Abstract
The World Health Organization classification of acute myeloid leukemia (AML) is hierarchically structured and integrates genetics, data on patients' history, and multilineage dysplasia (MLD). The category “AML with myelodysplastic syndrome (MDS)–related changes” (AML-MRC) is separated from “AML not otherwise specified” (AML-NOS) by presence of MLD, MDS-related cytogenetics, or history of MDS or MDS/myeloproliferative neoplasm (MPN). We analyzed 408 adult patients categorized as AML-MRC or AML-NOS. Three-year event-free survival (EFS; median, 13.8 vs 16.0 months) and 3-year overall survival (OS; 45.8% vs 53.9%) did not differ significantly between patients with MLD versus without. However, MLD correlated with preexisting MDS (P < .001) and MDS-related cytogenetics (P = .035). Patients with MLD as sole AML-MRC criterion (AML-MLD-sole; n = 90) had less frequently FLT3 internal tandem duplication (P = .032) and lower median age than AML-NOS (n = 232). Contrarily, patients with AML-NOS combined with AML-MLD-sole (n = 323) had better 3-year EFS (16.9 vs 10.7 months; P = .005) and 3-year OS (55.8% vs 32.5%; P = .001) than patients with history of MDS or MDS/MPN or MDS-related cytogenetics (n = 85). Gene expression analysis showed distinct clusters for AML-MLD-sole combined with AML-NOS versus AML with MDS-related cytogenetics or MDS history. Thus, MLD alone showed no independent clinical effect, whereas cytogenetics and MDS history were prognostically relevant.
Introduction
In 2008, the World Health Organization (WHO) published its revised classification of acute myeloid leukemia (AML).1 By paying attention to the clinical relevance of cytogenetic alterations2-4 and to the molecular heterogeneity of this complex disorder,5,6 the first category of AML was assigned to patients with recurrent genetic alterations.1 CEBPA- and NPM1-mutated AML were acknowledged as separate provisional entities. Because of its prognostic importance, it was recommended to always define the FLT3 mutation status, particularly in cytogenetically normal AML.7
Thus, genetic alterations play a major role in the WHO classification, but morphologic, cytochemical, immunophenotypic, and clinical information are also required for classification.7 One of these morphologic criteria is multilineage dysplasia (MLD), referring to cases with dysplastic features in ≥ 2 hematopoietic lineages. According to the WHO, cases with ≥ 20% of bone marrow or peripheral blasts that do not fulfill the criteria of the categories “AML with recurrent genetic abnormalities” or “therapy-related AML (t-AML),” are classified as AML with myelodysplasia-related changes (AML-MRC) when at least one of the following prerequisites is fulfilled: a history of secondary AML arising from a previous MDS (myelodysplastic syndrome) or MDS/myeloproliferative neoplasm (MPN; with overlapping myeloproliferative features), AML with MDS-related cytogenetic abnormalities, and/or AML with MLD. If none of these aspects is fulfilled, cases are classified as “AML, not otherwise specified” (AML-NOS). AML with < 20% of myeloid blasts and > 50% of erythroid precursors (the former French-American-British subtype M6)8,9 are classified as AML-NOS, regardless of the presence of dysplastic features.1 Therefore, in the new WHO classification, dysplasia alone can be sufficient to assign a case to the AML-MRC category.
Some previous studies in AML considered dysplasia to be prognostically relevant,10-14 whereas others were not able to confirm an independent clinical relevance of MLD.15 Our former analysis in 614 patients with de novo AML confirmed cytogenetics, age, and high lactate dehydrogenase level as independent prognostic parameters, whereas the presence of dysplasia had no independent clinical effect.16 More recently, we investigated MLD for its prognostic effect in NPM1-mutated AML and, again, observed no independent influence on outcomes, whereas a mutated FLT3 status represented a prognostically adverse parameter.17
Thus, whether the presence of MLD has an independent prognostic meaning in AML or not, is still controversial,18,19 and it remains to be clarified whether the categorization of AML cases as myelodysplasia-related or “MRC” solely based on the presence of MLD is justified according to biologic and clinical aspects.
Here, we studied the clinical relevance of MLD20 in a cohort of 408 patients with AML at diagnosis who were corresponding to the AML-MRC or AML-NOS categories according to the WHO classification1 therewith excluding cases with the above-mentioned criteria such as recurrent genetic alterations or t-AML. Patients were characterized by a comprehensive diagnostic assessment, consisting of detailed cytomorphologic investigation, immunophenotyping, cytogenetics, molecular genetics, and gene expression profiling, to evaluate whether presence of MLD was associated with specific phenotype or genotype features.
Methods
Patients
Our retrospective study was based on 408 adult patients with AML diagnosed between August 2005 and February 2009 as AML-MRC or AML-NOS according to the WHO guidelines.1 All patients were referred to the MLL Munich Leukemia Laboratory for diagnosis of AML. Prerequisites to entry into this study were adequate material for morphologic assessment, complete cytogenetic and molecular genetic information, and availability of clinical data. Thus, 461 patients were assigned to the AML-MRC or AML-NOS categories according to the WHO. After in-depth morphologic analysis, 53 cases had to be excluded because dysplastic features were not evaluable following WHO guidelines due to insufficient numbers of evaluable cells in the respective hematopoietic lineages as a result of a very high degree of blasts percentage, insufficient smears not allowing adequate analysis, or hypocellularity. Thus, 408 AML patients with complete clinical and laboratory data were remaining. Patients with a classification as “AML with mutated NPM1” or “mutated CEBPA,” both representing provisional separate entities according to the WHO classification, were not excluded from analysis. The study was performed in accordance with the Declaration of Helsinki, and informed consent was obtained in all cases. The study was approved by the Institutional Review Board of Munich Leukemia Laboratory. A subgroup of patients (n = 208 of 408) had been included in our previous study on MLD in NPM1-mutated AML.17
Cytomorphology
Morphologic analysis of all 408 AML cases was independently performed by different investigators (M.M., K.M., and U.B.) and subsequently validated (T.H.). Bone marrow and peripheral blood smears were stained with the May-Grünwald-Giemsa method. Cytochemistry was done with myeloperoxidase (MPO) and nonspecific esterase.21 Dysplasia was assessed following the definitions of Goasguen et al10 and the WHO 2008 classification1 : dysgranulopoiesis was defined as ≥ 50% of ≥ 10 polymorphonuclear neutrophils (PMNs) being agranular or hypogranular, or with hyposegmented nuclei (pseudo Pelger-Huët anomaly). At least 25 cells were evaluated, but usually 100 cells were counted. MPO deficiency in the PMNs was defined as ≥ 50% MPO-negative cells in ≥ 10 PMNs after strong positivity of eosinophils or other PMNs was confirmed. Dyserythropoiesis was defined as ≥ 50% of dysplastic features in ≥ 25 erythroid precursors: megaloblastoid aspects, karyorrhexis, nuclear particles, or multinuclearity. Dysmegakaryopoiesis was diagnosed when ≥ 3 megakaryocytes or ≥ 50% in ≥ 6 cells showed dysplastic features such as microkaryocytes, multiple separated nuclei, or very large single nuclei. According to the WHO, MLD was defined by dysplastic features in ≥ 2 hematopoietic lineages. Trilineage dysplasia (TLD) was diagnosed when dysgranulopoiesis, dyserythropoiesis, and dysmegakaryopoiesis in combination were detectable.1
Cases that were evaluable in only 1 or 2 hematopoietic lineages (and, for example, one was called dysplastic and the other one nondysplastic) had to be excluded from analysis (n = 53). Because the WHO 2008 classification does not differentiate between MLD and TLD, we included cases in which 2 or more lineages were evaluable and both were dysplastic or both were not, because they still were evaluable for the presence of MLD.
