Abstract
To identify cytogenetic risk factors predicting outcome in children with advanced myelodysplastic syndrome, overall survival of 192 children prospectively enrolled in European Working Group of Myelodysplastic Syndrome in Childhood studies was evaluated with regard to karyotypic complexity. Structurally complex constitutes a new definition of complex karyotype characterized by more than or equal to 3 chromosomal aberrations, including at least one structural aberration. Five-year overall survival in patients with more than or equal to 3 clonal aberrations, which were not structurally complex, did not differ from that observed in patients with normal karyotype. Cox regression analysis revealed the presence of a monosomal and structurally complex karyotype to be strongly associated with poor prognosis (hazard ratio = 4.6, P < .01). Notably, a structurally complex karyotype without a monosomy was associated with a very short 2-year overall survival probability of only 14% (hazard ratio = 14.5; P < .01). The presence of a structurally complex karyotype was the strongest independent prognostic marker predicting poor outcome in children with advanced myelodysplastic syndrome.
Introduction
Karyotypic complexity has been reported to be associated with a poor prognosis in myeloid neoplasia.1-3 However, the definition of a complex karyotype remains a matter of debate. Most studies defined a complex karyotype as more than or equal to 3 independent abnormalities.2,4-6 In the Medical Research Council Acute Myeloid Leukemia 10 (MRC AML10) trial, more than or equal to 5 independent abnormalities were required3 because acute myeloid leukemia (AML) patients with more than or equal to 5 abnormalities had a significantly worse 5-year overall survival (OS) than those with 3 or 4 abnormalities.1 Increased numbers of chromosomal abnormalities were also found to adversely influence median survival in adult myelodysplastic syndrome (MDS) patients.7 Breems et al recently investigated the prognostic value of different cytogenetic components of a complex karyotype in adult AML and identified a monosomal karyotype (ie, either one single autosomal monosomy in the presence of at least one structural aberration or at least 2 distinct autosomal monosomies) as a highly unfavorable risk category.8 To better stratify children with advanced MDS, we evaluated the outcome in 192 children prospectively diagnosed and treated within the studies of the European Working Group of MDS in Childhood (EWOG-MDS) related to karyotypic complexity. This study is registered at www.clinicaltrials.gov as #NCT00047268 and #NCT00662090.
Methods
All patients younger than 18 years with adequate cytogenetic studies and advanced MDS (ie, refractory anemia with excess blasts or refractory anemia with excess blasts in transformation),9,10 enrolled in studies EWOG-MDS 98 (www.clinicaltrials.gov; #NCT00047268) or EWOG-MDS 2006 (#NCT00662090) between July 1, 1998 and June 30, 2008 were included in this analysis. In both studies, therapy recommendation consisted of upfront hematopoietic stem cell transplantation (HSCT). Institutional review board approval was obtained for both EWOG studies from all participating institutions.
Cytogenetic analyses of bone marrow cells were performed according to standard procedures.11 Karyotypes were described according to International System for Human Cytogenetic Nomenclature 2009.12 At least 10 metaphases were analyzed (supplemental Tables 1-3, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Cytogenetic findings were centrally reviewed (G.G., H.B.B., D.B.). All cytogenetic studies were performed before administration of MDS-specific therapy. A structurally complex karyotype was defined as more than or equal to 3 chromosomal aberrations, including at least one structural aberration (supplemental Table 4). Because we have never been able to detect differences in OS between patients with monosomy 7 as sole aberration, monsomy 7 and clonal evolution, or monosomy 7 and other aberrations (excluding those with structurally complex karyotypes, supplemental Table 5), we grouped all patients with monosomy 7. Thus, the monosomy 7 group included those with clonal evolution and additional aberrations.
The Kaplan-Meier method was used to estimate OS probabilities.13 The log-rank test was used to compare survival between subgroups. For multivariate analysis, the Cox proportional hazard regression model was used.14 HSCT was included as a time-dependent covariate in the model. The different definitions of complex karyotype coded in a variable with k-categories were transformed into k-1 dummy variables and added to the model.15 All P values were 2-sided, and values less than .05 were considered statistically significant. Statistical analysis was performed using SPSS for Windows Version 15.0.1.
