, and they appeared to have been endocytosed (Figure 5A). To assess cell-surface localization of the added Dll4, cell-surface proteins were biotinylated and purified. A small amount of Dll4 was present at the membrane in HUVECs under normal culture conditions; however, after the addition of Dll4-containing exosomes, the amount present at the membrane increased substantially (Figure 5B). Incorporation of Dll4 into the membrane was also apparent in U87GM cells after incubation with Dll4-containing exosomes (Figure 5B).
Dll4 is transferred to other cells via exosomes. (A) U87GM cells were seeded onto 6-well plates at 3 × 105/well. The cells were then incubated with either control exosomes (50 μg/mL) or Dll4-containing exosomes (50 μg/mL) in Opti-Mem for 24 hours before extensive washing with PBS and lysis in radioimmunoprecipitation assay buffer. Lysates were subjected to Western blotting and were probed with either anti-Dll4 or β-tubulin as a loading control. Exo indicates exosomes. (B) The same experiment was repeated, except the cells were fixed in acetone after the 24-hour incubation. They were then stained with anti-hDll4. Images were taken on a Nikon Eclipse E800 microscope at 40× magnification. (C) U87 cells incubated with exosomes (50 μg/mL) for 24 hours were pelleted, embedded, and sectioned. The sections were then stained with anti-hDll4, and images were taken at 50× magnification with a Nikon Eclipse E800 microscope.
Dll4 is transferred to other cells via exosomes. (A) U87GM cells were seeded onto 6-well plates at 3 × 105/well. The cells were then incubated with either control exosomes (50 μg/mL) or Dll4-containing exosomes (50 μg/mL) in Opti-Mem for 24 hours before extensive washing with PBS and lysis in radioimmunoprecipitation assay buffer. Lysates were subjected to Western blotting and were probed with either anti-Dll4 or β-tubulin as a loading control. Exo indicates exosomes. (B) The same experiment was repeated, except the cells were fixed in acetone after the 24-hour incubation. They were then stained with anti-hDll4. Images were taken on a Nikon Eclipse E800 microscope at 40× magnification. (C) U87 cells incubated with exosomes (50 μg/mL) for 24 hours were pelleted, embedded, and sectioned. The sections were then stained with anti-hDll4, and images were taken at 50× magnification with a Nikon Eclipse E800 microscope.
Dll4-containing exosomes transfer Dll4 to the membrane, reduce Notch1 receptor levels, and are endocytosed. (A) Dll4-containing exosomes were labeled with a red fluorescent dye (PKH67) as described in “Methods.” HUVECs were incubated with Dll4-containing exosomes (50 μg/mL) for 16 hours before images were acquired with a Zeiss LSM 510 confocal microscope at 63× magnification. Images shown are a midsection slice through the cells showing intracellular localization of the exosomes and exclusion from the nucleus. Phase-contrast image and merged image show that fluorescence did not occur at the membrane. (B) HUVECs or U87GM cells were seeded at a density of 3 × 106 cells/150 cm2 dish. Each dish was incubated for 24 hours with control exosomes (50 μg/mL) or Dll4-containing exosomes (50 μg/mL) before the cell-surface proteins were biotinylated and purified as described in “Methods.” The purified proteins were subjected to Western blotting to detect Dll4 or Notch 1 at the cell membrane, with CD31 probed as a loading control. (C) The same experiment was repeated, except the cells were lysed to isolate total protein. The lysates were subjected to Western blotting to detect total Notch1, with β-actin used as a loading control. Exo indicates exosomes.
Dll4-containing exosomes transfer Dll4 to the membrane, reduce Notch1 receptor levels, and are endocytosed. (A) Dll4-containing exosomes were labeled with a red fluorescent dye (PKH67) as described in “Methods.” HUVECs were incubated with Dll4-containing exosomes (50 μg/mL) for 16 hours before images were acquired with a Zeiss LSM 510 confocal microscope at 63× magnification. Images shown are a midsection slice through the cells showing intracellular localization of the exosomes and exclusion from the nucleus. Phase-contrast image and merged image show that fluorescence did not occur at the membrane. (B) HUVECs or U87GM cells were seeded at a density of 3 × 106 cells/150 cm2 dish. Each dish was incubated for 24 hours with control exosomes (50 μg/mL) or Dll4-containing exosomes (50 μg/mL) before the cell-surface proteins were biotinylated and purified as described in “Methods.” The purified proteins were subjected to Western blotting to detect Dll4 or Notch 1 at the cell membrane, with CD31 probed as a loading control. (C) The same experiment was repeated, except the cells were lysed to isolate total protein. The lysates were subjected to Western blotting to detect total Notch1, with β-actin used as a loading control. Exo indicates exosomes.
Transfer of Dll4 occurs in vivo in tumor endothelium
Next, we addressed whether Dll4 can be transferred to mouse endothelial cells in vivo using a tumor xenograft model. U87 cells that overexpressed Dll4 or control U87 cells were implanted subcutaneously into BALB/c severe combined immunodeficiency (SCID) mice. The tumors were grown until they reached maximum size, at which time the mice were killed and the tumors harvested for immunohistochemistry. The tumor sections were stained with an antibody specific for hDll4, confirmation of which is shown in supplemental Figure 3A-B. We further confirmed that we were unable to detect mouse Dll4 in control tumor vessels (Figure 6A); however, hDll4 was easily detectable in tumors derived from U87 Dll4 cells and was detectable in the mouse vessels (Figure 6A). To confirm that this phenomenon was a result of ligand transfer and not vascular mimicry by the tumor cells, we stained serial sections of vessels with anti-hDll4 and anti–mouse CD31 antibodies. The cells surrounding the vessel were positive for both mouse CD31 and hDll4, which confirms that the mouse endothelial cells had acquired the hDll4 (Figure 6B). Higher-magnification images revealed that the transferred hDll4 was found intracellularly and at the membrane in the vessels. Surprisingly, there was also specific staining of hDll4 on the mouse red blood cells (RBCs; supplemental Figure 3C), which suggests that the hDll4 can transfer to the trapped RBCs. RBCs in control tumors were completely negative for this staining (supplemental Figure 3D).
Dll4 can be transferred to mouse endothelial cells in vivo. (A) Immunohistochemical staining of hDll4 in control tumors derived from U87 cells or tumors derived from U87 overexpressing Dll4. hDll4 staining was not detectable in mouse tissue (red arrow); however, hDll4 staining was detectable in tumor tissue (white arrow) and in mouse vessels (yellow arrow). (B) Immunohistochemical staining of serial sections derived from a U87 Dll4 tumor to detect mouse CD31 and human Dll4. The cells surrounding the vessel were positive for CD31 and hDll4, which confirms that the mouse endothelial cells had acquired the hDll4 from the tumor cells. Images were taken with a Nikon Eclipse E800 microscope at 40× magnification.
Dll4 can be transferred to mouse endothelial cells in vivo. (A) Immunohistochemical staining of hDll4 in control tumors derived from U87 cells or tumors derived from U87 overexpressing Dll4. hDll4 staining was not detectable in mouse tissue (red arrow); however, hDll4 staining was detectable in tumor tissue (white arrow) and in mouse vessels (yellow arrow). (B) Immunohistochemical staining of serial sections derived from a U87 Dll4 tumor to detect mouse CD31 and human Dll4. The cells surrounding the vessel were positive for CD31 and hDll4, which confirms that the mouse endothelial cells had acquired the hDll4 from the tumor cells. Images were taken with a Nikon Eclipse E800 microscope at 40× magnification.
Dll4-containing exosomes reduce the level of Notch receptors at the cell surface
It is clear that Dll4 is transferred to target cells, and it can be incorporated into the plasma membrane; however, it is unclear how this transfer inhibits Notch signaling. One possibility is that Dll4 on the exosomes binds to the Notch receptor and targets it for internalization with the rest of the exosomes. To explore this hypothesis, the membrane proteins were biotinylated and purified after incubation with exosomes. The level of Notch1 extracellular domain was assessed by Western blotting. Notch1 receptor was present at the membrane in HUVECs incubated with control exosomes; however, the level was almost completely ablated by the presence of Dll4-containing exosomes (Figure 5B). Total cellular Notch1 was also reduced in cell lysates after incubation with Dll4-containing exosomes, which suggests that Notch receptor is internalized and degraded rather than internalized and recycled to the surface at a later time point (Figure 5C).
Dll4-containing exosomes can reduce the level of stalk cell markers and induce a tip cell phenotype
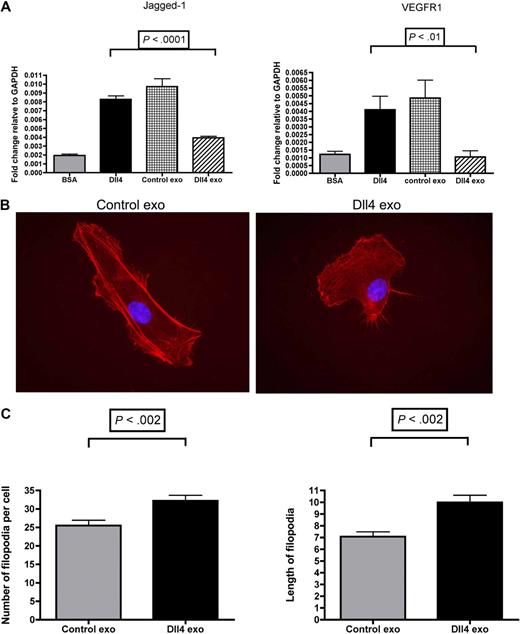
The default phenotype of endothelial cells is a tip cell, whereas activation of Notch signaling confers a stalk cell phenotype. We and others have demonstrated previously that activation of Notch signaling in endothelial cells using recombinant Dll4-coated plates results in the up-regulation of the stalk cell markers Jagged-1 and vascular endothelial growth factor receptor-1.2,14 To assess whether Dll4-containing exosomes are capable of reversing the expression of stalk cell markers, endothelial cells were incubated with 50 μg/mL of Dll4-containing exosomes or control exosomes for 24 hours in the presence of recombinant Dll4. Control exosomes had no effect on Jagged-1 and vascular endothelial growth factor receptor-1 expression; however, Dll4-containing exosomes inhibited their expression (Figure 7A). The presence of filopodia is a tip cell hallmark.2 To assess the effects of Dll4-containing exosomes on filopodia formation, HUVECs were incubated for 24 hours in the presence of 3 different batches of 50 μg/mL of control or Dll4-containing exosomes before being stained with tetramethylrhodamine B isothiocyanate–phalloidin. The filopodia were counted, and their number was found to be significantly higher on the Dll4-containing exosome–treated cells. Interestingly, the filopodia were also significantly longer (Figure 7B-C). These data also suggest that the Dll4-containing exosomes can confer a tip cell phenotype.
Dll4-containing exosomes reduce the level of stalk cell markers and confer a tip cell phenotype. (A) HUVECs were plated onto bovine serum albumin (BSA) or Dll4-coated 6-well plates at a density of 3 × 105/well in the presence of either control exosomes (50 μg/mL) or Dll4-containing exosomes (50 μg/mL). After 24 hours, the cells were extracted in TRI reagent to isolate the RNA. Quantitative polymerase chain reaction protocol was performed to examine the stalk cell markers Jagged-1 and vascular endothelial growth factor receptor-1 (VEGFR1). Samples were analyzed in triplicate. One-way analysis of variance with a Tukey multiple-comparison post hoc test was performed with GraphPad Prism 4.0b software. Exo indicates exosomes. (B) HUVECs were incubated with 3 different batches of control or Dll4-containing exosomes for 24 hours before staining with tetramethylrhodamine B isothiocyanate–phalloidin to visualize the filopodia. (C) The filopodia were counted on 40 cells from each condition, and the length of the longest 3 filopodia on each cell was assessed with ImageJ. Images were taken on a Zeiss Axioskop 2 microscope at 63× magnification. Data were analyzed by t test with GraphPad Prism 4.0b software and are shown as mean with SD.
Dll4-containing exosomes reduce the level of stalk cell markers and confer a tip cell phenotype. (A) HUVECs were plated onto bovine serum albumin (BSA) or Dll4-coated 6-well plates at a density of 3 × 105/well in the presence of either control exosomes (50 μg/mL) or Dll4-containing exosomes (50 μg/mL). After 24 hours, the cells were extracted in TRI reagent to isolate the RNA. Quantitative polymerase chain reaction protocol was performed to examine the stalk cell markers Jagged-1 and vascular endothelial growth factor receptor-1 (VEGFR1). Samples were analyzed in triplicate. One-way analysis of variance with a Tukey multiple-comparison post hoc test was performed with GraphPad Prism 4.0b software. Exo indicates exosomes. (B) HUVECs were incubated with 3 different batches of control or Dll4-containing exosomes for 24 hours before staining with tetramethylrhodamine B isothiocyanate–phalloidin to visualize the filopodia. (C) The filopodia were counted on 40 cells from each condition, and the length of the longest 3 filopodia on each cell was assessed with ImageJ. Images were taken on a Zeiss Axioskop 2 microscope at 63× magnification. Data were analyzed by t test with GraphPad Prism 4.0b software and are shown as mean with SD.
Discussion
It has been speculated that Dll4 is incorporated into exosomes.8 This stems from several studies that have identified endocytosis of DLLs as a key step during Notch signaling in Drosophila. We demonstrated that upon induction of Dll4 in HUVECs, the protein is indeed incorporated into exosomes. This also occurs in the U87 overexpression model. The exosomes have a proteomic profile that matches published data and are only slightly larger than the suggested size of exosomes (modal size of 114 nm for HUVECs and 120 nm for U87 cells, compared with published sizes of 50-100 nm). Exosome size is normally estimated with electron microscopy, which has its limitations. With that technique, exosomes are fixed and dehydrated, which may result in shrinkage and hence an underestimation of their actual size. Furthermore, the analysis of images of exosomes obtained by electron microscopy is highly subjective. Nanoparticle tracking analysis (NanoSight Ltd) overcomes these issues, because exosomes can be measured in a biologic buffer such as PBS, thus eliminating any shrinkage of the particles. Moreover, nanoparticle tracking analysis is an objective technique in which large numbers of individual particles are tracked, sized, and displayed in the analysis profile. It has also been shown by atomic force microscopy that light-scattering techniques are superior to electron microscopy in determining the size of particles in solution.26 We therefore conclude that we obtained good-quality exosome preparations.
We have demonstrated that Dll4 can be transferred from exosomes to cells after coincubation. Dll4 was internalized with the exosomes as visualized by immunocytochemistry, and the protein was found to be located intracellularly and at the cell membrane by both immunohistochemistry and biotin labeling. Cellular uptake of exosomes requires clathrin-mediated endocytosis, which has been reported to result in the transfer of cytosol and membrane from the exosome to the cell in the proper topology.27 This type of transfer has been reported previously with an oncogenic mutant form of epidermal growth factor receptor (EGFRvIII), which is incorporated into microvesicles of cultured U373 glioma cells that overexpress the EGFRvIII mutant. These vesicles can fuse with cells that lack the mutant receptor and confer a transformed phenotype.28 However, the glioblastoma cell line used in the present study does not express the mutant EGFR, and we isolated exosomes, not microvesicles, which may contain larger plasma membrane–derived particles.
We also demonstrated the transfer of Dll4 in vivo. We postulate that this transfer occurs via exosomes. Exosomes have been shown to interact with target cells to regulate cell-cell communication. This has been demonstrated in the immune system, where dendritic cells release exosomes that contain major histocompatibility complex to stimulate T-cell activation.18 Glioblastoma and ovarian tumor cells have also been shown to release exosomes that are capable of stimulating tube formation in vitro.29,30 These studies used microvesicles, which contain larger plasma membrane–derived particles, which may be responsible for the enhanced tube formation. We reported Dll4 expression in colon cancer tumor cells, so there is a potential for human cancers to give rise to Dll4-containing exosomes.31 A surprising finding was hDll4 staining on mouse RBCs in the U87 Dll4 tumors but not the controls. This will need further study, (eg, isolation of mouse RBC membranes) but does suggest that other cell types may take up exosomes.
We have demonstrated that Dll4-containing exosomes are also capable of effecting tube formation, increasing branching and overall vessel density, in an in vitro tube assay. An increase in vessel branching was also observed in vivo after injection of Dll4-containing exosomes into tumor xenografts. This is indicative of Notch inhibition, because enhanced tip cell formation and endothelial sprouting have been shown to occur when other inhibitors of Notch signaling are used, such as soluble Dll4-Fc, anti-Dll4 antibodies, or γ-secretase inhibitors.32,33 An increase in vessel length and lumen size was also observed in the present in vivo model. This phenotype has been described previously in our laboratory when Dll4 was overexpressed in the U87 model13 ; however, the vessel density was reduced because of Dll4 overexpression and increased Notch signaling.
We demonstrated that the Dll4-containing exosomes inhibited Notch signaling upon their uptake into endothelial cells, which could be responsible for the increased branching seen in vivo; however, we speculate that their eventual role is to promote a tip cell phenotype. During normal angiogenesis, a tip cell will promote the growth of new vessels and activate Notch signaling in neighboring cells, which may explain why the Dll4-containing exosomes also have the ability to form longer, larger vessels. Other angiogenic proteins have also been identified in the Dll4 exosomes and may contribute to these effects on tube formation; however, these proteins have not been reported to enhance branching and tip formation.
There are several potential mechanisms that may be involved in switching HUVECs to a tip cell phenotype. First, the Dll4-containing exosomes can reduce Notch signaling and reduce the expression of stalk cell markers. Dll4-containing exosomes may bind to the Notch receptor at the cell surface and compete with membrane-bound ligands. Soluble ligands have been reported to act in a similar manner and to block signaling induced by Notch trans ligands.6 However, we have strikingly demonstrated that Dll4-containing exosomes remove the Notch1 receptor from the cell surface. Because our confocal microscopy experiments demonstrated that the exosomes were internalized, this suggests that the Dll4 on the exosomes binds to the receptor at the cell surface and internalizes it with the rest of the exosome. The Notch receptor is ultimately degraded, as demonstrated by a total loss of cellular receptor. It may also be “trapping” newly synthesized Notch receptor in endosomal compartments before it reaches the cell surface, because intracellular interaction of Delta and Notch has been reported to reduce the ability of Notch to interact with Delta on neighboring cells.34
The concentration of Dll4 in the cell membrane may also be responsible in part for the inhibition of Notch signaling. Altering the ratio of DLL to Notch receptor has been shown to change the cell signaling response. Overexpression of DLL in Drosophila inhibits Notch signaling by making the signal-receiving cell refractory to the signal; conversely, reducing the level of DLL in the receiving cell makes the cell more receptive.35 DLLs are capable of inhibiting Notch signaling through interaction with Notch in the same cell.34 This cis-interaction may stimulate Notch endocytosis or stop Notch from reaching the cell surface; however, cell-surface expression of ligand has also been shown to be required for cis-inhibition.36,37 Notch-ligand interactions within the same cell may compete with trans-ligand interactions, because cells that overexpress Notch1 and Dll1 are unable to bind ligands on another cell.34 Increases of ligand at the cell surface may also promote ligand-ligand interactions and decrease available ligand for trans-activation of Notch, although ligand-ligand interactions are weaker than Notch-ligand interactions.38-40 None of these effects would be expected to produce the marked depletion of cell-surface Notch. The presence of Dll4 on the cell surface with low Notch signaling, however, is another tip cell–like phenotype.
If endothelial cells secrete exosomes that contain Dll4 in vivo, how could this affect surrounding cells? Dll4 is expressed on tip cells, and it controls tip/stalk cell differentiation and inhibits the angiogenic response of adjacent endothelial cells to vascular endothelial growth factor stimulation through Notch signaling.32 Dll4 on the tip cell activates Notch on an adjacent endothelial cell, which commits the cell to a stalk cell phenotype. Notch signaling in the stalk cell then up-regulates the expression of Dll4, which can activate Notch on the next cell, hence producing a line of Notch-signaling stalk cells. Evidence for this was described in the developmental retinal angiogenesis model reported by Hellstrom et al.41 In this system, Dll4 was expressed and seen to accumulate intracellularly in the tip cells and in many of the stalk cells immediately in contact with the tip cells. Further away from the tip, the intracellular Dll4 signal decreased. This intracellular accumulation of DLLs has been described previously and is thought to correlate well with Notch signaling7,42 ; however, this accumulation could also be an indication of exosome formation. Dll4-containing exosomes produced by these cells could diffuse and be taken up by endothelial cells in their immediate vicinity. We have demonstrated that uptake of Dll4-containing exosomes by endothelial cells results in the cells having a higher Dll4/Notch-receptor ratio and a lower Notch-signaling capacity. Dll4-containing exosomes are also capable of reducing the expression of the stalk cell markers Jagged-1 and vascular endothelial growth factor receptor-12 and increasing the level of filopodia. The exosomes, therefore, appear to be able to confer a tip cell phenotype on the receiving cells and may represent a mechanism by which stalk cell differentiation is restricted to allow tip cell formation to reoccur to produce an even network of vessels.
In summary, we present evidence that Dll4 is incorporated into the exosomes of endothelial cells and tumor cells. Dll4 can be transferred from one cell type to another by exosomes in vitro and in vivo. D114-containing exosomes increase angiogenesis by inhibition of Notch signaling and consequently confer a tip cell phenotype on endothelial cells. Thus, our results suggest a new mechanism of DLL/Notch signal regulation that does not require classic cell-cell contact.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Sandra Peak, Del Watling, Rachel Hayes, Sarah Scott, and Christine White (Cancer Research UK Biological Resources Unit) for assistance with xenograft experiments. Regeneron 242 anti–human Dll4 was a kind gift from Dr Gavin Thurston (Regeneron Pharmaceuticals).
H.S., H.T., P.T., R.C.A.S., J.-L.L., and A.L.H. are supported by Cancer Research UK. B.K. is supported by the NIHR Biomedical Research Center, Oxford, United Kingdom. C.E.O. is supported by the Ministry of Higher Education, Malaysia. I.S. was supported by the Oxford Partnership Comprehensive Biomedical Research Center with funding from the Department of Health's NIHR Biomedical Research Centres funding scheme. A Wellcome Trust technology development grant (No. GR087730) funded the NanoSight analyzer.
Wellcome Trust
Authorship
Contribution: H.S. designed and performed the research, analyzed the data, and wrote the paper; E.H., H.T., R.D., P.T., C.E.O., M.E., and B.K. performed research and analyzed data; R.L. and R.C.A.S. assisted in experimental design and data analysis; I.S. provided expertise in exosome analysis; J.-L.L. coordinated animal studies; and A.L.H. initiated and directed the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof A. L. Harris, Weatherall Institute of Molecular Medicine, University of Oxford, John Radcliffe Hospital, Oxford OX3 9DS, United Kingdom; e-mail: aharris.lab@imm.ox.ac.uk.