Cytogenetic analysis
Chromosome banding analysis in combination with fluorescence in situ hybridization techniques was performed in all 408 patients according to standard procedures.22 Patients were categorized as having “AML with an MDS-related cytogenetic abnormality” according to WHO definitions: complex aberrant, −7/del(7q), −5/del(5q), isochromosome i(17q), −13/del(13q), del(11q), del(12p), t(12p) (ie, translocations involving 12p), del(9q), idic(X)(q13), t(11;16), t(3,21), t(1;3), t(2;11), t(5;7), t(5;17), t(5;10), t(3;5).1 For prognostic evaluation that was based on cytogenetics, all cases were assigned to the “favorable,” “intermediate,“ or “adverse” risk categories following the revised MRC (Medical Research Council) criteria according to Grimwade and Hills23 and Grimwade et al.24
Molecular mutation screening and immunophenotyping
In addition, polymerase chain reaction screening was done for the following molecular aberrations: NPM1 (nucleophosmin) mutations,25 FLT3-ITD (internal tandem duplications of the FLT3 gene),26 NRAS mutations,27 partial tandem duplications of the MLL gene (MLL-PTD),28 and CEBPA mutations.29 Immunophenotyping with multiparameter flow cytometry was performed as previously described.30
Gene expression profiling
Microarray analysis was performed on 96 cases with the use of HG-U133 Plus 2.0 microarrays (Affymetrix). The samples were prepared as previously reported.31,32 Gene expression raw data were processed according to the manufacturer's recommendations. After quality control, raw data were normalized with the robust multiple array average normalization algorithm as implemented in the R package affy version 1.22.0.33 For supervised statistical analyses, samples were grouped accordingly, and for each disease entity differentially expressed genes were calculated by t statistics. Resulting P values were adjusted for multiple testing.34 To visualize similarity of gene expression patterns, principal component analyses were applied. Transformed gene expression data were analyzed with Partek Genomics Suite Version 6.4 (Partek Inc). Microarray data can be downloaded at Gene Expression Omnibus (National Center for Biotechnology Information) under accession number GSE21261.35
Statistical analysis
Mean differences were analyzed with the t test. The χ2 test and Fisher exact test were applied in case of contingency tables. Overall survival (OS) was defined as the time from diagnosis to death or date of the last follow-up. Event-free survival (EFS) was defined as the time from diagnosis to date of failure (no complete remission [CR], relapse, death) or date of the last follow-up. The probabilities of OS and EFS were estimated with the Kaplan-Meier method. The log-rank test was used to compare risk factor categories in survival analysis. Cox proportional hazard regression models were performed to investigate treatment results and the risk factors affecting time to event. All tests were 2-sided, accepting P values of .05 or less as indicating a statistically significant difference. These P values were not adjusted for multiple testing. Statistical analyses were performed with SPSS software Version 14.0.1.
Results
Clinical characteristics and outcomes
Cases were equally distributed by sex (males, n = 212; 52.0%; females, n = 196; 48.0%). The median age was 69.8 years (range, 18.3-88.1 years); the median white blood cell (WBC) count was 8.9 × 109/L (range, 0.14-600.0 × 109/L). Median follow-up was 13.9 months (95% confidence interval [CI], 11.8-16.0 months).
Classification following the WHO classification of 2008
For classification of cases following the 2008 WHO classification,1 the following parameters had to be evaluated in all 408 cases: total blast count ≥ 20%, presence of MDS-related cytogenetic changes, a history of previous MDS (or MDS/MPN), and the presence of MLD. Taking these parameters into account, this resulted in 175 cases that were assigned to the category AML-MRC (42.9%) and the remaining 233 patients were categorized as AML-NOS (57.1%). Most cases qualified by one criterion for classification as AML-MRC: 90 cases (90 of 175; 51.4%) showed solely MLD (AML-MLD-sole), 25 cases (14.3%) had MDS-related cytogenetics, and 11 cases (6.3%) had a history of MDS (n = 9) or MDS/MPN (n = 2). The remaining 49 cases (28.0%) showed more than one criterion for the category AML-MRC: 44 cases (44 of 49; 89.8%) had a combination of MLD plus MDS-related cytogenetics or plus an MDS or MDS/MPN history, whereas 5 cases showed other combinations (Table 1).
Patients with AML-MRC: breakdown by WHO qualification criteria
| Parameter . | No of cases (%) . |
|---|---|
| MLD sole | 90 (51.4) |
| MDS related cytogenetics (MRC) sole | 25 (14.3) |
| History of MDS or MDS/MPN-sole | 11 (6.3) |
| MLD + preexisting MDS | 22 (12.6) |
| MLD + MDS-related cytogenetics | 22 (12.6) |
| MLD + MDS-related cytogenetics + preexisting MDS/MPN | 2 (1.1) |
| MDS related cytogenetics + preexisting MDS/ MPN | 3 (1.7) |
| Total | 175 (100.0) |
| Parameter . | No of cases (%) . |
|---|---|
| MLD sole | 90 (51.4) |
| MDS related cytogenetics (MRC) sole | 25 (14.3) |
| History of MDS or MDS/MPN-sole | 11 (6.3) |
| MLD + preexisting MDS | 22 (12.6) |
| MLD + MDS-related cytogenetics | 22 (12.6) |
| MLD + MDS-related cytogenetics + preexisting MDS/MPN | 2 (1.1) |
| MDS related cytogenetics + preexisting MDS/ MPN | 3 (1.7) |
| Total | 175 (100.0) |
Morphologic findings
Dysplasia in one hematopoietic lineage was seen in 130 of 408 cases (31.9%). In 110 cases (27.0%), we detected dysplasia in 2 cell lineages, whereas 36 of 408 cases (8.8%) had TLD. Thus, a total of 146 cases (35.8% from the whole cohort) had MLD.
Cytogenetics and molecular genetics
Cytogenetics was available in all 408 cases. According to the refined MRC criteria,23,24 most cases (n = 366; 89.7%) were assigned to the intermediate prognostic group which was due to the high proportion of normal karyotypes (n = 281; 68.9% of all cases), whereas 42 cases (10.3%) were classified as adverse risk. Cytogenetics of the cohort was detailed in Table 2. “Other” chromosomal abnormalities present were those different from deletions of the chromosomes 5 or 7, translocations, or complex karyotypes.4 Because patients from the category AML with recurrent genetic abnormalities according to the WHO1 had to be excluded from our study, there were no favorable risk cases (Table 2).
| Refined Medical Research Council criteria23,24 . | No. of cases (%) . |
|---|---|
| Intermediate risk group | 366 (89.7) |
| Normal karyotype | 281 (68.9) |
| Other non-complex | 85 (20.8) |
| Adverse risk group | 42 (10.3) |
| Complex alterations | 21 (5.1) |
| −7/add(7q) | 11 (2.7) |
| del(5q)/−5/add(5q) | 6 (1.5) |
| Others* | 4 (1.0) |
| Total* | 408 (100) |
| Refined Medical Research Council criteria23,24 . | No. of cases (%) . |
|---|---|
| Intermediate risk group | 366 (89.7) |
| Normal karyotype | 281 (68.9) |
| Other non-complex | 85 (20.8) |
| Adverse risk group | 42 (10.3) |
| Complex alterations | 21 (5.1) |
| −7/add(7q) | 11 (2.7) |
| del(5q)/−5/add(5q) | 6 (1.5) |
| Others* | 4 (1.0) |
| Total* | 408 (100) |
Cases with “others” were represented by deletions, other aberrations, and cases with independent clones as classified according to Medical Research Council criteria.
Molecular screening showed NPM1 mutations as the most frequent aberrations (203 of 397; 51.1%). This was followed by mutations of FLT3-ITD (89 of 398; 22.4%), NRAS (24 of 187; 12.8%), CEBPA (17 of 225; 7.6%), and MLL-PTD (24 of 399; 6.0%) of cases investigated, respectively (Table 3).
Incidence of molecular mutations in AML subgroups AML-NOS, AML-MRC with MDS-related cytogenetics and/or MDS/MPN history, and AML-MLD-sole
| Mutation (no. of patients analyzed*) . | Patients with mutation, n (%) . | AML-NOS n = 233 (%) . | AML-MRC due to cytogenetics and/or MDS or MDS/MPN history n = 85 (%) . | AML-MLD-sole n = 90 (%) . | P . |
|---|---|---|---|---|---|
| NPM1 (n = 397) | 203 (51.1) | 144 (62.3) | 10 (12.3) | 49 (57.6) | < .001 |
| FLT3-ITD (n = 398) | 89 (22.4) | 68 (29.4) | 6 (7.4) | 15 (17.4) | < .001 |
| NRAS (n = 187) | 24 (12.8) | 8 (8.7) | 8 (16.0) | 8 (17.8) | NS |
| MLL-PTD (n = 399) | 24 (6.0) | 10 (4.3) | 6 (7.4) | 8 (9.3) | NS |
| CEBPA (n = 225) | 17 (7.6) | 9 (7.1) | 3 (6.7) | 5 (9.3) | NS |
| Mutation (no. of patients analyzed*) . | Patients with mutation, n (%) . | AML-NOS n = 233 (%) . | AML-MRC due to cytogenetics and/or MDS or MDS/MPN history n = 85 (%) . | AML-MLD-sole n = 90 (%) . | P . |
|---|---|---|---|---|---|
| NPM1 (n = 397) | 203 (51.1) | 144 (62.3) | 10 (12.3) | 49 (57.6) | < .001 |
| FLT3-ITD (n = 398) | 89 (22.4) | 68 (29.4) | 6 (7.4) | 15 (17.4) | < .001 |
| NRAS (n = 187) | 24 (12.8) | 8 (8.7) | 8 (16.0) | 8 (17.8) | NS |
| MLL-PTD (n = 399) | 24 (6.0) | 10 (4.3) | 6 (7.4) | 8 (9.3) | NS |
| CEBPA (n = 225) | 17 (7.6) | 9 (7.1) | 3 (6.7) | 5 (9.3) | NS |
The P values result from the comparison of the frequency of molecular markers between the 3 cohorts (patients with AML-NOS, AML-MRC because of cytogenetics, and MDS history, or AML-MLD-sole).
NS indicates not significant.
From our cohort of 408 patients.
Immunophenotyping
Immunophenotyping was performed in 324 of 408 cases (155 AML cases with MLD and 169 cases without MLD, respectively).30 Although the expression of most antigens did not differ between both groups, human leukocyte antigen–DR (HLA-DR) showed lower levels of expression in cases with MLD (positive cells, 31.13% ± 1.71% vs 40.19% ± 1.79%; P < .001). Other antigens with weaker expression in MLD cases included CD117 (25.49% ± 1.92% vs 31.53% ± 1.75%; P = .023), CD135 (8.92% ± 1.27% vs 13.47% ± 1.53%; P = .023), CD38 (66.71% ± 2.01% vs 72.26% ± 1.57%; P = .029), and terminal deoxynucleotidyl transferase (4.28% ± 0.86% vs 8.43% ± 1.26%; P = .07), whereas lactoferrin was more strongly expressed in MLD cases (14.63% ± 1.36% vs 8.79% ± 0.85%; P < .001).
Comparison of different subgroups
To determine whether the different subgroups as defined earlier were clinically relevant, we performed comparisons of different parameters. These included biologic characteristics (age, WBC count), genetic features as assessed by chromosome banding/fluorescence in situ hybridization, molecular alterations, and the immunophenotype, as well as gene expression studies. Clinical outcomes were compared for OS and EFS.
Comparison of patients according to the presence and number of dysplastic cell lineages
We compared EFS and OS between the different subgroups as defined by the presence and number of dysplastic cell lineages. The median EFS did not differ significantly between the different cohorts with 18.4 months (95% CI, 14.1-22.7 months) in those without dysplasia, and 12.2 months (95% CI, 7.1-17.2 months), 11.6 months (95% CI, 6.2-16.9 months), and 17.4 months (95% CI, 10.3-24.5 months) in those with dysplasia present in 1, 2, or 3 hematopoietic lineages, respectively. Similar, OS did not differ significantly between these cohorts (Table 4).
Dysplastic features in 408 patients with AML
| Dysplastic lineages/parameter . | No. of cases (%) . | No. of cases . | MLD . | No. of cases (%) . | P for EFS . | P for OS . |
|---|---|---|---|---|---|---|
| No dysplasia | 132 (32.3) | No | 261 (64.0)* | .333 | .202 | |
| One dysplastic cell lineage | 129 (31.9) | No | ||||
| DysG | 34 | |||||
| DysE | 12 | |||||
| Dys M | 83 | |||||
| Two dysplastic cell lineages | 111 (30.0) | Yes | ||||
| DysG + DysE | 12 | 147 (36.0)† | ||||
| DysG + DysM | 79 | |||||
| DysE + DysM | 20 | |||||
| Trilineage dysplasia | 36 (8.8) | Yes | ||||
| Total | 408 (100) | 408 (100) |
| Dysplastic lineages/parameter . | No. of cases (%) . | No. of cases . | MLD . | No. of cases (%) . | P for EFS . | P for OS . |
|---|---|---|---|---|---|---|
| No dysplasia | 132 (32.3) | No | 261 (64.0)* | .333 | .202 | |
| One dysplastic cell lineage | 129 (31.9) | No | ||||
| DysG | 34 | |||||
| DysE | 12 | |||||
| Dys M | 83 | |||||
| Two dysplastic cell lineages | 111 (30.0) | Yes | ||||
| DysG + DysE | 12 | 147 (36.0)† | ||||
| DysG + DysM | 79 | |||||
| DysE + DysM | 20 | |||||
| Trilineage dysplasia | 36 (8.8) | Yes | ||||
| Total | 408 (100) | 408 (100) |
DysG indicates dysgranulopoiesis; DysE, dyserythropoiesis; DysM, dysmegakaryopoiesis; MLD, multilineage dysplasia; ie, 2 or 3 lineages show dysplastic features.
Includes no dysplasia and one dysplastic cell lineage.
Includes 2 dysplastic cell lineages and trilineage dysplasia.
MLD was seen in a higher frequency in AML cases with a history of MDS or MDS/MPN compared with others (28 of 42; 66.7% vs 119 of 366; 32.5%; P < .001). In addition, we observed a higher frequency of MLD in cases with MDS-related cytogenetic abnormalities compared with others (27 of 55, 49.1% vs 120 of 353; 34.0%; P = .035).
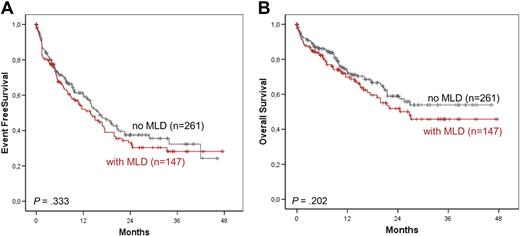
Outcomes were compared between patients with AML with MLD (MLD+; n = 147) versus patients without MLD (MLD−; n = 261). The median EFS (MLD+: 13.8, 95% CI, 9.4-18.2 months; MLD−: 16.0 months; 95% CI, 12.3-19.7 months) and 3-year OS (MLD+: 45.8%, MLD−: 53.9%) showed no significant differences (Figure 1A-B). We performed the same analysis in the subgroup of patients with normal karyotype. In addition, no differences in outcome of patients with AML with MLD (MLD+; n = 91) versus patients without MLD (MLD− n = 190) were observed. The median EFS (MLD+: 17.3, 95% CI, 12.7-21.8 months; MLD−: 18.4 months; 95% CI, 13.5-23.3 months; P = .333.) and 3-year-OS (MLD+: 59.2%, MLD−: 57.0; P = .202.) also did not differ.
Kaplan-Meier survival rates. EFS (A) and OS (B) in AML with and without MLD according to Kaplan-Meier.
Kaplan-Meier survival rates. EFS (A) and OS (B) in AML with and without MLD according to Kaplan-Meier.
Comparison of patients with AML-MRC solely because of MLD versus patients with AML-NOS
Next, we analyzed whether MLD as a sole criterion for the classification of cases as AML-MRC (AML-MLD-sole) was clinically relevant. Patients from this subgroup (n = 90) were characterized by ≥ 20% blasts, presence of MLD, but no history of MDS or MDS/MPN, and no evidence of MDS-related cytogenetic abnormalities. This subgroup was compared with the patients with AML-NOS (referring to cases with blast counts ≥ 20% or patients with > 50% of erythroid cells in combination with the presence of ≥ 20% of blasts of total bone marrow cells36 regardless of MLD presence, and to cases without the MRC criteria; n = 233).
The patients with AML-MLD-sole showed a significantly lower median age (62.8 years; range, 20.4-85.0 years) than those with AML-NOS (66.0 years) between both subgroups (AML-MLD-sole: 21.8 × 109/L; range, 0.4-370.0 × 109/L; AML-NOS: 11.4 × 109/L; range, 0.14-600.0 × 109/L; P = .279).
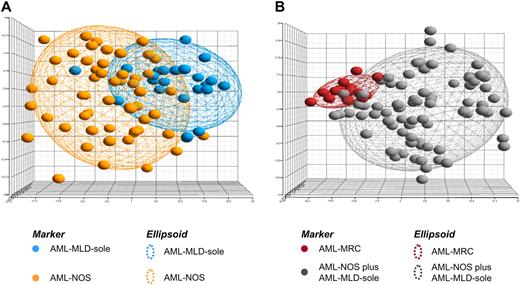
In gene expression analysis, 24 cases with AML-MLD-sole were compared with 56 cases with AML-NOS. With the use of the top 500 genes sorted according to the t statistic, no significant differences were observed according to the underlying expression profile for these 2 groups. As observed in a principal component analysis, the patients largely intercalated with each other (Figure 2A; supplemental data, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), meaning that there were no clear expression signatures to distinguish between AML-MLD-sole and AML-NOS.
Microarray analysis. In this supervised principal component analysis (PCA) each patient is represented by a colored sphere. Ellipsoids are drawn with 2-fold standard deviations. (A) The gene expression signature is given for the top 500 probe sets differentially detected between 24 cases with AML-MLD-sole and 56 cases displaying AML-NOS. (B) The gene expression signature is given for the top 500 probe sets differentially expressed between 80 cases AML-NOS plus AML-MLD-sole and 16 AML-MRC cases on the basis of cytogenetics or a MDS history. Detailed information on the significantly differentially expressed probe sets and their functional annotation is available in supplemental data.
Microarray analysis. In this supervised principal component analysis (PCA) each patient is represented by a colored sphere. Ellipsoids are drawn with 2-fold standard deviations. (A) The gene expression signature is given for the top 500 probe sets differentially detected between 24 cases with AML-MLD-sole and 56 cases displaying AML-NOS. (B) The gene expression signature is given for the top 500 probe sets differentially expressed between 80 cases AML-NOS plus AML-MLD-sole and 16 AML-MRC cases on the basis of cytogenetics or a MDS history. Detailed information on the significantly differentially expressed probe sets and their functional annotation is available in supplemental data.
For molecular mutations, the AML-MLD-sole cohort exhibited an equal incidence of NPM1 mutations (49 of 203, 57.6%; vs 144 of 203, 62.3%, NS), whereas there was a significantly lower frequency of FLT3-ITD (15 of 89, 17.4% vs 68 of 89, 29.4%; P = .032) than in those with AML-NOS. NRAS mutations were slightly more frequent in the AML-MLD-sole cohort than in the AML-NOS cohort (8 of 24, 17.8% vs 8 of 24 8.7%; NS). MLL-PTD and CEBPA mutations were similarly distributed (Table 3).
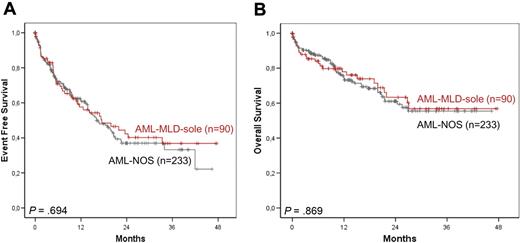
Median EFS did not differ significantly between the AML-MLD-sole and AML-NOS subgroups (17.5 months, 95% CI, 7.8-27.2 months vs 16.1 months, 95% CI, 11.7-20.6 months) or did 3-year OS (56.8% vs 55.4%); indeed, survival curves were overlapping (Figure 3A-B). Thus, the only parameter differing significantly between both cohorts was the frequency of FLT3-ITD mutations, whereas biologic characteristics and clinical outcomes showed no significant differences.
Survival rates of AML-MRC solely because of MLD. EFS (A) and OS (B) of AML-MLD-sole versus AML-NOS.
Survival rates of AML-MRC solely because of MLD. EFS (A) and OS (B) of AML-MLD-sole versus AML-NOS.
Comparison of patients with AML-MRC because of cytogenetics or because of MDS or MDS/MPN history with the remaining patients of the total cohort
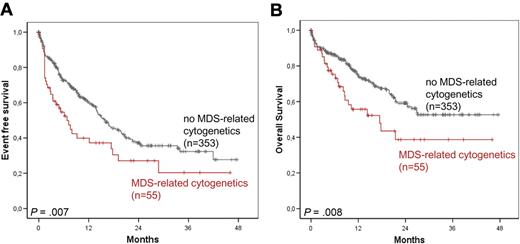
Subsequently, patients with MDS-related cytogenetic abnormalities (n = 55) and those with AML arising from previous MDS or MDS/MPN (n = 42) were each compared with the respective remaining patients of the total cohort (n = 353 and n = 366, respectively). Here, the presence of MDS-related cytogenetics was associated with shorter median EFS (6.8 vs 16.0 months; P = .007) and OS (17.4 months vs not reached; P = .008; Figure 4A-B). In addition, MDS or MDS/MPN history was associated with shorter median EFS (12.7 vs 16.6 months; P = .075) and OS (15.0 months vs not reached; P = .018, respectively) compared with all others.
Survival rates in AML with MDS-related cytogenetics. EFS (A) and OS (B) in AML with MDS-related cytogenetics versus without MDS-related cytogenetics.
Survival rates in AML with MDS-related cytogenetics. EFS (A) and OS (B) in AML with MDS-related cytogenetics versus without MDS-related cytogenetics.
Comparison of patients with AML-MRC because of cytogenetics or MDS or MDS/MPN history to patients classified as AML-MRC only because of the presence of MLD
Next, we combined patients with AML-MRC because of cytogenetics or MDS or MDS/MPN history to one cohort (n = 85). Compared with the subgroup of AML-MLD-sole (n = 90; ie, those with MLD but without MDS-related cytogenetic changes and without MDS or MDS/MPN history), the cohort with the characteristic cytogenetics or MDS or MDS/MPN history had significantly higher age (median, 69.8 vs 62.8 years; P = .002) and lower WBC count (median, 6.3 vs 21.8 × 109/L; P = .001). They further had a lower frequency of NPM1 mutations (10 of 81; 12.3% vs 49 of 85; 57.6%; P < .001). No significant differences were detected for all other mutations. Median EFS was significantly worse in the subgroup of AML-MRC because of cytogenetics or MDS or MDS/MPN history compared with the AML-MLD-sole group (10.7 months vs 17.5 months; P = .019); the same effect was seen for median OS (16.8 months vs not reached; P = .009; Figure 5A-B).
Survival rates with the presence of MLD. EFS (A) and OS (B) in AML-MRC because of cytogenetics or MDS or MDS/MPN history versus AML-MRC-sole because of the presence of MLD.
Survival rates with the presence of MLD. EFS (A) and OS (B) in AML-MRC because of cytogenetics or MDS or MDS/MPN history versus AML-MRC-sole because of the presence of MLD.