Results and discussion
Cytogenetic data from 192 patients with refractory anemia with excess blasts and refractory anemia with excess blasts in transformation were analyzed (supplemental Table 3). HSCT was performed in 143 patients (74%) resulting in a probability of OS at 5 years of 0.58 for all children with primary MDS and 0.47 for those with secondary MDS (P = .09). Here we analyze the prognostic significance of a novel simple definition of a complex karyotype termed structurally complex (ie, ≥ 3 chromosomal aberrations, including at least one structural aberration), introduced for the following reasons: The most frequent chromosomal aberrations of highly complex clones are deletions, unbalanced translocations and dicentric chromosomes. Monosomies are also often noted in karyotypes with multiple aberrations. However, it may be difficult to be certain that a particular chromosome is lost. Using more advanced molecular cytogenetic techniques, such as fluorescence in situ hybridization or array comparative genomic hybridization, it often transpires that parts of “missing” chromosomes are involved in structural aberrations. In addition, structural aberrations, such as deletion 5q, deletion 7q, and deletion 17p, or aberrations, involving 3q, 11q, and 12p, are known to be associated with a poor prognosis in AML and MDS.1,16,17
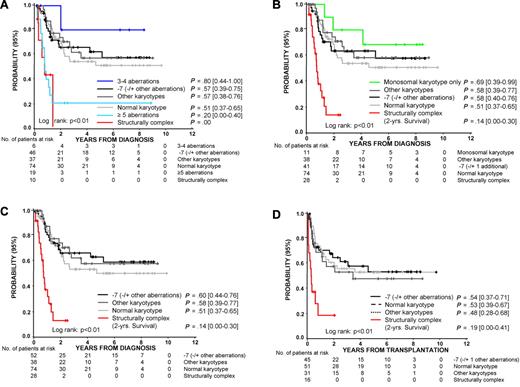
The number of patients identified by the traditional definition of complex karyotype with more than or equal to 3 clonal aberrations, complex karyotype with more than or equal to 5 clonal aberrations, and structurally complex karyotype was 35 (18%), 19 (10%), and 28 (15%), respectively (supplemental Table 6). With the exception of one patient, all patients with more than or equal to 5 clonal aberrations were also recognized as structurally complex. Seven patients with more than or equal to 3 clonal aberrations did not fulfill the criteria of a structurally complex karyotype, resulting from the absence of structural aberrations by standard cytogenetics. Notably, 5 of these 7 patients are still alive at a median time of 6.9 years (range, 0.2-8.7 years) after diagnosis (Figure 1A). Children with a structurally complex karyotype had a highly unfavorable prognosis with a 2-year OS probability of 0.14 (range, 0.00-0.30; Figure 1B-C). The group of patients with a structurally complex karyotype given HSCT had a significantly shorter event-free survival (P < .01, Figure 1D) and OS (data not shown) than children with other karyotypes resulting from a higher risk of relapse after HSCT (0.48, range, 0.27-0.82 vs 0.28, 0.20-0.38, P < .01). Whether novel therapy approaches before HSCT can improve outcome after HSCT for children with structurally complex karyotype remains to be determined.
Probabilities of 5-year survival according to cytogenetic stratifications. (A) Probability of 5-year OS for children with advanced primary or secondary MDS according to cytogenetic stratification. Traditionally, a complex karyotype is defined by the presence of more than or equal to 3 or more than or equal to 5 aberrations. Structurally complex karyotype is defined by at least 3 chromosomal aberrations, including at least one structural aberration, excluding those with clonal evolution of monosomy 7. Karyotypes were grouped in the following hierarchical order: more than or equal to 5 aberrations, 3 or 4 aberrations, structurally complex, monosomy 7 (with or without other aberrations), normal karyotype, other karyotypes. (B) The probability of 5-year OS for children with: advanced primary or secondary MDS with structurally complex karyotype; monosomal karyotype with at least one autosomal monosomy and one or more structural aberrations that did not fulfill the criteria of a structurally complex karoytpe (“monosomal karyotype only”); monosomy 7 with or without additional aberrations, including clonal evolution of monosomy 7; normal karyotype; other karyotypes. (C) The probability of 5-year OS for patients according to cytogenetic subgroup. Patients classified as monosomal karyotype only in panel B are now included in the group of monosomy 7 (with or without additional aberrations) or other karyotypes. (D) Probability of 5-year event-free survival for patients who received allogeneic HSCT (N = 143).
Probabilities of 5-year survival according to cytogenetic stratifications. (A) Probability of 5-year OS for children with advanced primary or secondary MDS according to cytogenetic stratification. Traditionally, a complex karyotype is defined by the presence of more than or equal to 3 or more than or equal to 5 aberrations. Structurally complex karyotype is defined by at least 3 chromosomal aberrations, including at least one structural aberration, excluding those with clonal evolution of monosomy 7. Karyotypes were grouped in the following hierarchical order: more than or equal to 5 aberrations, 3 or 4 aberrations, structurally complex, monosomy 7 (with or without other aberrations), normal karyotype, other karyotypes. (B) The probability of 5-year OS for children with: advanced primary or secondary MDS with structurally complex karyotype; monosomal karyotype with at least one autosomal monosomy and one or more structural aberrations that did not fulfill the criteria of a structurally complex karoytpe (“monosomal karyotype only”); monosomy 7 with or without additional aberrations, including clonal evolution of monosomy 7; normal karyotype; other karyotypes. (C) The probability of 5-year OS for patients according to cytogenetic subgroup. Patients classified as monosomal karyotype only in panel B are now included in the group of monosomy 7 (with or without additional aberrations) or other karyotypes. (D) Probability of 5-year event-free survival for patients who received allogeneic HSCT (N = 143).