Comparison of patients with AML-MRC because of cytogenetics or MDS or MDS/MPN history to patients classified as AML-NOS
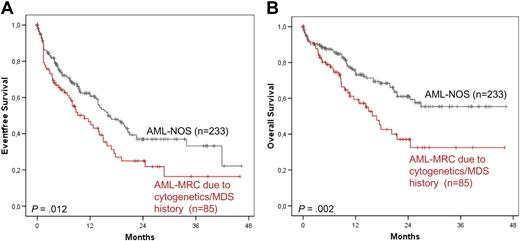
The comparison of the above-established new cohort with AML-MRC because of cytogenetics or MDS or MDS/MPN history (n = 85) to the AML-NOS cohort showed a trend to higher age (median, 69.8 vs 66.0; P = .049) and significantly lower WBC count (median, 6.3 vs 11.4 × 109/L; P < .001). For molecular markers, we found a significantly lower frequency of NPM1 mutations and FLT3-ITD in this group compared with the MLD-NOS group (P < .001). All other molecular markers did not differ significantly. Patients with AML-MRC because of cytogenetics or MDS or MDS/MPN history had both a worse EFS and OS (10.7 months vs 16.1 months; P = .012; 16.8 months vs not reached, 0.002, respectively) compared with the patients in the AML-NOS group (Figure 6A-B).
Survival rates compared with AML-NOS. EFS (A) and OS (B) in AML-MRC because of cytogenetics or MDS or MDS/MPN history versus AML-NOS.
Survival rates compared with AML-NOS. EFS (A) and OS (B) in AML-MRC because of cytogenetics or MDS or MDS/MPN history versus AML-NOS.
Comparison of patients with AML-NOS combined with patients with MLD as sole MRC criterion to AML-MRC because of cytogenetics or MDS or MDS/MPN history
As explained earlier, we had not been able to define any clinical and prognostic differences between patients with AML-MLD-sole and patients with AML-NOS, but we observed significant differences when both are combined and then compared with the cohort resulting from the combination of patients with MDS-related cytogenetics and MDS or MDS/MPN history. Therefore, we established a novel cohort (n = 323) that was composed of patients with AML-NOS (n = 233) and with AML-MLD-sole of the MRC group (n = 90). We tested this newly defined group against all remaining MRC cases (n = 85) being composed of AML-MRC according to myelodysplasia-related cytogenetics (n = 47) and patients with a preceding MDS or MDS/MPN (n = 33) or both criteria in combination (n = 5).
First, significant differences were observed with respect to age and WBC count. Median age of the new cohort composed of AML-NOS plus AML-MLD-sole was significantly lower (65.6 years; range, 18.3-88.1 years) compared with AML-MRC according to cytogenetics or an MDS or MDS/MPN history (69.8 years; range, 28.5-85.1 years; P = .011). Median WBC count was 13.8 × 109/L (range, 0.14-600.0 × 109/L) in the AML-NOS plus AML-MLD-sole group, being significantly higher compared with the second cohort (6.3 × 109/L; range, 0.8-11.5 × 109/L; P < .001).
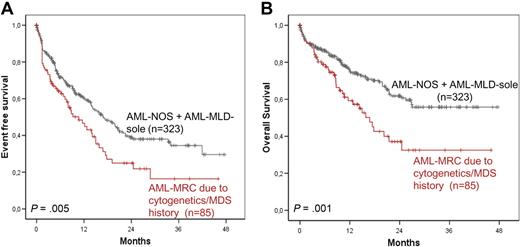
Median EFS was significantly better in the AML-NOS plus AML-MLD-sole group compared with the second cohort (16.9 months vs 10.7 months; P = .005); the same effect was seen for median OS (not reached vs 16.8 months; P = .001; Figure 7A-B).
Survival rates compared with AML-NOS combined with AML-MLD-sole. EFS (A) and OS (B) in AML-MRC because of cytogenetics or MDS or MDS/MPN history versus AML-NOS combined with AML-MLD-sole.
Survival rates compared with AML-NOS combined with AML-MLD-sole. EFS (A) and OS (B) in AML-MRC because of cytogenetics or MDS or MDS/MPN history versus AML-NOS combined with AML-MLD-sole.
In gene expression analysis, 80 cases of the combined group AML-NOS plus AML-MLD-sole were compared against 16 AML-MRC cases on the basis of cytogenetics or a MDS history or both. According to the t statistic and adjusted for multiple testing, the underlying expression profiles for these 2 groups showed significant differences (supplemental Spreadsheet). Although we had not been able to define any significant differences of gene expression patterns of patients with AML-MLD-sole versus AML-NOS (Figure 2A), these newly defined groups (AML-NOS + AML-MLD-sole vs AML-MRC because of cytogenetics/MDS history) formed 2 separate clusters on the basis of statistically significant differential gene expression (Figure 2B). Genes with higher expression in AML-MRC cases were involved in cellular processes such as regulation of transcription (PSIP1, SOX15, LYL1), signal transduction (KIT, JAK3, SYDE1, TIGD5), or chromatin modification (HDAC7). Genes with lower expression in AML-MRC included genes with known relevance in cell development, DNA-dependent regulation of transcription, and nuclear mRNA splicing (HDAC8, POU4F2, HIPK2, SFRS11, MEIS1, HOXA1, HOXA5, HOXB6, HOXB7, NKX3-1). Additional information on the top 500 significantly differentially expressed probe sets and their functional annotation is available in supplemental data.
Analysis of prognostic factors
Univariate analysis was performed for WBC count, sex, age, presence of MLD, history of MDS or MDS/MPN, MDS-related cytogenetics, and mutations of NPM1, FLT3-ITD, CEBPA, MLL-PTD, and NRAS to evaluate the prognostic significance of these distinct risk parameters. Among the above-mentioned parameters only higher age, a history of MDS or MDS/MPN, the presence of MDS-related cytogenetics, and the absence of an NPM1 mutation were associated with significantly worse EFS. A significantly worse OS in univariate analysis was only seen for cases with higher age, the presence of MDS-related cytogenetics, and cases without an NPM1 mutation. In multivariate analysis, higher age was the only parameter independently related to worse EFS (P < .001) and OS (P < .001), whereas absence of an NPM1 mutation only was associated with a significantly shorter OS (P = .037; Table 5).
Univariate and multivariate analysis of risk factors in study cohort
| Parameter . | P for EFS . | P for OS . | ||
|---|---|---|---|---|
| Univariate . | Multivariate . | Univariate . | Multivariate . | |
| WBC count | .674 | .789 | ||
| Sex | .746 | .548 | ||
| Age | < .001 | < .001 | < .001 | < .001 |
| Presence of MLD | .334 | .204 | ||
| History of MDS or MDS/MPN | .078 | .020 | .592 | |
| MDS-related cytogenetic changes | .008 | .136 | .009 | .362 |
| NPM1 mutated | .005 | .110 | < .001 | .037 |
| FLT3-ITD mutated | .457 | .410 | ||
| CEBPA mutated | .853 | .650 | ||
| MLL-PTD mutated | .094 | .071 | ||
| NRAS mutated | .807 | .759 | ||
| Parameter . | P for EFS . | P for OS . | ||
|---|---|---|---|---|
| Univariate . | Multivariate . | Univariate . | Multivariate . | |
| WBC count | .674 | .789 | ||
| Sex | .746 | .548 | ||
| Age | < .001 | < .001 | < .001 | < .001 |
| Presence of MLD | .334 | .204 | ||
| History of MDS or MDS/MPN | .078 | .020 | .592 | |
| MDS-related cytogenetic changes | .008 | .136 | .009 | .362 |
| NPM1 mutated | .005 | .110 | < .001 | .037 |
| FLT3-ITD mutated | .457 | .410 | ||
| CEBPA mutated | .853 | .650 | ||
| MLL-PTD mutated | .094 | .071 | ||
| NRAS mutated | .807 | .759 | ||
EFS indicates event-free survival; OS, overall survival; WBC, white blood cell; MLD, multilineage dysplasia; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasm; ITD, internal tandem duplication; and PTD, partial tandem duplication.
Discussion
It was one major achievement of the WHO classification of 2001 to consider genetic features as a main category for the classification of AML. In addition, the WHO focused on morphologic, immunophenotypic, and other clinical data. MLD was introduced to encompass AML cases with MDS-like features, including unfavorable cytogenetic abnormalities and an unfavorable response to therapy.7 On the basis of the difficulties to define whether MLD has independent prognostic significance in AML13,14 or not,15,16 this subgroup was renamed in the updated WHO classification of 2008 as AML-MRC, and the criteria were expanded to include a history of MDS or MDS/MPN or specific cytogenetic findings.1,7
However, the categorization of AML cases solely on the basis of dysplastic features is still being controversially discussed: Wandt et al15 performed an investigation of 1766 patients with de novo, secondary AML, and t-AML (excluding patients with acute promyelocytic leukemia) and showed a significant association of MLD with unfavorable cytogenetic profiles. Moreover, the presence of MLD was negatively correlated with the achievement of complete remission in univariate analysis. Nevertheless, in multivariate analysis, age, cytogenetics, and the NPM1 mutant/FLT3-ITD status retained prognostic significance, whereas MLD had no independent prognostic power.15 Similarly, we had previously analyzed an independent series of 614 patients with de novo AML recruited between 1992 and 1999 for the prognostic implications of dysplasia. TLD correlated with unfavorable cytogenetics, but the presence or absence of dysplasia failed to show independent prognostic relevance. In multivariate analysis, only cytogenetics, age, and elevated lactate dehydrogenase had an independent prognostic effect.16 In the analysis of Yanada et al,37 including 109 adult AML cases, significant differences were observed between outcomes of patients with recurrent genetic abnormalities, AML with MLD, t-AML, and AML-NOS according to the WHO (2001) criteria. Cytogenetic risk and patient age maintained their prognostic value in multivariate analysis, whereas the prognostic significance of MLD and prior therapy was lost after adjusting for cytogenetic risk and age.37 More recently, we investigated the prognostic meaning of MLD in a cohort of 318 patients all with NPM1 mutated AML. Again, we were not able to determine any independent influence of MLD for outcomes in this distinct subgroup, whereas the FLT3-ITD status remained the sole prognostically relevant parameter in multivariate analysis.17 Thus, these studies led to the conclusion that dysplasia showed correlations with other adverse parameters, such as unfavorable karyotypes, but had no independent effect on outcomes when other clinically relevant parameters were included in multivariate analyses.
Others, in contrast, were able to determine a significant adverse effect of morphologic features in AML on clinical outcomes: The morphologic and histopathologic studies from Arber et al13 in 300 patients with AML with ≥ 20% of bone marrow myeloblasts (AML and refractory anemia with excess blasts in transformation) showed significantly worse survival in patients with MLD, whereas there were no significant differences in prognosis between AML arising from MDS and de novo AML with MLD. However, control cases in the Arber et al13 study were more heterogeneous (including recurrent cytogenetic aberrations) compared with the present study in which control cases were recruited from AML-NOS and AML-MRC strictly in accordance with the WHO classification.1 Earlier, Goasguen et al10 suggested an association of dysgranulopoiesis with lower complete remission rates in de novo AML, whereas Gahn et al11 reported an unfavorable prognostic effect of dysplastic features in AML, at least in patients with favorable or intermediate cytogenetic risk profile. Very recently, Weinberg et al14 evaluated the clinical, pathologic, cytogenetic, and molecular features in a relatively small cohort of 100 patients with AML with the use of 2008 WHO criteria and described significantly worse survival outcomes and CR rates for patients from the AML-MRC cohort (n = 48) than for patients with AML-NOS.14 According to the investigators, these results gave support to the previously observed clinical significance of MLD when strictly defined by WHO 2008 criteria.1,7,14
At this time the reasons for these diverging results,14 which are in contrast to the studies from Wandt et al,15 Yanada et al,37 and our own previous works16,17 (all failing to show an independent prognostic effect for dysplasia in AML) remain unclear. We aimed at clarifying whether the recent WHO category AML with MRC if based solely on the criterion of MLD was justified from biologic and clinical aspects. Therefore, we followed an algorithm that was based on the WHO classification from 20081 and first excluded from our analysis patients with recurrent genetic alterations or t-AML and were then investigating a study cohort of 408 AML cases. According to the WHO 2008 definitions, these were assigned to the AML-MRC category because of MLD, or MDS or MDS/MPN history or myelodysplasia-related cytogenetics either as sole criterion or in combinations. All remaining patients were grouped to the AML-NOS category.
Because we were not able to determine any prognostic effect of the presence of MLD, we compared patients who were categorized as AML-MRC solely based on MLD with patients with AML-NOS: no significant differences about the biologic profile (age, WBC count), and the clinical outcomes in terms of OS and EFS were detectable. Also with gene expression profiling, no significant differences were found between both patient cohorts. The only significant difference was a lower frequency of FLT3-ITD in the cohort being classified as AML-MRC solely on the basis of the presence of MLD (P = .032), but this does not translate into clinical different behavior.
Our results were in contrast to the results of 2 previous studies (Weinberg et al14 and Gahn et al11 ). In detail, Weinberg et al14 performed separation of 100 patients with AML according to WHO 2008 criteria: The AML-MRC cohort (n = 48) was based on the presence of MLD (n = 41), or MDS-related cytogenetic abnormalities (n = 14), or MDS history (n = 16), whereas 3 patients had therapy-related myeloid neoplasms. compared with the patients with AML-NOS (n = 39), the patients with AML-MRC showed significantly worse OS, PFS, and lower CR rate. In a multivariate analysis, the cytogenetic risk group (P = .001), age (P = .037), evidence of FLT3-ITD (P = .047), and the AML-MRC category (P = .041) had an independent prognostic effect. However, Weinberg et al14 did not consider the presence of MLD separately in statistical analysis but focused only on the more comprehensive AML-MRC category. Therefore, it is difficult to draw final conclusions whether the presence of MLD alone would have sustained an independent prognostically relevant position in multivariate analysis in a larger AML-MRC cohort.
Earlier, Gahn et al11 analyzed a total of 102 patients with newly diagnosed AML for a potential clinical effect of dysplasia. Patients with dysgranulopoiesis had significantly shorter EFS than those without (P = .025) and a CR rate of 56% versus 74% (P = .04). In patients with unfavorable karyotypes, the presence or absence of dysgranulopoiesis had no additional prognostic impact, whereas those with favorable karyotypes and evidence of dysgranulopoiesis experienced a significantly lower continuous complete remission rate compared with those without (P = .03). Of note, evidence of dysplastic features in megakaryopoiesis or erythropoiesis had no significant effect on remission rates. The presence of TLD was correlated with a nonsignificant trend to lower CR rate only (55% in patients with TLD vs 65% in those without), whereas MLD was not considered in this analysis. Apparently, multivariate analysis had not been performed in the study from Gahn et al,11 which renders a direct comparison with our data difficult.
Because we here have been unable to find significant differences between patients with AML-MRC solely based on MLD and patients with AML-NOS, we subsequently combined patients from the AML-NOS and the AML-MRC solely because of MLD categories and compared them to the patients with an MDS history or myelodysplasia-related cytogenetics. Comparison of these newly established cohorts showed significant differences for age (P = .011) and WBC count (P < .001) and found as well different EFS (P = .005) and OS (P = .001). Significant differences were additionally shown by gene expression profiling, because distinct clusters were observed for combined patients with AML-MLD-sole with AML-NOS compared with those with MDS-related cytogenetics or MDS or MDS/MPN history. Finally, patients with an MDS history or with MDS-related cytogenetic abnormalities showed significantly worse EFS compared with the other remaining cases of the total cohort.
Thus, although we were not able to determine a significant effect of the presence of MLD as sole criterion for the AML-MRC category, myelodysplasia-related cytogenetics and a history of previous MDS or MDS/MPN were biologically and prognostically highly relevant. These results suggest that separate categories AML with MRC solely based on MLD and AML-NOS has no fundamental underlying biologic basis. Instead, we would like to suggest restricting the AML-MRC category to cases with a history of MDS or MDS/MPN or myelodysplasia-related cytogenetics completely irrespective of MLD findings, whereas cases solely being defined by morphologic criteria (MLD) should be combined with the AML-NOS category. As a consequence, this would result in only 2 distinct groups and avoid the interobserver variability of grading dysplastic features. This will finally represent the differences in the biology and prognosis of these AML cohorts and therefore would improve the reproducibility of risk stratification in the setting of clinical trials. We would like to suggest considering these aspects for further evaluations and implement data in a revised WHO classification of AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank all coworkers in our laboratory for their excellent technical assistance as well as all physicians for referring sample material to our center. We list the centers and investigators (contributing 5 or more cases) in order of the number of cases provided: Klinikum Augsburg (D. Oruzio), St Johannes Krankenhaus, Dortmund (H. Pielken), St Antonius Krankenhaus, Eschweiler (P. Staib, F. Schlegel), Krankenhaus Düren (M. Flaßhove), Klinikum Bremen-Mitte (B. Hertenstein), Knappschaftskrankenhaus Bottrop (G. Trenn), Städtisches Krankenhaus Gütersloh (G. Massenkeil), Katholisches Krankenhaus, Hagen (H.-W. Lindemann), Vivantes Klinikum Neukölln, Berlin (M. de Wit), Universitätsklinikum Mannheim (W.-K. Hofmann, E. Lengfelder), Klinikum Fulda (H.-G. Höffkes), Paracelsus Klinik, Osnabrück (O. Koch), Städtisches Klinikum Kassel (B. Ritter, M. Wolf), Franziskus Hospital, Bielefeld (H.-J. Weh), Klinikum Krefeld (Th. Freiling), Kreiskrankenhaus Gummersbach (M. Sieber), Universitätsklinikum Regensburg (A. Reichle), Asklepios Klinik Altona, Hamburg (D. Braumann), Evangelisches Krankenhaus Hamm (J. Schubert), Gemeinschaftspraxis Drs. med. A. Bethge, H.-W. Tessen, Goslar, Harzkliniken/Dr Herbert Nieper Krankenhaus, Goslar (A. Hoyer), Kliniken der Stadt Köln (A. Dormann), Onkologisches Schwerpunktpraxis Leer (L. Müller), and Praxis Dr S. Prenniger (Passau).
Authorship
Contribution: M.M., U.B., and T.W. analyzed the data and wrote the manuscript; M.M., U.B., and K.M. did morphologic analysis; T.W. collected clinical data and contributed to statistics and figures; C.H. was responsible for chromosome banding analysis and fluorescence in situ hybridization; S.S. was responsible for molecular analysis; W.K. was responsible for immunophenotyping and was involved in statistical analysis; A.K. performed GEP studies; biomathematics analyses were done by H.-U.K. and M.D.; and T.H. was the principal investigator of the study and validated cytomorphologic analysis and clinical data. All authors contributed to write this manuscript and agreed with the final version before submission.
Conflict-of-interest disclosure: C.H., W.K., S.S., and T.H. are part owners of the Munich Leukemia Laboratory (MLL). M.M., T.W., K.M., and A.K. are employed by MLL. The remaining authors declare no competing financial interests.
Correspondence: Torsten Haferlach, MLL Munich Leukemia Laboratory, Max-Lebsche-Platz 31, 81377 Munich, Germany; e-mail: torsten.haferlach@mll-online.com.