Recently, patients with a monosomal karyotype were shown to have a dismal prognosis.8 Here, a monosomal karyotype with at least 2 autosomal monosomies was seen in 12 (6%) patients. Of these 12 patients, all but one were included in the group with structurally complex karyotype, whereas 17 of the 28 patients with a structurally complex karyotype did not have 2 autosomal monosomies. A monosomal karyotype with at least one autosomal monosomy and one/or more structural aberrations was noted in 30 (16%) patients (supplemental Tables 7, 8). In summary, of the 30 monosomal karyotypes with at least one autosomal monosomy and one or more structural aberrations, 19 fulfilled the definition of a structurally complex karyotype. The 5-year OS of these patients was significantly worse than that of the 11 monosomal patients not coded as structurally complex (P < .01, Figure 1B). Thus, a monosomal karyotype in childhood MDS identifies a group of patients with a heterogeneous prognosis. In contrast, using the new definition of a structurally complex karyotype, only children with a highly unfavorable prognosis were identified.
The Cox model included demographic data (gender, age) and all variables with a P < .1 in the univariate analysis. Age more than or equal to 15 years was associated with an increased risk of patient death (hazard ratio [HR] = 1.7; P = .05, Table 1). HSCT improved the outcome significantly (HR = 0.4; P = .05). Statistical analysis of the different definitions of complex karyotype had to take into account that some patients fulfilled the criteria of more than one definition. The presence of a monosomal karyotype was an independent adverse prognostic factor only when it also fulfilled the criteria of a structurally complex karyotype (HR = 4.6; P < .01, Table 1). In contrast, a monosomal karyotype that did not fulfill the criteria of a structurally complex karyotype (supplemental Table 8, “monosomal karyotype only”) did not identify a group with an inferior outcome. Notably, a structurally complex karyotype without a monosomy (“structurally complex only”) was associated with a poor probability of OS (HR = 14.4; P < .01). This held for both primary and secondary MDS (primary MDS, n = 9; secondary MDS, n = 19; supplemental Table 6). Thus, the presence of a structurally complex karyotype was a better predictor of a very unfavorable prognosis in children with MDS than the presence of more than or equal to 3 clonal aberrations or a monosomal karyotype.
Multivariate analysis for probability of overall survival in 192 consecutive children with advanced MDS and different karyotypic complexities
| Variable . | Relative risk of death . | 95% CI . | P . |
|---|---|---|---|
| Age at diagnosis ≥ 15 y | 1.7 | 1.0-2.9 | .05 |
| Male sex | 1.2 | 0.8-2.0 | NS |
| Secondary MDS | 1.0 | 0.6-1.8 | NS |
| Different karyotypic complexities | |||
| Monosomal karyotype but not structurally complex (n = 11) | 0.6 | 0.2-1.9 | NS |
| Structurally complex and monosomal karyotype (n = 19) | 4.6 | 2.2-9.5 | < .01 |
| Structurally complex but not monosomal (n = 9) | 14.5 | 5.1-41.2 | < .01 |
| HSCT | 0.4 | 0.2-1.0 | .05 |
| Variable . | Relative risk of death . | 95% CI . | P . |
|---|---|---|---|
| Age at diagnosis ≥ 15 y | 1.7 | 1.0-2.9 | .05 |
| Male sex | 1.2 | 0.8-2.0 | NS |
| Secondary MDS | 1.0 | 0.6-1.8 | NS |
| Different karyotypic complexities | |||
| Monosomal karyotype but not structurally complex (n = 11) | 0.6 | 0.2-1.9 | NS |
| Structurally complex and monosomal karyotype (n = 19) | 4.6 | 2.2-9.5 | < .01 |
| Structurally complex but not monosomal (n = 9) | 14.5 | 5.1-41.2 | < .01 |
| HSCT | 0.4 | 0.2-1.0 | .05 |
CI indicates confidence interval; and NS, not significant.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all participants of the EWOG-MDS study group.
This work was supported by COST Action BM0801 (EuGESMA) and by the Czech Ministry of Education (MSM0021620813).
Authorship
Contribution: G.G., B. Schlegelberger, F.L., and C.M.N. designed the study and wrote the manuscript; G.G., B. Schlegelberger, K.M., H.B.B., D.B., J.H., O.A.H., G.K., L.S., and E.R.v.W. performed the cytogenetic analysis; E.B., H.H., J.S., M.T., M.M.v.d.H.-E., and M.Z. are the Regional Coordinators of the EWOG MDS Study Group and performed the collection of the clinical data; P.N., A.F., G.G., B. Schlegelberger, B. Strahm, F.L., and C.M.N. analyzed the data; and all authors critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Charlotte M. Niemeyer, Department of Adolescent and Pediatric Medicine, University Hospital, Mathildenstr 1, 79106 Freiburg, Germany; e-mail: charlotte.niemeyer@uniklinik-freiburg.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal