Abstract
Acute myeloid leukemia (AML) with mutated NPM1 shows distinctive biologic and clinical features, including absent/low CD34 expression, the significance of which remains unclear. Therefore, we analyzed CD34+ cells from 41 NPM1-mutated AML. At flow cytometry, 31 of 41 samples contained less than 10% cells showing low intensity CD34 positivity and variable expression of CD38. Mutational analysis and/or Western blotting of purified CD34+ cells from 17 patients revealed NPM1-mutated gene and/or protein in all. Immunohistochemistry of trephine bone marrow biopsies and/or flow cytometry proved CD34+ leukemia cells from NPM1-mutated AML had aberrant nucleophosmin expression in cytoplasm. NPM1-mutated gene and/or protein was also confirmed in a CD34+ subfraction exhibiting the phenotype (CD34+/CD38−/CD123+/CD33+/CD90−) of leukemic stem cells. When transplanted into immunocompromised mice, CD34+ cells generated a leukemia recapitulating, both morphologically and immunohistochemically (aberrant cytoplasmic nucleophosmin, CD34 negativity), the original patient's disease. These results indicate that the CD34+ fraction in NPM1-mutated AML belongs to the leukemic clone and contains NPM1-mutated cells exhibiting properties typical of leukemia-initiating cells. CD34− cells from few cases (2/15) also showed significant leukemia-initiating cell potential in immunocompromised mice. This study provides further evidence that NPM1 mutation is a founder genetic lesion and has potential implications for the cell-of-origin and targeted therapy of NPM1-mutated AML.
Introduction
Acute myeloid leukemia (AML), with mutated nucleophosmin (NPM1) and aberrant cytoplasmic expression of nucleophosmin (NPMc+ AML),1 accounts for approximately one-third of AML. Because of its distinctive molecular, clinical, and prognostic features.2-5 AML with mutated NPM1 was included as a new provisional entity in the 2008 World Health Organization classification of myeloid neoplasms.6
Its unique gene expression profile is characterized by up-regulation of most HOX genes and down-regulation of CD34 and CD133.7,8 Because HOX genes are involved in stem-cell phenotype maintenance,9 gene expression profile findings strongly suggest that NPM1-mutated AML originates from an early hematopoietic progenitor. This view is also supported by immunohistochemistry with antibodies specific for NPM1 mutants and by mutational analysis of laser-microdissected bone marrow cells showing that NPM1-mutated AML frequently displays multilineage involvement,10 with exclusion of lymphoid lineage.11 Conversely, the observation that leukemic cells in most NPM1-mutated AML show down-regulation of CD341,7,8 raises questions as to whether the NPM1 mutation occurs in a CD34− multipotent hemopoietic progenitor12,13 or whether a minimal pool of CD34+/CD38−NPM1-mutated progenitors exists.
CD34+/CD38− cells usually contain the so-called leukemia-initiating cells (LICs) or leukemic stem cells (LSCs) that exhibit long-term repopulating potential and the ability to propagate and maintain the AML phenotype in immunocompromised mice.14,15 Engraftment capability of AML cells has been also associated with prognosis.16
CD34+/CD38− hematopoietic stem cells (HSCs) are thought to be the cell of origin of most AML cases. Indeed, these cells were found to carry the same genetic lesion as the more mature CD34+/CD38+ and CD34− leukemic populations in various cytogenetic AML subtypes, including those with inv(16), t(6;9), and +8,17,18 but not in acute promyelocytic leukemia, which may derive from a more mature hemopoietic progenitor.19,20
Until the discovery of NPM1 mutation,1 the genetic and functional characterization of CD34+/CD38− cells in AML with normal karyotype was difficult because of the lack of reliable molecular markers. Molecular and/or immunohistochemical detection of NPM1 mutations allows tracking of the genetic lesion in leukemic cells at different hierarchical stages in approximately 60% of AML with normal cytogenetics.
Aims of this study were: (1) to search for NPM1-mutated gene and/or protein in CD34+ cells (including CD38− and CD38+ subsets) purified from NPM1-mutated AML patients; (2) to determine by immunohistochemistry and/or flow cytometry whether these CD34+ cells carried aberrant cytoplasmic NPM (a distinctive functional feature of NPM1-mutated AML); (3) to investigate the capability of purified CD34+ and CD34− cells from NPM1-mutated AML to engraft in immunocompromised mice and to evaluate the nature and topographic distribution of engrafted cells; and (4) to compare the morphologic, immunophenotypic, and molecular features of murine-engrafted and patients' primary AML cells.
We proved that the minor population of CD34+ hemopoietic progenitors in NPM1-mutated AML consistently carried the NPM1 mutation, at least when CD34+ cells represented more than 1% of the bulk cell population. In most cases, CD34+, but not CD34−, cells generated in immunocompromised mice a leukemia recapitulating, both morphologically and immunohistochemically (aberrant cytoplasmic NPM1 and CD34 negativity), the original patient's disease. As previously reported by Taussig et al,21 we also found that CD34− cells from a few NPM1-mutated AML patients had significant LIC potential in immunocompromised mice.
Methods
Samples from AML patients
We studied 41 leukemia samples from 38 consecutive NPM1-mutated AML patients, including 3 cases evaluated at diagnosis and relapse (Table 1; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Patients were from the Institutes of Hematology of the Universities of Perugia, Bari, and Catania (Italy), and Dresden (Germany). AML was defined as NPM1-mutated based on cytoplasmic expression of nucleophosmin at immunohistochemistry or flow cytometry, which is predictive of NPM1 mutations.22,23 Mutated NPM1 protein and/or gene was also confirmed by Western blotting (WB)24 and/or mutational analysis.1 The study was approved by the local ethical committees, and written informed consent for analysis of leukemic samples was obtained at each participating center.
Characteristics of samples from 21 patients with NPM1-mutated AML
| Patient code . | Disease status . | FAB . | IHC (NPM) . | WB . | Karyotype . | FLT3 status . | WBC/μL . | Sample source . | CD34,* % (of MNCs) . | IHC (CD34), % . | Post-MACS* purity (% CD34+) . | CD34+/CD38−, % (gated on CD34+) . | MFI* (CD34+/CD38−) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Diagnosis | M1 | NPMc+ | + | NA | NA | 20 000 | Pb | 23 | 20-30 | 93 | 25 | 2.62 |
| 2 | Diagnosis | M5 | NPMc+ | + | 46XX; t(2; 17)(p22;q25) | FLT3-ITD | 31 040 | Pb | 5 | − | 90.64 | 94 | 15 |
| 2R | Relapse | M5 | NPMc+ | + | 46XX; t(2; 17)(p22;q25) | FLT3-ITD | 67 080 | Pb | 34 | − | 99 | 98.4 | 18.5 |
| 4 | Diagnosis | NC | NPMc+ | + | Normal | wt | 26 870 | Pb | 60 | 40-50 | 96.3 | 0.16 | 16 |
| 7 | Relapse | M5b | NPMc+ | NA | Normal | wt | 30 170 | Pb | 7.4 | − | 94 | 15.6 | 14 |
| 8R | Relapse | M4 | NPMc+ | + | Normal | wt | 127 700 | Pb | 1.9 | − | 77 | 33.3 | 1.7 |
| 10 | Diagnosis | M2 | NPMc+ | + | NA | FLT3-ITD | 25 000 | Pb | 27 | Rare | 98.5 | 70.5 | 3.2 |
| 11 | Relapse | M2 | NPMc+ | + | Normal | wt | 66 790 | Pb | 11.8 | − | 96.2 | 70.1 | 5.1 |
| 12 | Diagnosis | M1 | NPMc+ | + | NA | NA | 116 000 | Pb | 21.1 | Rare | 96 | 75.7 | 2.8 |
| 15 | Diagnosis | M4 | NPMc+ | NA | Normal | wt | 28 730 | Pb | 1.5 | − | 70 | 37 | 7.47 |
| 17 | Relapse | M4 | NPMc+ | + | Normal | FLT3-ITD | 35 750 | Pb | 27 | Rare | 98.3 | 70 | 5.9 |
| 18 | Diagnosis | M4 | NPMc+ | + | Normal | FLT3-ITD | 62 440 | Pb | 8.7 | − | 95 | 92.5 | 4.5 |
| 19 | Relapse | M4 | NPMc+ | + | Normal | wt | 4070 | BM | 1.5 | − | 99.7§ | 39 | 5.2 |
| 22 | Diagnosis | M1 | NPMc+ | + | Normal | wt | 44 000 | Pb | 4.3 | − | 86.2‖ | 11 | 2 |
| 22R | Relapse | M1 | NPMc+ | + | NA | wt | 10 800 | Pb | 72 | 50-60 | 99.9 | 15 | 2.4 |
| 23 | Relapse | M2 | NPMc+ | + | Normal | wt | 19 780 | Pb | 1.7 | Rare | 18.5‖ | 45.5 | 2 |
| 25 | Diagnosis | M5b | NPMc+† | NA | NA | FLT3-ITD | 48 920 | BM | 2.0 | NA | 99¶ | 20 | 8.3 |
| 26 | Diagnosis | M2 | NPMc+† | NA | 46XX; 46XX; del(3)(p21) | FLT3-ITD | 2600 | BM | 2.6 | NA | 99¶ | 2.2 | 7.0 |
| 30 | Diagnosis | M4 | NPMc+ | + | 47XX; +8 | FLT3-D385 | 84 770 | Pb | 2.93 | − | 79 | 29.4 | 4.8 and 11.4# |
| 31 | Diagnosis | M4 | NPMc+ | + | Normal | wt | 23 000 | Pb | 4.76 | − | 92.4 | 45 | 11 |
| 32 | Diagnosis | M4 | NPMc+ | + | 46XX; t(2;12) | FLT3-ITD | 264 000 | Pb | 1.3 | − | 81/92.3§ | 88 | 6.2 and 12.4# |
| 34 | Diagnosis | M4 | NA‡ | + | NA | wt | 150 000 | Pb | 5 | NA | 96.5 | 2.48 | 3.8 and 13.6# |
| 35 | Diagnosis | M2 | NPMc+ | + | Normal | FLT3-ITD | 7600 | Pb | 22 | − | 96.4 | 68 | 4.8 |
| Patient code . | Disease status . | FAB . | IHC (NPM) . | WB . | Karyotype . | FLT3 status . | WBC/μL . | Sample source . | CD34,* % (of MNCs) . | IHC (CD34), % . | Post-MACS* purity (% CD34+) . | CD34+/CD38−, % (gated on CD34+) . | MFI* (CD34+/CD38−) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Diagnosis | M1 | NPMc+ | + | NA | NA | 20 000 | Pb | 23 | 20-30 | 93 | 25 | 2.62 |
| 2 | Diagnosis | M5 | NPMc+ | + | 46XX; t(2; 17)(p22;q25) | FLT3-ITD | 31 040 | Pb | 5 | − | 90.64 | 94 | 15 |
| 2R | Relapse | M5 | NPMc+ | + | 46XX; t(2; 17)(p22;q25) | FLT3-ITD | 67 080 | Pb | 34 | − | 99 | 98.4 | 18.5 |
| 4 | Diagnosis | NC | NPMc+ | + | Normal | wt | 26 870 | Pb | 60 | 40-50 | 96.3 | 0.16 | 16 |
| 7 | Relapse | M5b | NPMc+ | NA | Normal | wt | 30 170 | Pb | 7.4 | − | 94 | 15.6 | 14 |
| 8R | Relapse | M4 | NPMc+ | + | Normal | wt | 127 700 | Pb | 1.9 | − | 77 | 33.3 | 1.7 |
| 10 | Diagnosis | M2 | NPMc+ | + | NA | FLT3-ITD | 25 000 | Pb | 27 | Rare | 98.5 | 70.5 | 3.2 |
| 11 | Relapse | M2 | NPMc+ | + | Normal | wt | 66 790 | Pb | 11.8 | − | 96.2 | 70.1 | 5.1 |
| 12 | Diagnosis | M1 | NPMc+ | + | NA | NA | 116 000 | Pb | 21.1 | Rare | 96 | 75.7 | 2.8 |
| 15 | Diagnosis | M4 | NPMc+ | NA | Normal | wt | 28 730 | Pb | 1.5 | − | 70 | 37 | 7.47 |
| 17 | Relapse | M4 | NPMc+ | + | Normal | FLT3-ITD | 35 750 | Pb | 27 | Rare | 98.3 | 70 | 5.9 |
| 18 | Diagnosis | M4 | NPMc+ | + | Normal | FLT3-ITD | 62 440 | Pb | 8.7 | − | 95 | 92.5 | 4.5 |
| 19 | Relapse | M4 | NPMc+ | + | Normal | wt | 4070 | BM | 1.5 | − | 99.7§ | 39 | 5.2 |
| 22 | Diagnosis | M1 | NPMc+ | + | Normal | wt | 44 000 | Pb | 4.3 | − | 86.2‖ | 11 | 2 |
| 22R | Relapse | M1 | NPMc+ | + | NA | wt | 10 800 | Pb | 72 | 50-60 | 99.9 | 15 | 2.4 |
| 23 | Relapse | M2 | NPMc+ | + | Normal | wt | 19 780 | Pb | 1.7 | Rare | 18.5‖ | 45.5 | 2 |
| 25 | Diagnosis | M5b | NPMc+† | NA | NA | FLT3-ITD | 48 920 | BM | 2.0 | NA | 99¶ | 20 | 8.3 |
| 26 | Diagnosis | M2 | NPMc+† | NA | 46XX; 46XX; del(3)(p21) | FLT3-ITD | 2600 | BM | 2.6 | NA | 99¶ | 2.2 | 7.0 |
| 30 | Diagnosis | M4 | NPMc+ | + | 47XX; +8 | FLT3-D385 | 84 770 | Pb | 2.93 | − | 79 | 29.4 | 4.8 and 11.4# |
| 31 | Diagnosis | M4 | NPMc+ | + | Normal | wt | 23 000 | Pb | 4.76 | − | 92.4 | 45 | 11 |
| 32 | Diagnosis | M4 | NPMc+ | + | 46XX; t(2;12) | FLT3-ITD | 264 000 | Pb | 1.3 | − | 81/92.3§ | 88 | 6.2 and 12.4# |
| 34 | Diagnosis | M4 | NA‡ | + | NA | wt | 150 000 | Pb | 5 | NA | 96.5 | 2.48 | 3.8 and 13.6# |
| 35 | Diagnosis | M2 | NPMc+ | + | Normal | FLT3-ITD | 7600 | Pb | 22 | − | 96.4 | 68 | 4.8 |
Characteristics of other patients are shown in supplemental Table 1.
FAB indicates French-American-British classification; IHC, immunohistochemistry analysis; WB, Western blot with specific anti-NPM mutant antibodies24 ; WBC, white blood cell; MFI, mean fluorescence intensity; NC, not classified; NA, not available; wt, wild-type; Pb, peripheral blood; R, relapse; –, CD34+ cells not detectable at IHC; and BM, bone marrow.
Analyzed on Cytomics FC500 cytometer equipped with the CXP analysis software (Beckman Coulter).
Cytoplasmic NPM was detected by flow cytometry.23
Only studied by WB.
After 2 purification steps.
CD34+ MACS-enriched cell fraction used for FACS sorting.
CD34+ FACS sorted.
Two distinct cell populations (CD34low and CD34bright).
Flow cytometric immunophenotyping
Immunophenotyping was performed using the following antibodies: peridinin chlorophyll protein complex-conjugated anti-CD45 (CD45-peridinin chlorophyll protein complex), fluorescein isothiocyanate (FITC)–conjugated anti-CD34 (CD34−FITC), phycoerythrin (PE)–conjugated, or allophycocyanin (APC)–conjugated anti-CD38 (CD38-PE, CD38-APC), CD33-APC, and CD123-PE (Becton Dickinson [BD] Biosciences) or phycoerythrin-Texas Red (ECD)–conjugated anti-CD45 (CD45-ECD), CD34−FITC, phycoerythrin-cyanin 5 (PC5)–conjugated anti-CD38 (CD38-PC5), CD33-PE, phycoerythrin-cyanin 7 (PC7)–conjugated anti-CD19 (CD19-PC7), CD11b-FITC, CD90-FITC, and CD90-PC5 (Beckman Coulter). Cytoplasmic NPM1 in leukemic cells was detected by flow cytometry as previously described.23 Analysis was performed on either a Cytomics FC500 cytometer equipped with the CXP analysis software 2.0 (Beckman Coulter) or a FACSCalibur or FACSAria flow cytometers using the CellQuest Pro analysis software 6.0 (BD Biosciences). Gates were drawn to exclude nonviable cells and debris.
MACS cell sorting
All leukemia samples were subjected to CD34+ cell selection by magnetic-activated cell sorting (MACS) technology, according to manufacturer's instruction (CD34 MicroBeads; Miltenyi Biotec). Both positive and negative cell fractions were analyzed for CD34+ cells percentage.
FACS
The CD34+ cells MACS-enriched cell fraction from 1 case and leukemic bulk cells from 2 additional cases were subjected to sorting for specific cell subpopulations by the cell sorter FACSAria (BD Biosciences) equipped with blue, red, and violet lasers. Dead cells were excluded by analyzing forward scatter versus side scatter dot plots. Doublets were excluded by forward scatter-H versus forward scatter-A dot plots. Data compensation and analysis were obtained using the “logicle” display method.25
Detection of NPM1 mutated protein and gene
NPM1 mutant protein was detected on lysates from 1 to 2 × 106 cells by WB analysis with a rabbit polyclonal antibody specific for the mutated NPM1 protein.10,24 Lysate from the human leukemic cell line OCI/AML326 was used as positive control for NPM1 mutant A protein expression. In selected cases, NPM1 mutations were analyzed by either direct sequencing8 or genomic DNA fragment analysis.27
Immunohistochemical studies
Immunohistochemistry was performed on human and mice paraffin-embedded samples fixed in B5 (Bio-Optica) for 2 hours; bone tissues were also decalcified in ethylenediaminetetraacetic acid (Osteodec; Bio-Optica) for 5 to 6 hours. Antigen retrieval was carried out by microwaving in 0.1mM ethylenediaminetetraacetic acid, pH 8.0.
Cytoplasmic nucleophosmin was revealed using a mouse anti-NPM monoclonal antibody (mAb).1 Other antigens included: nucleolin (C23; mAb MS-3; Santa Cruz Biotechnology), myeloperoxidase (rabbit antimyeloperoxidase antibody; Dako Denmark), macrophage-restricted CD68 (mouse mAb PG-M1 generated by B.F.), CD20 (mouse mAb; L26; Dako Denmark), CD3 (rabbit mAb, SP7; Thermo Scientific), CD45 and glycophorin (Dako Denmark). The antibody/antigen interaction was revealed by the alkaline phosphatase anti-alkaline phosphatase (APAAP) technique.1
Double stainings for CD34/NPM and CD34/C23 were performed using a sequential immunoperoxidase/APAAP procedure.10
Leukemia-initiating ability of CD34+ versus CD34− cells from NPM1-mutated AML in immunocompromised mice
Isolated CD34+ and CD34− cells from NPM1-mutated AML were screened for their potential to engraft and generate leukemia in either nonobese diabetic/severe combined immunodeficient (NOD/SCID) or NOD/SCID/IL2rγnull (NOG)28 mice. Mice were originally obtained from The Jackson Laboratory. Mouse colonies were maintained in the certified Animal Facility of University of Perugia, Perugia, Italy, in accordance with national guidelines. They were kept in microisolator cages and fed sterile food and acidified water, containing 100 μg/mL ciprofloxacin. Mice 6 to 10 weeks of age were subjected to 3.5 Gy γ-irradiation up to 24 hours before intravenous (tail vein) injection of cells. Mice were killed at 3 to 21 weeks after transplantation; bone marrow was removed from one of the femurs and tibias by flushing with phosphate-buffered saline and analyzed for engraftment using a specific anti-hCD45 mAb. Other bones, including vertebral bodies, were fixed/decalcified and processed for paraffin embedding. Positive marrow samples (> 0.1% hCD45+ cells) were further analyzed by immunophenotyping, immunohistochemistry, WB, and molecular analysis for NPM1-mutated protein and/or gene to confirm and characterize the type of engraftment. In some mice, spleen, liver, lung, and brain were also studied by immunohistochemistry for leukemic infiltration. Self-renewal capacity of cells recovered from primary recipients was assessed by serial transplantations in mice.
CFC assay
A total of 10 to 50 × 103 cells from the CD34+ or CD34− fractions of patients 17, 32, and 34 were plated in triplicate in 1 mL of MethoCult GF H4434 (StemCell Technologies) in 35-mm tissue-culture dishes. Colony scoring was performed after 14 days of culture. Colonies were analyzed for NPM1 mutation by genomic DNA fragment analysis.
Statistics
Leukemia-initiating cells frequency in CD34+ and CD34− cells from NPM1-mutated AML was calculated in limiting-dilution experiments using the StemSoft's L-Calc software Version 1.1.1 (StemCell Technologies).
Results
Immunophenotypic characterization and isolation of CD34+ cells in NPM1-mutated AML
Flow cytometry of the 41 NPM1-mutated AML samples showed variable percentage of CD34+ cells (range, 0.02%-75%) with 31 of 41 (75.6%) samples expressing less than 10% CD34+ cells (Table 1; supplemental Table 1). Notably, 18 of 41 samples expressed less than 1% CD34+ cells (supplemental Table 1). Mean fluorescence intensity for CD34 was low (range, 1.7-18.5) in most of samples (Table 1; supplemental Table 1).
Immunohistochemistry for CD34 was carried out in bone marrow trephines from 37 of 41 cases. No or rare CD34+ cells (Table 1; supplemental Table 1) were detected in 34 of 37 samples, whereas in 3 cases (patients 1, 4, and 22R) the CD34+ cells ranged between 20% and 60% (Table 1). The lower percentage of CD34+ cells detectable by immunohistochemistry compared with flow cytometry is probably the result of the lower sensitivity of immunohistochemistry for detecting low-intensity CD34 expression.
CD34+ cells were purified by MACS (39 of 41 samples) or FACS (2 of 41 samples; Table 1; supplemental Table 1). The MACS-sorting approach was chosen because it allows reliable recovery of cells expressing CD34 at different levels, including low-intensity CD34+ cells, which are characteristically found in NPM1-mutated AML. Indeed, MACS-purified CD34+ cells showed the same mean fluorescence intensity for CD34 as the CD34+ cells in the original sample, with low, intermediate, or bright intensity cell populations similarly represented. The CD34+ cell fraction also showed the expected heterogeneity in terms of CD38 expression (Table 1; supplemental Table 1). In particular, early CD34+/CD38− hematopoietic progenitors were variously represented within the CD34+ population (range, 0.16%-98.4%; Table 1; supplemental Table 1).
These results clearly indicate that the minor population of CD34+ cells in NPM1-mutated AML usually express the CD34 molecule at low intensity and exhibit the expected heterogeneous phenotype of this cell fraction.
CD34+ cells from NPM1-mutated AML express mutated NPM1 gene and protein
We then asked whether CD34+ cells from NPM1-mutated AML patients harbor NPM1 mutation and, thus, belong to the leukemic clone. CD34+ sorted cells (at least 1 × 106 cells) from 13 of 41 samples (CD34+ cells ranging from 1.3% to 60% of mononuclear cells [MNCs]) were suitable for WB with anti-NPM mutant specific antibodies (Figure 1A). In all of them, a specific 37-kDa molecular weight band, corresponding to NPM1 mutant protein, was detected (Figure 1A). This signal could not be the result of contaminating CD34− leukemic cells because in 12 of 13 samples CD34+ cells accounted for more than 90% of the total analyzed population (2 × 106 cells), whereas our WB detection method is not sensitive enough to reveal NPM1 mutant protein from cells, which accounts for less than or equal to 10% (Figure 1B). Moreover, unlike that observed in AML cell dilution assay (Figure 1B), the WB signal intensity was not decreased in the CD34+ purified cells compared with leukemic bulk (Figure 1A). Thus, it is conceivable that, even in the case with lower CD34+ cell purity (patient 30, 79% CD34+ cell purity; Table 1), positive signal could not be ascribed to contaminating CD34− cells.
CD34+ cells from AML with mutated NPM1 express mutated NPM1 gene and protein. (A) WB analysis of CD34+ cells isolated from 13 NPM1-mutated AML patient samples. Patient sample codes are indicated on the left (Table 1). Lysates from 1 to 2 × 106 cells of either leukemic bulk (Bulk) or CD34+ MACS sorted (Sorted CD34+) cell population were loaded and run on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel, transferred to polyvinylidene difluoride membrane, and probed with an anti-NPM mutant (anti-NPMm) rabbit polyclonal antibody. A specific band corresponding to NPM mutant protein is detected at 37-kDa molecular weight (MW) in both bulk and CD34+ cell populations from patients. Columns on the right show percentage of CD34+ cells in the original patient sample and in the CD34-sorted cell population. Lysates from human myeloid leukemic cell lines OCI/AML3 and U937 were used, respectively, as positive and negative control for NPM1 mutant protein expression. (B) AML patient sample dilution test for WB with anti-NPM mutant specific antibody. Unpurified cell fraction from 3 representative NPM1-mutated AML patient samples (patients 30, 32, and 33) was progressively diluted (100%, 50%, 25%, 20%, 15%, 10%, 5%, and 0%) with cells of AML with unmutated NPM1. Lysates from a total of 2 × 106 cells were loaded in each lane and checked for NPM1 mutant protein detection by WB (anti-NPMm, top panels). Progressive dilution of signal indicates that saturation for NPM1 mutant protein detection is not reached in our experimental conditions. Signal is not anymore detectable when NPM1-mutated AML sample is less than 15% to 10% of the original sample. Equal protein lysate loading was demonstrated by blotting the same membranes with an anti–β-tubulin monoclonal antibody (bottom panels). (C) Chromatograms of direct sequencing of the NPM1 gene exon 12 (forward sequence reading) showing both wild-type sequence (coming from the wild-type allele) and TCTG insertion (type A mutation; coming from the mutated allele) in CD34+ cells isolated from bone marrow of 1 patient with NPM1-mutated AML (patient 19; bottom panel) and in the relative controls (leukemic bulk and CD34− cell populations; top and middle panels, respectively). Percentage of CD34+ cells in each sample is shown on the right. Pt. code indicates patient code; and mut A = NPM1 gene mutation A.
CD34+ cells from AML with mutated NPM1 express mutated NPM1 gene and protein. (A) WB analysis of CD34+ cells isolated from 13 NPM1-mutated AML patient samples. Patient sample codes are indicated on the left (Table 1). Lysates from 1 to 2 × 106 cells of either leukemic bulk (Bulk) or CD34+ MACS sorted (Sorted CD34+) cell population were loaded and run on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel, transferred to polyvinylidene difluoride membrane, and probed with an anti-NPM mutant (anti-NPMm) rabbit polyclonal antibody. A specific band corresponding to NPM mutant protein is detected at 37-kDa molecular weight (MW) in both bulk and CD34+ cell populations from patients. Columns on the right show percentage of CD34+ cells in the original patient sample and in the CD34-sorted cell population. Lysates from human myeloid leukemic cell lines OCI/AML3 and U937 were used, respectively, as positive and negative control for NPM1 mutant protein expression. (B) AML patient sample dilution test for WB with anti-NPM mutant specific antibody. Unpurified cell fraction from 3 representative NPM1-mutated AML patient samples (patients 30, 32, and 33) was progressively diluted (100%, 50%, 25%, 20%, 15%, 10%, 5%, and 0%) with cells of AML with unmutated NPM1. Lysates from a total of 2 × 106 cells were loaded in each lane and checked for NPM1 mutant protein detection by WB (anti-NPMm, top panels). Progressive dilution of signal indicates that saturation for NPM1 mutant protein detection is not reached in our experimental conditions. Signal is not anymore detectable when NPM1-mutated AML sample is less than 15% to 10% of the original sample. Equal protein lysate loading was demonstrated by blotting the same membranes with an anti–β-tubulin monoclonal antibody (bottom panels). (C) Chromatograms of direct sequencing of the NPM1 gene exon 12 (forward sequence reading) showing both wild-type sequence (coming from the wild-type allele) and TCTG insertion (type A mutation; coming from the mutated allele) in CD34+ cells isolated from bone marrow of 1 patient with NPM1-mutated AML (patient 19; bottom panel) and in the relative controls (leukemic bulk and CD34− cell populations; top and middle panels, respectively). Percentage of CD34+ cells in each sample is shown on the right. Pt. code indicates patient code; and mut A = NPM1 gene mutation A.
In 1 leukopenic patient (patient 19), only 0.1 × 106 CD34+ bone marrow cells (99.7% pure) could be recovered, which were not enough for WB analysis. In this case, NPM1 mutation A was detected by direct gene sequencing in purified CD34+, CD34−, and presorting cell populations (Figure 1C). Again, detection of NPM1 mutation in the CD34+ cells could not be the result of the minority of contaminating CD34− cells (0.3%) because the latter was under the detection threshold (∼ 25%) of our direct gene sequencing assay (supplemental Figure 1).
These findings clearly show that the CD34+ fraction from NPM1-mutated AML with CD34+ cells representing more than 1% of the bulk cell population is mutated both at gene and protein level, thus indicating it belongs to the leukemic clone.
The CD34+ fraction from NPM1-mutated AML contains CD34+/CD38− cells harboring NPM1 mutated gene and protein
Given the heterogeneity of CD34+ cells and to better track the NPM1 mutation in the context of hematopoietic hierarchy of NPM1-mutated AML, we next investigated whether CD34+/CD38− cells carry the genetic lesion. The CD34/CD38 flow cytometric staining patterns of the 5 cases investigated (Table 1: patients 2, 18, 22, 25, and 26) are shown in Figure 2A.
CD34+/CD38− cells from AML with mutated NPM1 express mutated NPM1 gene and protein and display immunophenotypic features of LSCs. (A) CD34/CD38 flow cytometry staining patterns of peripheral blood MNCs (patients 2, 18, and 22) or whole bone marrow (patients 25 and 26) from the 5 NPM1-mutated AML patients studied for involvement of CD34+/CD38− cells by NPM1 mutation. Flow cytometric analysis was performed on either FACSAria (BD Biosciences; patients 22, 25, and 26) or Cytomics FC500 cytometer (Beckman Coulter; patients 2 and 18), as indicated in “Flow cytometric immunophenotyping.” Concomitant expression of CD123 and CD33 on the CD34+/CD38− cell population is shown in a representative example (patient 18, bottom right panels). Here, within the CD34+/CD38− cell population, only a very small cell fraction (gate Q: ∼ 0.8%) was negative for both CD123 and CD33 that could represent normal residual HSCs. (B) WB showing expression of NPM1 mutant protein in MACS-sorted CD34+ cells from 2 selected cases from Figure 1 (patients 2 and 18) where CD34+/CD38− cells represented almost the whole CD34+ cell population. Here, percentages of CD34+/CD38− and CD34+/CD38+ cells are shown. (C-D) CD34+/CD38− and CD34+/CD38+ cells were FACS-sorted from bone marrow of a patient (patient 25) with AML with cytoplasmic NPM1 with 0.3% CD34+ cells. Expression of CD123 and CD33 antigens in the 2 cell subpopulations is shown (C bottom panels). Molecular analysis for NPM1 and FLT3-ITD mutations was performed on genomic DNA by high-resolution fragment analysis (D). Results obtained on sorted CD34+/CD38− (purity of 99%; i) and CD34+/CD38+ (purity of 99%; ii) cells were compared with the total bone marrow mononuclear cells (BM-MNC; iii). As shown in green, the typical double peak indicating the NPM1 mutation (green arrows) was detectable at similar levels in both populations as well as in the MNCs. Peaks indicating FLT3 gene status (wild-type [wt] and FLT3-ITD [ITD] mutation) are in blue. FLT3-ITD mutation/FLT3-wt ratio (mut/wt) is shown on the right. (E) Flow cytometric analysis of a representative NPM1-mutated AML patient sample (patient 30) showing an evident double-cell population with distinct immunophenotypic features within CD34+/CD38− progenitor cells: (i) CD34+ cell percentage (2.93%) in the leukemic bulk from the patient's peripheral blood (MNCs); (ii) expression of CD38 on the purified CD34+ cells (CD34+ after sorting); and detection of 2 distinct cell subpopulations within CD34+/CD38− cells: CD34bright/CD38−/CD123−/CD33− (iii) versus CD34low/CD38−/CD123+/CD33+ (iv) cells.
CD34+/CD38− cells from AML with mutated NPM1 express mutated NPM1 gene and protein and display immunophenotypic features of LSCs. (A) CD34/CD38 flow cytometry staining patterns of peripheral blood MNCs (patients 2, 18, and 22) or whole bone marrow (patients 25 and 26) from the 5 NPM1-mutated AML patients studied for involvement of CD34+/CD38− cells by NPM1 mutation. Flow cytometric analysis was performed on either FACSAria (BD Biosciences; patients 22, 25, and 26) or Cytomics FC500 cytometer (Beckman Coulter; patients 2 and 18), as indicated in “Flow cytometric immunophenotyping.” Concomitant expression of CD123 and CD33 on the CD34+/CD38− cell population is shown in a representative example (patient 18, bottom right panels). Here, within the CD34+/CD38− cell population, only a very small cell fraction (gate Q: ∼ 0.8%) was negative for both CD123 and CD33 that could represent normal residual HSCs. (B) WB showing expression of NPM1 mutant protein in MACS-sorted CD34+ cells from 2 selected cases from Figure 1 (patients 2 and 18) where CD34+/CD38− cells represented almost the whole CD34+ cell population. Here, percentages of CD34+/CD38− and CD34+/CD38+ cells are shown. (C-D) CD34+/CD38− and CD34+/CD38+ cells were FACS-sorted from bone marrow of a patient (patient 25) with AML with cytoplasmic NPM1 with 0.3% CD34+ cells. Expression of CD123 and CD33 antigens in the 2 cell subpopulations is shown (C bottom panels). Molecular analysis for NPM1 and FLT3-ITD mutations was performed on genomic DNA by high-resolution fragment analysis (D). Results obtained on sorted CD34+/CD38− (purity of 99%; i) and CD34+/CD38+ (purity of 99%; ii) cells were compared with the total bone marrow mononuclear cells (BM-MNC; iii). As shown in green, the typical double peak indicating the NPM1 mutation (green arrows) was detectable at similar levels in both populations as well as in the MNCs. Peaks indicating FLT3 gene status (wild-type [wt] and FLT3-ITD [ITD] mutation) are in blue. FLT3-ITD mutation/FLT3-wt ratio (mut/wt) is shown on the right. (E) Flow cytometric analysis of a representative NPM1-mutated AML patient sample (patient 30) showing an evident double-cell population with distinct immunophenotypic features within CD34+/CD38− progenitor cells: (i) CD34+ cell percentage (2.93%) in the leukemic bulk from the patient's peripheral blood (MNCs); (ii) expression of CD38 on the purified CD34+ cells (CD34+ after sorting); and detection of 2 distinct cell subpopulations within CD34+/CD38− cells: CD34bright/CD38−/CD123−/CD33− (iii) versus CD34low/CD38−/CD123+/CD33+ (iv) cells.
In 2 of 5 cases (Table 1: patients 2 and 18) whose MACS-sorted CD34+ cell fraction was represented almost exclusively by CD34+/CD38− cells (purity of 94% and 92.5%, respectively), expression of mutated NPM1 protein was detected by WB (Figure 2B).
In the remaining 3 cases (patients 22, 25, and 26), the FACS-sorted CD34+/CD38− subpopulation (purity of 96%, 99%, and 99%, respectively) was NPM1-mutated by either direct gene sequencing (patient 22, harboring NPM1 mutation 73 ) or genomic DNA fragment analysis (patients 25 and 26). The finding of NPM1 mutation in these samples could not be the result of the small number of contaminating CD34− cells (range, 1%-4%) because this is below the detection threshold of our molecular assays (supplemental Figure 1). A representative case (patient 25) harboring the NPM1 mutation in the CD34+/CD38− and CD34+/CD38+ cell fractions (both 99% pure), as well as in the bulk leukemic population, is shown in Figure 2C-D. In this case, the CD34+/CD38− cells also carried FLT3-ITD mutation (Figure 2Di), further confirming that they belong to the leukemic clone. FLT3-ITD showed the highest prevalence in the CD34+/CD38− cells (Figure 2Di), whereas, in the more differentiated cells (CD34+/CD38+), the FLT3-ITD mut/wt ratio was significantly lower (Figure 2Dii) and, in the bulk leukemic population, FLT3-ITD was only present in a small fraction of cells (Figure 2Diii).
In conclusion, our results clearly indicate that, in the 5 cases studied, the CD34+/CD38− subpopulation carried the mutated NPM1 gene and protein.
Immunophenotypic features of CD34+/CD38− cells from NPM1-mutated AML
We then asked whether CD34+/CD38− cells from NPM1-mutated AML displayed the immunophenotype (CD123+/CD33+/CD90−)29-31 typical of LSCs. Most or all of CD34+/CD38− cells were also CD123+ in 21 of 24 samples and CD33+ (generally at high intensity) in 14 of 15 samples (eg, patient 18 of Figure 2A; supplemental Table 2). CD90 was negative in all cases (data not shown). In 9 of 41 (21.9%) samples (Table 1; supplemental Table 1), a double CD34low and CD34bright cell population was observed (Figure 2E). Interestingly, CD34low and CD34bright cells within the CD34+/CD38− fractions showed a different phenotype (Figure 2E; supplemental Table 2). CD34low were CD123+/CD33+ (a phenotype resembling that of LSCs), whereas CD34bright cells were negative for both antigens, probably representing residual circulating normal CD34+ HSCs. The latter have been previously described in some AML patients at diagnosis.32
Our results clearly indicate that CD34+/CD38− cells from NPM1-mutated AML show the genotypic and phenotypic features of LSCs (ie, NPM1-mutated gene/protein and expression of CD123 and CD33). In some cases, a variable fraction of CD34bright cells was detected, possibly representing residual circulating normal HSCs.
CD34+ cells in NPM1-mutated AML show cytoplasmic expression of nucleophosmin
Then we asked whether CD34+NPM1-mutated AML cells expressed aberrantly nucleophosmin in their cytoplasm, which is the most distinctive functional consequence of NPM1 mutations.33-36
To address this issue, we double-stained for CD34 and NPM bone marrow paraffin sections from 5 NPM1-mutated AML containing a percentage of CD34+ cells ranging between 2% and 20%. In all cases, the CD34+ cells showed aberrant cytoplasmic positivity for nucleophosmin (NPMc+) and nucleus-restricted positivity for another nucleolar protein (C23/nucleolin) that was used as control (Figure 3A-D). Double-stained CD34+/NPMc+ cells were randomly scattered in bone marrow biopsies from patients, without predilection for areas close to bone trabeculae or marrow vessels. Expression of cytoplasmic NPM1 in CD34+ cells (both CD38− and CD38+ subsets) was also found by flow cytometry (Figure 3E-F).
CD34+ cells in NPM1-mutated AML show cytoplasmic expression of nucleophosmin. (A-B) CD34+ leukemic cells (brown) from a patient with NPM1-mutated AML showing cytoplasmic expression of NPM (blue; A arrows) and nucleus-restricted positivity for C23/nucleolin (B arrows). (C-D) Another NPM1-mutated patient showing rare CD34+ cells (C) that double stain for CD34 (brown) and cytoplasmic NPM (blue; D single arrow); endothelium of a vessel express CD34 (D double arrow). CD34+ leukemic cell (brown) from the same case show nucleus-restricted positivity for C23/nucleolin (blue; D inset). (A-B,D) Sequential immunoperoxidase/APAAP staining; no counterstaining. (C) APAAP technique (hematoxylin counterstaining). (A-D) Paraffin sections from bone marrow biopsies. All images were collected using an Olympus B61 microscope and a UPlan FI 100×/1.3 NA oil objective; Camedia 4040, Dp_soft Version 3.2; and Adobe Photoshop 7.0. (E-F) Flow cytometric detection of NPM-cytoplasmic expression (patient 25) in CD34+ and CD34− blasts (E), and after additional gating on CD34+/CD38− or CD34+/CD38+ cells, respectively (F).
CD34+ cells in NPM1-mutated AML show cytoplasmic expression of nucleophosmin. (A-B) CD34+ leukemic cells (brown) from a patient with NPM1-mutated AML showing cytoplasmic expression of NPM (blue; A arrows) and nucleus-restricted positivity for C23/nucleolin (B arrows). (C-D) Another NPM1-mutated patient showing rare CD34+ cells (C) that double stain for CD34 (brown) and cytoplasmic NPM (blue; D single arrow); endothelium of a vessel express CD34 (D double arrow). CD34+ leukemic cell (brown) from the same case show nucleus-restricted positivity for C23/nucleolin (blue; D inset). (A-B,D) Sequential immunoperoxidase/APAAP staining; no counterstaining. (C) APAAP technique (hematoxylin counterstaining). (A-D) Paraffin sections from bone marrow biopsies. All images were collected using an Olympus B61 microscope and a UPlan FI 100×/1.3 NA oil objective; Camedia 4040, Dp_soft Version 3.2; and Adobe Photoshop 7.0. (E-F) Flow cytometric detection of NPM-cytoplasmic expression (patient 25) in CD34+ and CD34− blasts (E), and after additional gating on CD34+/CD38− or CD34+/CD38+ cells, respectively (F).
Thus, CD34+ leukemic cells (including CD34+/CD38− and CD34+/CD38+ subpopulations) not only carry the mutated NPM1 gene and protein but also show aberrant nucleophosmin expression in cytoplasm, which is a distinguishing feature of AML with mutated NPM1.33-36
CD34+ cells from NPM1-mutated AML generate leukemia in immunocompromised mice
Isolated CD34+ cells from NPM1-mutated AML were screened for their potential to engraft and generate leukemia in immunocompromised mice. Engraftment was observed in 10 of 16 samples (Table 2; supplemental Tables 3-4), as documented by positivity for hCD45 (at flow cytometry and immunohistochemistry). The leukemic nature of hCD45+ engrafted cells was clearly demonstrated by the combined morphologic and immunohistochemical analysis of bone marrow sections that showed 3 main infiltration patterns: (1) sparse aggregates of human leukemic cells mainly adjacent to the bone (suggestive of initial marrow involvement, supplemental Figure 2); (2) leukemic infiltration of marrow areas near to and distant from the bone (the most common pattern, Figure 4); and (3) massive marrow infiltration by leukemic cells (supplemental Figure 3).
Leukemic engraftment of CD34+ cells from 10 NPM1-mutated AML in immunocompromised mice
| Patient code . | % CD34+ in original sample . | Purity of inoculum . | No./mice . | No. of cells/mice, ×106 . | No. of mice with AML/no. of mice evaluated . | Time of evaluation, w . | % hCD45 in mice BM . | % CD34+ in mice (gated on hCD45) . | % hCD45+/CD19+ (or CD20+) . | WB (NPMm) . | IHC (NPM) . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 60 | 96.3 | 1/NOD | 5 | 1/1 | 4 | 27 | 50 | −‡‡ | + | c+ |
| 3/NOD | 2 | 2/3 | 8 | 60.4/80§ | ND/55 | −‡‡ | + | c+ | |||
| 10 | 27 | 98.5 | 2/NOD | 5 | 1/2 | 16 | 0.62 | ND | ND | ND | ND |
| 4/NOD | 2 | 3/4 | 16/16/20/20 | 12.8‖/9.7/0/17.4§ | 1.9/ND/—/2.4 | −‡‡ | + | c+ | |||
| 11 | 11.8 | 96.2 | 2/NOD | 2 | 2/2 | 20 | 39.8§/19.8 | 1 | −‡‡ | ND | c+ |
| 13 | 0.8 | 90 | 1/NOD | 2.5 | 1/1 | 19 | 32.3 | CD34−‡‡ | −‡‡ | ND | c+ |
| 22R* | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 |
| 17*† | 27 | 98.3 | 4/NOG | 2 | 4/4 | 9/9/12/20 | 4.5/15.8/52‖/80§ | ND/52/35/14 | ND/0/3.2**/0 | + | c+ |
| 4/NOG | 1 | 4/4 | 9/9/13/20 | 1.5/1.8/5.8/24.7 | ND/ND/21/23.7 | 0 | + | c+ | |||
| sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | |||
| 23† | |||||||||||
| CD34+ | 1.7 | 99 | 1/NOG | 0.2 | 1/1 | 15 | 21 | 0.57 | 0 | + | c+ |
| 2/NOG | 0.001 | 0/2 | 21 | 0 | — | — | — | — | |||
| CD34− | 99.9 | 1/NOG | 0.2 | 0/1 | 15 | 0 | — | — | — | — | |
| 2/NOG | 0.001 | 2/2 | 21 | 53.8¶/95.7§ | 0 | 0 | + | c+ | |||
| 30† | |||||||||||
| CD34+ | 2.93 | 79 | 4/NOG | 1 | 2/4 | 7/12/18/18 | 28#/27.3#/33**††/47.3**†† | 13/15.2/6.9/10.8 | 25/85/22/52.6 | − | c−/c+§§ |
| CD34− | 99.5 | 4/NOG | 1 | 0/4 | 7/12/18/18 | 0 | — | — | — | — | |
| 32† | |||||||||||
| CD34+ | 1.3 | 92.3 | 3/NOG | 0.5 | 2/3 | 12/18/20 | 8.2#/1.25**/1.54** | ND/0.42/1 | 81.5/15/1 | ND | c−/c+‖‖ |
| 3/NOG | 0.2 | 1/3 | 3/6/18 | 80.8#/32.8#/59.6**†† | 1.2/2.57/0 | 0/36/0.2 | −/+ | c−/c+‖‖ | |||
| 3/NOG | 0.1 | 1/3 | 17/19/20 | 0.72#/0.6††/0.14# | 0.1/2.7/0.3 | 50/5.2/0 | — | c−/c+‖‖ | |||
| 4/NOG | 0.01 | 0/4 | 3/3/15/20 | 0 | — | — | — | — | |||
| CD34− | 98.8 | 3/NOG | 0.5 | 0/3 | 13/18/20 | 0 | — | — | — | — | |
| 3/NOG | 0.2 | 0/3 | 3/6/15/20 | 0 | — | — | — | — | |||
| 4/NOG | 0.1 | 0/4 | 12/15/18/20 | 0 | — | — | — | — | |||
| 4/NOG | 0.01 | 0/4 | 6/7/20/20 | 0 | — | — | — | — | |||
| 34*†‡ | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 |
| Patient code . | % CD34+ in original sample . | Purity of inoculum . | No./mice . | No. of cells/mice, ×106 . | No. of mice with AML/no. of mice evaluated . | Time of evaluation, w . | % hCD45 in mice BM . | % CD34+ in mice (gated on hCD45) . | % hCD45+/CD19+ (or CD20+) . | WB (NPMm) . | IHC (NPM) . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 60 | 96.3 | 1/NOD | 5 | 1/1 | 4 | 27 | 50 | −‡‡ | + | c+ |
| 3/NOD | 2 | 2/3 | 8 | 60.4/80§ | ND/55 | −‡‡ | + | c+ | |||
| 10 | 27 | 98.5 | 2/NOD | 5 | 1/2 | 16 | 0.62 | ND | ND | ND | ND |
| 4/NOD | 2 | 3/4 | 16/16/20/20 | 12.8‖/9.7/0/17.4§ | 1.9/ND/—/2.4 | −‡‡ | + | c+ | |||
| 11 | 11.8 | 96.2 | 2/NOD | 2 | 2/2 | 20 | 39.8§/19.8 | 1 | −‡‡ | ND | c+ |
| 13 | 0.8 | 90 | 1/NOD | 2.5 | 1/1 | 19 | 32.3 | CD34−‡‡ | −‡‡ | ND | c+ |
| 22R* | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 |
| 17*† | 27 | 98.3 | 4/NOG | 2 | 4/4 | 9/9/12/20 | 4.5/15.8/52‖/80§ | ND/52/35/14 | ND/0/3.2**/0 | + | c+ |
| 4/NOG | 1 | 4/4 | 9/9/13/20 | 1.5/1.8/5.8/24.7 | ND/ND/21/23.7 | 0 | + | c+ | |||
| sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | |||
| 23† | |||||||||||
| CD34+ | 1.7 | 99 | 1/NOG | 0.2 | 1/1 | 15 | 21 | 0.57 | 0 | + | c+ |
| 2/NOG | 0.001 | 0/2 | 21 | 0 | — | — | — | — | |||
| CD34− | 99.9 | 1/NOG | 0.2 | 0/1 | 15 | 0 | — | — | — | — | |
| 2/NOG | 0.001 | 2/2 | 21 | 53.8¶/95.7§ | 0 | 0 | + | c+ | |||
| 30† | |||||||||||
| CD34+ | 2.93 | 79 | 4/NOG | 1 | 2/4 | 7/12/18/18 | 28#/27.3#/33**††/47.3**†† | 13/15.2/6.9/10.8 | 25/85/22/52.6 | − | c−/c+§§ |
| CD34− | 99.5 | 4/NOG | 1 | 0/4 | 7/12/18/18 | 0 | — | — | — | — | |
| 32† | |||||||||||
| CD34+ | 1.3 | 92.3 | 3/NOG | 0.5 | 2/3 | 12/18/20 | 8.2#/1.25**/1.54** | ND/0.42/1 | 81.5/15/1 | ND | c−/c+‖‖ |
| 3/NOG | 0.2 | 1/3 | 3/6/18 | 80.8#/32.8#/59.6**†† | 1.2/2.57/0 | 0/36/0.2 | −/+ | c−/c+‖‖ | |||
| 3/NOG | 0.1 | 1/3 | 17/19/20 | 0.72#/0.6††/0.14# | 0.1/2.7/0.3 | 50/5.2/0 | — | c−/c+‖‖ | |||
| 4/NOG | 0.01 | 0/4 | 3/3/15/20 | 0 | — | — | — | — | |||
| CD34− | 98.8 | 3/NOG | 0.5 | 0/3 | 13/18/20 | 0 | — | — | — | — | |
| 3/NOG | 0.2 | 0/3 | 3/6/15/20 | 0 | — | — | — | — | |||
| 4/NOG | 0.1 | 0/4 | 12/15/18/20 | 0 | — | — | — | — | |||
| 4/NOG | 0.01 | 0/4 | 6/7/20/20 | 0 | — | — | — | — | |||
| 34*†‡ | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 | sT4 |
BM indicates bone marrow; WB (NPMm), Western blot analysis with anti-NPM mutant specific antibody; IHC (NPM), immunohistochemical analysis for NPM1 subcellular localization on hCD45+ cells; —, not applicable; ND, not done; sT4, full details on the characteristics of the engraftment are provided in supplemental Table 4; NOD, NOD/SCID; and NOG, NOD/SCID/IL2Rγnull.
Experiments to calculate frequency of LIC were performed with these samples (see Table 5).
Paired experiments using corresponding doses of CD34+ versus CD34− cells were performed with these samples (Table 5, details of patients 17 and 34).
Higher cell doses of CD34− cells gave rise to engraftment as pattern 4 (see “Patterns of engraftment of CD34− cells from NPM1-mutated AML in immunocompromised mice” for description; Table 4).
Used for serial transplantation experiments.
FLT3-ITD+.
FLT3-ITD−.
Normal engraftment.
Mixed normal-leukemic engraftment.
Positive for NPM1 mutation by genomic DNA fragment analysis.
Evaluated only at immunohistochemistry.
In the same mice areas of leukemia (NPMc+) and normal engraftment (NPMc−) were detected.
In the same mice foci of NPMc+, cells were detected within areas of normal hemopoiesis.
Leukemic engraftment of CD34+ cells from NPM1-mutated AML in immunodeficient mice. (A-D) Bone marrow paraffin sections from a NOG mouse, inoculated (12 weeks before) with 2 × 106 CD34+ cells (purity 98.3%) isolated from 1 patient (patient 17) with NPM1-mutated AML, showing infiltration by human cells, which are positive at immunostaining with a specific anti–human CD45 antibody (A), display a myeloid phenotype (myeloperoxidase, MPO-positive; B) and aberrant cytoplasmic expression of nucleophosmin (NPM; C arrow). Double arrows in panel C indicate a normal osteoblast with nucleus-restricted NPM on the endosteal surface of bone. As expected, nucleolin/C23 was also restricted to the nucleus (D, arrow). (A-D) APAAP; hematoxylin counterstaining. Images were collected using an Olympus B61 microscope and a UPlanApo 20×/0.70 (A-B) and a UPlan FI 100×/1.3 NA oil objective (C-D) Camedia 4040, Dp_soft Version 3.2; and Adobe Photoshop 7.0. (E) Flow cytometric analysis of murine bone marrow confirmed engraftment of human cells (hCD45, 52%) which are prevalently CD33+ (92.7%) and express the monocytic marker CD11b (18.6%). CD34/CD38 staining pattern of engrafted hCD45+ cells is also shown (E right panels). CD34+/CD38− cells (gate on F4) were also CD123+ (E right bottom panel). (F) May-Grünwald-Giemsa staining of a cell cytospin preparation from murine bone marrow of the same case (patient 17) showing large-size leukemic cells with monocytoid appearance (cleaved nucleus, basophilic cytoplasm; arrow) admixed with cells of murine origin (double arrow indicate a normal murine polymorphonucleated cell). (G) WB analysis with anti-NPM mutant specific antibodies (anti-NPMm) of mouse bone marrow cells confirmed expression of NPM1 mutated protein in cells engrafted in mice (lane 2) as well as in the original AML patient sample (lane 3). Positive (OCI/AML3, lane 1) and negative (U937, lane 4) controls are shown.
Leukemic engraftment of CD34+ cells from NPM1-mutated AML in immunodeficient mice. (A-D) Bone marrow paraffin sections from a NOG mouse, inoculated (12 weeks before) with 2 × 106 CD34+ cells (purity 98.3%) isolated from 1 patient (patient 17) with NPM1-mutated AML, showing infiltration by human cells, which are positive at immunostaining with a specific anti–human CD45 antibody (A), display a myeloid phenotype (myeloperoxidase, MPO-positive; B) and aberrant cytoplasmic expression of nucleophosmin (NPM; C arrow). Double arrows in panel C indicate a normal osteoblast with nucleus-restricted NPM on the endosteal surface of bone. As expected, nucleolin/C23 was also restricted to the nucleus (D, arrow). (A-D) APAAP; hematoxylin counterstaining. Images were collected using an Olympus B61 microscope and a UPlanApo 20×/0.70 (A-B) and a UPlan FI 100×/1.3 NA oil objective (C-D) Camedia 4040, Dp_soft Version 3.2; and Adobe Photoshop 7.0. (E) Flow cytometric analysis of murine bone marrow confirmed engraftment of human cells (hCD45, 52%) which are prevalently CD33+ (92.7%) and express the monocytic marker CD11b (18.6%). CD34/CD38 staining pattern of engrafted hCD45+ cells is also shown (E right panels). CD34+/CD38− cells (gate on F4) were also CD123+ (E right bottom panel). (F) May-Grünwald-Giemsa staining of a cell cytospin preparation from murine bone marrow of the same case (patient 17) showing large-size leukemic cells with monocytoid appearance (cleaved nucleus, basophilic cytoplasm; arrow) admixed with cells of murine origin (double arrow indicate a normal murine polymorphonucleated cell). (G) WB analysis with anti-NPM mutant specific antibodies (anti-NPMm) of mouse bone marrow cells confirmed expression of NPM1 mutated protein in cells engrafted in mice (lane 2) as well as in the original AML patient sample (lane 3). Positive (OCI/AML3, lane 1) and negative (U937, lane 4) controls are shown.
The leukemic nature of all hCD45+ engrafted cells was further supported in 7 of 10 cases where the morphology and immunophenotype of engrafted cells paralleled that of patient's primary AML (Table 2; supplemental Table 4). In these cases, all engrafted cells were myeloid (hCD45+/CD33+ and myeloperoxidase-positive; Figure 4B,E-F). In contrast, no hCD45+ cells coexpressing CD19 and/or CD20 (suggestive of normal engraftment) were detected in the great majority of evaluated mice (Table 2; supplemental Table 4). Conclusive evidence that all human engrafted cells belonged to the leukemic clone came from the immunohistochemical demonstration that they expressed cytoplasmic NPM/nuclear C23 (Figure 4C-D), the hallmark feature of human NPM1-mutated AML cells. Moreover, in 5 of 7 cases (patients 4, 10, 17, 22R, and 23), the WB and/or molecular analysis showed a mutated NPM1 protein and/or gene (Figure 4G; Table 2; supplemental Table 4). Although CD34+ cells from 1 patient (patient 17) did not show the ability to produce AML colonies in in vitro colony-forming cell (CFC) assay (data not shown), cells recovered from primary mice recipients demonstrated self-renewal capacity in serial transplantations (Table 3: patients 4, 10, 11, 17, and 22R).
Summary of serial transplantations experiments
| Patient code . | Engrafted cell fraction in primary recipients* . | No./mice . | No. of cells/mice, ×106† . | No. of mice with AML/no. of mice evaluated . | Time of evaluation, w . | % hCD45 in mice BM‡ . |
|---|---|---|---|---|---|---|
| 4 | CD34+ | 4/NOD | 2 | 4/4§ | 6 | 87/15/89/87.5 |
| 10 | CD34+ | 1/NOD | 0.6 | 1/1 | 18 | 94 |
| 11 | CD34+ | 1/NOD | 1.6 | 1/1 | 18 | 3 |
| 17‖ | CD34+ | 2/NOG¶ | 0.125¶ | 1/2 | 25 | 0.12 |
| 17‖ | CD34+ | 4/NOG# | 0.125# | 0/4 | 25 | 0 |
| 22R | CD34+ | 4/NOG | 0.52 | 4/4 | 11 | 7.1/6/8.3/7 |
| 8R | CD34− (pattern 2) | 3/NOD | 1 | 3/3 | 6.5 | 6.7/1.2/0.5 |
| 23 | CD34− (pattern 2) | 3/NOG | 4/4/2 | 3/3 | 12 | 60.5/79/69.5 |
| 22 | CD34− (pattern 3) | 2/NOG | 2.5 | 0/2 | 8 | 0 |
| 22 | CD34− (pattern 3) | 2/NOG | 1 | 0/2 | 12 | 0 |
| 27 | CD34− (pattern 3) | 1/NOG | 0.5 | 0/1 | 12 | 0 |
| Patient code . | Engrafted cell fraction in primary recipients* . | No./mice . | No. of cells/mice, ×106† . | No. of mice with AML/no. of mice evaluated . | Time of evaluation, w . | % hCD45 in mice BM‡ . |
|---|---|---|---|---|---|---|
| 4 | CD34+ | 4/NOD | 2 | 4/4§ | 6 | 87/15/89/87.5 |
| 10 | CD34+ | 1/NOD | 0.6 | 1/1 | 18 | 94 |
| 11 | CD34+ | 1/NOD | 1.6 | 1/1 | 18 | 3 |
| 17‖ | CD34+ | 2/NOG¶ | 0.125¶ | 1/2 | 25 | 0.12 |
| 17‖ | CD34+ | 4/NOG# | 0.125# | 0/4 | 25 | 0 |
| 22R | CD34+ | 4/NOG | 0.52 | 4/4 | 11 | 7.1/6/8.3/7 |
| 8R | CD34− (pattern 2) | 3/NOD | 1 | 3/3 | 6.5 | 6.7/1.2/0.5 |
| 23 | CD34− (pattern 2) | 3/NOG | 4/4/2 | 3/3 | 12 | 60.5/79/69.5 |
| 22 | CD34− (pattern 3) | 2/NOG | 2.5 | 0/2 | 8 | 0 |
| 22 | CD34− (pattern 3) | 2/NOG | 1 | 0/2 | 12 | 0 |
| 27 | CD34− (pattern 3) | 1/NOG | 0.5 | 0/1 | 12 | 0 |
R indicates relapse.
hCD45+ cell equivalent.
Human cells were all hCD45+/CD33+ and expressed cytoplasmic NPM1 at immunohistochemical analysis.
From these mice, we obtained engraftment up to quaternary recipients.
Human cells recovered from engrafted mice were sorted in CD34+ and CD34− cell fractions and inoculated separately.
CD34+.
CD34−.
Interestingly, in 3 of 10 cases (patients 30, 32, and 34), immunohistochemistry and flow cytometry showed either normal or mixed (normal plus leukemic) engraftment. The latter pattern of engraftment was characterized by areas of bone marrow infiltrated by human NPMc+ AML cells adjacent to areas of normal human trilineage hematopoiesis (Table 2; Figure 5). Molecular analysis (performed on the whole cell population recovered by bone flushing) confirmed NPM1-mutation in all cases (Table 2). AML development was usually seen later than normal human hematopoietic engraftment (Table 2). Interestingly, flow cytometry of original MNCs from patients 30, 32, and 34 showed a relevant subpopulation of CD34bright/CD38−/CD123−/CD33− cells, which probably represent normal HSCs (Figure 2E; supplemental Table 2). Furthermore, in CFC assay, CD34+ cells from 2 of these cases (patients 32 and 34) produced mixed and erythroid colonies, which, because of germline NPM1 gene (data not shown), are normal in origin.
Mixed (normal and leukemic) engraftment of CD34+ cells from NPM1-mutated AML-immunodeficient mice. (A-F) Bone marrow paraffin sections from a vertebral body of an NOG mouse, inoculated (18 weeks before) with 1 × 106 CD34+ cells (purity 79%) from patient 30 (Table 2). An area in the bottom part of panel A (square) is packed with leukemic cells expressing hCD45 (B left square), cytoplasmic NPM1 (C) and nucleus-restricted C23 (D). Another area from the same section shows involvement by normal human hemopoietic cells (square in the top part of panel A), expressing CD45 (B right square), nucleus-restricted NPM1 and C23 (E-F single and double arrows). (A) Hematoxylin-eosin. (B-F) APAAP, hematoxylin counterstaining. Images were collected using an Olympus B61 microscope and a UPlanApo 20×/0.70 (A-B), UPlanApo 40×/0.85 (C-F); Olympus E330-ADU1.2× camera; and Adobe Photoshop 7.0.
Mixed (normal and leukemic) engraftment of CD34+ cells from NPM1-mutated AML-immunodeficient mice. (A-F) Bone marrow paraffin sections from a vertebral body of an NOG mouse, inoculated (18 weeks before) with 1 × 106 CD34+ cells (purity 79%) from patient 30 (Table 2). An area in the bottom part of panel A (square) is packed with leukemic cells expressing hCD45 (B left square), cytoplasmic NPM1 (C) and nucleus-restricted C23 (D). Another area from the same section shows involvement by normal human hemopoietic cells (square in the top part of panel A), expressing CD45 (B right square), nucleus-restricted NPM1 and C23 (E-F single and double arrows). (A) Hematoxylin-eosin. (B-F) APAAP, hematoxylin counterstaining. Images were collected using an Olympus B61 microscope and a UPlanApo 20×/0.70 (A-B), UPlanApo 40×/0.85 (C-F); Olympus E330-ADU1.2× camera; and Adobe Photoshop 7.0.
Our findings clearly indicate that the small fraction of CD34+ cells from most NPM1-mutated AML cases generate a leukemia showing the same morphologic and immunohistochemical features (aberrant cytoplasmic NPM1) as the original patient's AML.
Patterns of engraftment of CD34− cells from NPM1-mutated AML in immunocompromised mice
The main aim of this study was to characterize the small fraction of CD34+ cells in NPM1-mutated AML. However, there is experimental evidence that, in some AML cases37,38 (including NPM1-mutated AML21 ), also the CD34− population may contain the LICs. Therefore, we assessed the engraftment capability of CD34− cells from 15 NPM1-mutated AMLs (Table 4; supplemental Table 5). Inoculation of these cells in immunocompromised mice led to 4 different patterns, which varied also according to the injected cell dose:
Heterogeneous patterns of engraftment of CD34− cells from NPM1-mutated AML in immunocompromised mice
| Patient code . | % CD34+ in original sample . | Purity of CD34− cells . | No./type mice evaluated . | No. of cells/mice, × 106 . | No. of mice engrafted/no. evaluated . | Time of evaluation, w . | % hCD45+ in mice BM* . | % hCD45+/CD33+ (gated on hCD45)* . | Pattern of engraftment† . | WB (NPMm) . | NPM1 gene status‡ . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 8R | 1.9 | 98.5 | 2/NOD | 2 | 2/2 | 16 | 36/73‖ | 97.5/99 | 2 | + | ND |
| 23 | 1.7 | 99 | 1/NOG | 0.2 | 1/1 | 15 | 21 | 0.57 | 2 | + | Mutated |
| 21 | 0.02 | 99.99 | 2/NOG | 2 | 2/2 | 6 | 29/7.5¶ | 13.8/95 | 4 | ND | ND |
| 22§ | 4.5 | 99.9 | 6/NOG | 10 | 6/6 | 4/5/6/7/8/9 | 75/72/86‖/82/87#/94‖ | 82/85/93/90/89/97 | 3 | + | Mutated |
| 27 | 0.02 | 99.98 | 3/NOG | 10 | 3/3 | 4/6/8 | 33/64‖/42 | 73/86/56 | 3 | + | Mutated |
| 28§ | 0.07 | 99.98 | 1/NOG | 10 | 1/1 | 5 | 42 | 46 | 3 | ND | Mutated |
| 3/NOG | 2 | 3/3 | 6/9/13 | 4.3/12/5¶ | 30/35/82 | 3 | ND | Mutated | |||
| 29§ | 0.5 | 99.9 | 1/NOG | 10 | 1/1 | 6 | 14¶ | 0.7 | 4 | NA | NA |
| 4/NOG | 4 | 4/4 | 5/6/7/14 | 16/ND/2.7/10.7¶ | 2.2/–/1.7/1 | 4 | NA | NA | |||
| 2/NOG | 2 | 2/2 | 7/14 | 1.6/16 | 4/2.2 | 4 | NA | NA | |||
| 34§ | 5 | 96 | 2/NOG | 10 | 2/2 | 4 | 0.61/4.9 | –/0.2 | 4 | NA | NA |
| 36 | 0.26 | 99.98 | 3/NOG | 10 | 3/3 | 6 | 4.7/2.3/5.2¶ | 0.3/5.9/9.9 | 4 | NA | NA |
| 37§ | 0.2 | 99.8 | 4/NOG | 10 | 3/4 | 5/7/8/11 | 1.8/3.2/0/1.65 | 95.8/98.7/–/96 | 4 | NA | NA |
| 38§ | 0.16 | 99.9 | 3/NOG | 10 | 3/3 | 7 | 0.68/1.2/20¶ | 1.8/2.5/1 | 4 | NA | NA |
| Patient code . | % CD34+ in original sample . | Purity of CD34− cells . | No./type mice evaluated . | No. of cells/mice, × 106 . | No. of mice engrafted/no. evaluated . | Time of evaluation, w . | % hCD45+ in mice BM* . | % hCD45+/CD33+ (gated on hCD45)* . | Pattern of engraftment† . | WB (NPMm) . | NPM1 gene status‡ . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 8R | 1.9 | 98.5 | 2/NOD | 2 | 2/2 | 16 | 36/73‖ | 97.5/99 | 2 | + | ND |
| 23 | 1.7 | 99 | 1/NOG | 0.2 | 1/1 | 15 | 21 | 0.57 | 2 | + | Mutated |
| 21 | 0.02 | 99.99 | 2/NOG | 2 | 2/2 | 6 | 29/7.5¶ | 13.8/95 | 4 | ND | ND |
| 22§ | 4.5 | 99.9 | 6/NOG | 10 | 6/6 | 4/5/6/7/8/9 | 75/72/86‖/82/87#/94‖ | 82/85/93/90/89/97 | 3 | + | Mutated |
| 27 | 0.02 | 99.98 | 3/NOG | 10 | 3/3 | 4/6/8 | 33/64‖/42 | 73/86/56 | 3 | + | Mutated |
| 28§ | 0.07 | 99.98 | 1/NOG | 10 | 1/1 | 5 | 42 | 46 | 3 | ND | Mutated |
| 3/NOG | 2 | 3/3 | 6/9/13 | 4.3/12/5¶ | 30/35/82 | 3 | ND | Mutated | |||
| 29§ | 0.5 | 99.9 | 1/NOG | 10 | 1/1 | 6 | 14¶ | 0.7 | 4 | NA | NA |
| 4/NOG | 4 | 4/4 | 5/6/7/14 | 16/ND/2.7/10.7¶ | 2.2/–/1.7/1 | 4 | NA | NA | |||
| 2/NOG | 2 | 2/2 | 7/14 | 1.6/16 | 4/2.2 | 4 | NA | NA | |||
| 34§ | 5 | 96 | 2/NOG | 10 | 2/2 | 4 | 0.61/4.9 | –/0.2 | 4 | NA | NA |
| 36 | 0.26 | 99.98 | 3/NOG | 10 | 3/3 | 6 | 4.7/2.3/5.2¶ | 0.3/5.9/9.9 | 4 | NA | NA |
| 37§ | 0.2 | 99.8 | 4/NOG | 10 | 3/4 | 5/7/8/11 | 1.8/3.2/0/1.65 | 95.8/98.7/–/96 | 4 | NA | NA |
| 38§ | 0.16 | 99.9 | 3/NOG | 10 | 3/3 | 7 | 0.68/1.2/20¶ | 1.8/2.5/1 | 4 | NA | NA |
BM indicates bone marrow; NOD, NOD/SCID; NOG, NOD/SCID/IL2Rγnull; WB (NPMm), Western blot analysis with anti-NPM mutant specific antibody; R, relapse; ND, not done; and NA, not applicable
Evaluated by flow cytometry on cells recovered from femurs or tibias.
See “Patterns of engraftment of CD34− cells from NPM1-mutated AML in immunocompromised mice” for description of patterns of engraftment.
Evaluated by genomic DNA fragment analysis.
Lower cell doses did not engraft (supplemental Table 5).
Used for serial transplantation experiments.
Human cell infiltration of mice BM was much higher when evaluated at immunohistochemistry.
FLT3-ITD-negative.
Pattern 1.
CD34− cells from 10 patients did not engraft in mice when inoculated at doses less than or equal to 1 × 106 (supplemental Table 5). In contrast, CD34+ cells from 4 of these cases generated typical NPMc+ AML, when injected in mice at the same doses (Table 2: patients 17, 30, 32, and 34; supplemental Table 4). Moreover, CD34− cells from patients 17, 32, and 34 did not outgrow into AML colonies in CFC assays (data not shown).
Pattern 2.
Engraftment of CD34− as typical NPMc+ AML (similar to that observed injecting mice with purified CD34+ cells) was observed in 2 cases (not shown). In both of them, CD34− cells were purified from NPM1-mutated AML at relapse (Table 4: patients 8R and 23). Self-renewal capability of CD34− cells from these cases was demonstrated after transfer to secondary recipients (Table 3).
Pattern 3.
Inoculation of CD34− fraction (≥ 2 × 106 cells) from 3 cases (Table 4: patients 22, 27, and 28) resulted into marrow engraftment by hCD45+/hCD33+ cells (Figure 6A), which, in tissue sections, consisted of 2 populations (Figure 6B-D): (1) myeloperoxidase-positive cells located close to bone trabeculae that exhibited weak cytoplasmic NPM1 positivity (not shown); and (2) mature CD68+ histiocytes that were located in the central area of bone marrow. In all cases, detection of NPM1 mutant protein and gene (Figure 6 E-F) proved the leukemic nature of engrafted cells. These findings possibly reflect short-term engraftment by leukemic cells devoid of self-renewal potential that differentiated into mature elements. Indeed, serial transplantation experiments performed with cells from 2 of 3 cases (Table 3: patients 22 and 27) did not result in further engraftment in secondary recipients.
Patterns of engraftment of CD34− cells from NPM1-mutated AML in immunodeficient mice. (A-F) Example of engraftment (pattern 3) in NOG mice inoculated with 10 × 106 CD34− cells (purity 99.9%) from patient 22. (A) Flow cytometric analysis of bone marrow (6 weeks after inoculum) showing engraftment of human myeloid (93% hCD45+/CD33+) cells, which appeared CD34− (0.3% CD34+ cells) and CD11b+ (70.8% hCD45+/CD11b+). A small percentage of T cells (5.9% hCD45+/CD3+) was also detected. (B-D) Tibia paraffin sections showing massive marrow infiltration by a double population: (1) mononuclear cells close to bone trabeculae (B double arrows; inset, from a different section), which are MPO+ and PGM1(CD68)+ (C-D double arrows); (2) mature histiocytes located in the central area of bone marrow (B single arrow), which are MPO− and PGM1+ (C-D single arrows). Asterisk in panel B inset and in panel C indicates bone area. (E-F) Leukemic origin of these cells was confirmed by WB with anti-NPM mutant (anti-NPMm) antibody (E) and genomic DNA fragment analysis showing double peaks (F). (G-I) Example of engraftment (pattern 4) in NOG mice inoculated 6 weeks before with 2 × 106 CD34− cells (purity 99.99%) isolated from patient 21. Tibia paraffin sections showing marrow infiltration by a mixed population of mature histiocytes (single arrow) and lymphocytes (double arrows). Immunostaining for hCD45 confirmed the human origin of these cells. (B,G) Hematoxylin and eosin. (C-D,H-I) APAAP; hematoxylin counterstaining. Images were collected using an Olympus B61 microscope and a UPlanApo 40×/0.85 (B-D,G-H) and a UPlan FI 100×/1.3 NA oil objective (B inset, I); Olympus E330-ADU1.2× camera; and Adobe Photoshop 7.0.
Patterns of engraftment of CD34− cells from NPM1-mutated AML in immunodeficient mice. (A-F) Example of engraftment (pattern 3) in NOG mice inoculated with 10 × 106 CD34− cells (purity 99.9%) from patient 22. (A) Flow cytometric analysis of bone marrow (6 weeks after inoculum) showing engraftment of human myeloid (93% hCD45+/CD33+) cells, which appeared CD34− (0.3% CD34+ cells) and CD11b+ (70.8% hCD45+/CD11b+). A small percentage of T cells (5.9% hCD45+/CD3+) was also detected. (B-D) Tibia paraffin sections showing massive marrow infiltration by a double population: (1) mononuclear cells close to bone trabeculae (B double arrows; inset, from a different section), which are MPO+ and PGM1(CD68)+ (C-D double arrows); (2) mature histiocytes located in the central area of bone marrow (B single arrow), which are MPO− and PGM1+ (C-D single arrows). Asterisk in panel B inset and in panel C indicates bone area. (E-F) Leukemic origin of these cells was confirmed by WB with anti-NPM mutant (anti-NPMm) antibody (E) and genomic DNA fragment analysis showing double peaks (F). (G-I) Example of engraftment (pattern 4) in NOG mice inoculated 6 weeks before with 2 × 106 CD34− cells (purity 99.99%) isolated from patient 21. Tibia paraffin sections showing marrow infiltration by a mixed population of mature histiocytes (single arrow) and lymphocytes (double arrows). Immunostaining for hCD45 confirmed the human origin of these cells. (B,G) Hematoxylin and eosin. (C-D,H-I) APAAP; hematoxylin counterstaining. Images were collected using an Olympus B61 microscope and a UPlanApo 40×/0.85 (B-D,G-H) and a UPlan FI 100×/1.3 NA oil objective (B inset, I); Olympus E330-ADU1.2× camera; and Adobe Photoshop 7.0.
Pattern 4.
Engraftment of CD34− cells as a mixed population of mature-looking human CD68+ histiocytes (Table 4; Figure 6G-I) and CD3+ lymphocytes (not shown) in the absence of cells with clear blastic appearance was observed in 6 cases inoculated with more than or equal to 2 × 106 cells. The inability to recover enough histiocytes because of their high cohesivity precluded molecular studies. Thus, the nature (leukemic vs normal) of these cells remains uncertain.
Frequency of LICs in CD34+ and CD34− cell fractions in NPM1-mutated AML
The presence and frequency of LICs in specific cell fractions were assessed by limiting-dilution transplantation assay in 3 cases (patients 17, 22R, and 34). Results are shown in Table 5.
LICs frequency in CD34+ and CD34− cell fractions of NPM1-mutated AML
| Patient code . | Fraction (purity) . | Dose, ×106 . | LIC frequency (95% CI) . | LICs/106 MNCs . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 . | 1 . | 0.5 . | 0.2 . | 0.1 . | 0.01 . | 0.001 . | ||||
| 17 | CD34+ | — | — | — | 3/4 | 4/4 | 3/4 | 0/2 | 1 in 50 400 | 5.4 |
| (98.3%) | (19 967–127 223) | |||||||||
| 17 | CD34− | — | — | — | 0/3 | 0/4 | 0/4 | — | — | — |
| (90%) | ||||||||||
| 22R | CD34+ | 3/3 | 2/2 | — | — | 4/4 | 0/4 | — | 1 in 41704 | 18 |
| (99.9%) | (13 820–125 855) | |||||||||
| 34 | CD34+ | — | 2/3 | 3/5 | 4/4 | 0/4 | — | — | 1 in 448 907 | 0.11 |
| (96.5%) | (214 463–939 640 | |||||||||
| 34 | CD34− | — | 0/4 | 0/4 | 0/4 | 0/4 | — | — | — | — |
| (96%) | ||||||||||
| Patient code . | Fraction (purity) . | Dose, ×106 . | LIC frequency (95% CI) . | LICs/106 MNCs . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 . | 1 . | 0.5 . | 0.2 . | 0.1 . | 0.01 . | 0.001 . | ||||
| 17 | CD34+ | — | — | — | 3/4 | 4/4 | 3/4 | 0/2 | 1 in 50 400 | 5.4 |
| (98.3%) | (19 967–127 223) | |||||||||
| 17 | CD34− | — | — | — | 0/3 | 0/4 | 0/4 | — | — | — |
| (90%) | ||||||||||
| 22R | CD34+ | 3/3 | 2/2 | — | — | 4/4 | 0/4 | — | 1 in 41704 | 18 |
| (99.9%) | (13 820–125 855) | |||||||||
| 34 | CD34+ | — | 2/3 | 3/5 | 4/4 | 0/4 | — | — | 1 in 448 907 | 0.11 |
| (96.5%) | (214 463–939 640 | |||||||||
| 34 | CD34− | — | 0/4 | 0/4 | 0/4 | 0/4 | — | — | — | — |
| (96%) | ||||||||||
Mice used were NOD/SCID/IL2Rγnull. Values indicate no. of mice with AML/no. of mice evaluated. Details on immunophenotypic and immunohistochemical analysis of engrafted mice are shown in supplemental Table 4. Time of evaluation (weeks): +17 (patient 17); +10 (patient 22R); +4 (patient 34).
R indicates relapse; and —, not done.
CD34+ cells generate a CD34−NPM1-mutated AML in immunocompromised mice
Because down-regulation of CD34 at both RNA7,8 and protein1 level is a unique characteristic of NPM1-mutated AML, we monitored CD34 expression after engraftment of purified hCD34+ leukemic cells in immunocompromised mice.
Notably, transplanted hCD34+ cells gave rise to an NPM1-mutated AML mainly consisting of CD34− cells. In 6 of 9 evaluable cases (patients 4, 13, 17, 23, 30, and 32, Table 2), a small pool of CD34+ cells similar to that observed in the original patient's sample was also detected. In 3 cases (patients 10, 11, and 22R, Table 2), the percentage of CD34+ leukemic cells (and the proportion of CD34+/CD38− and CD34+/CD38+ subsets) in engrafted tumors was even lower that that detected in the original patient's sample, probably reflecting the different influence of mice microenvironment in maintaining stem cell phenotype and pool size. As an example, the bone marrow sample from patient 10 (Table 2) at diagnosis contained 27% AML cells expressing CD34 at low intensity (Figure 7A), whereas only 1.9% of hCD45+/CD33+ leukemic cells engrafted in mice (16 weeks after the inoculum) expressed CD34 (Table 2; Figure 7B). Thus, most of mice leukemic bulk consisted of NPM1-mutated (Figure 7C), CD34− cells expressing cytoplasmic NPM1 (Figure 7D-F). Interestingly, in some mice, leukemic cells showing cytoplasmic (dot-like) CD34 were seen (supplemental Figure 4).
CD34+ cells from NPM1-mutated AML generate CD34− NPMc+ AML in mice. (A-F) Flow cytometry and immunohistochemistry analysis of CD34 antigen expression in leukemia developed in immunodeficient mice (NOD/SCID) inoculated with CD34+ cells (purity 98.5%) isolated from 1 patient (patient 10) with NPM1-mutated AML (27% CD34+ cells in the original sample; A). Flow cytometric analysis of mice bone marrow (BM;16 weeks after the inoculum) showing engraftment of myeloid (CD33+ and CD117+) human cells, which appeared mainly CD34− (1.9% CD34+ cells; B). The leukemic nature of these cells was confirmed by WB with anti-NPM mutant antibody (C), as well as morphologic analysis (D hematoxylin and eosin) and expression of nucleophosmin (NPM) in the cytoplasm of human leukemic cells on bone marrow paraffin sections (E, arrow). As staining control, NPM was characteristically nucleus-restricted in normal murine cells (E, double arrows). The asterisk in (E) indicates an empty space originally filled by bone. CD34 immunostaining was negative in leukemic cells (F arrow). (E-F: APAAP; hematoxylin counterstaining). All images were collected using an Olympus B61 microscope and a UPlan FI 100×/1.3 NA oil objective; Camedia 4040, Dp_soft Version 3.2; and Adobe Photoshop 7.0. A vertical line has been inserted in panel C to indicate a repositioned gel lane. (G-I) Serial flow cytometric evaluation of CD34 antigen expression in human leukemic cells grown in immunodeficient mice (NOD/SCID/IL2Rγnull) killed at 9 (H) and 12 (I) weeks after injection of the same number of CD34+ cells (2 × 106, purity 98.3%) isolated from 1 patient (patient 17) with NPM1-mutated AML (27% CD34+ cells in the original sample; G).
CD34+ cells from NPM1-mutated AML generate CD34− NPMc+ AML in mice. (A-F) Flow cytometry and immunohistochemistry analysis of CD34 antigen expression in leukemia developed in immunodeficient mice (NOD/SCID) inoculated with CD34+ cells (purity 98.5%) isolated from 1 patient (patient 10) with NPM1-mutated AML (27% CD34+ cells in the original sample; A). Flow cytometric analysis of mice bone marrow (BM;16 weeks after the inoculum) showing engraftment of myeloid (CD33+ and CD117+) human cells, which appeared mainly CD34− (1.9% CD34+ cells; B). The leukemic nature of these cells was confirmed by WB with anti-NPM mutant antibody (C), as well as morphologic analysis (D hematoxylin and eosin) and expression of nucleophosmin (NPM) in the cytoplasm of human leukemic cells on bone marrow paraffin sections (E, arrow). As staining control, NPM was characteristically nucleus-restricted in normal murine cells (E, double arrows). The asterisk in (E) indicates an empty space originally filled by bone. CD34 immunostaining was negative in leukemic cells (F arrow). (E-F: APAAP; hematoxylin counterstaining). All images were collected using an Olympus B61 microscope and a UPlan FI 100×/1.3 NA oil objective; Camedia 4040, Dp_soft Version 3.2; and Adobe Photoshop 7.0. A vertical line has been inserted in panel C to indicate a repositioned gel lane. (G-I) Serial flow cytometric evaluation of CD34 antigen expression in human leukemic cells grown in immunodeficient mice (NOD/SCID/IL2Rγnull) killed at 9 (H) and 12 (I) weeks after injection of the same number of CD34+ cells (2 × 106, purity 98.3%) isolated from 1 patient (patient 17) with NPM1-mutated AML (27% CD34+ cells in the original sample; G).
In patients 17 and 22R, we were able to track CD34 expression in engrafted leukemia at different time points. Interestingly, starting from CD34+ cells with a purity of 98.3% and 99.9%, respectively, we observed a progressive reduction of the percentage of CD34+ cells within the leukemic bulk with time (eg, patient 17, from 52% at 9 weeks to 35% at 12 weeks and 14% at 20 weeks; Figure 5G-I; Table 2) and outgrowth of leukemia from 15.8% to 52% and 80% at correspondent time points (Table 2).
Our findings suggest the CD34− phenotype of NPM1-mutated AML developing in mice may be the result of CD34 down-regulation.
Discussion
Most NPM1-mutated AML patients are CD34−.1 Here, we demonstrate that the small fraction of CD34+ cells in NPM1-mutated AML expresses CD34 at low intensity, exhibits variable expression of CD38, carries the mutated NPM1 gene/protein, and shows aberrant NPM1 cytoplasmic expression.
As the mutated NPM1 gene and protein were detected in the CD34+/CD38− and CD34+/CD38+ subsets, they both belong to the leukemic clone. CD34+/CD38− cells from NPM1-mutated AML usually displayed LSC immunophenotypic features (ie, CD123 and CD33 expression29,30 and absence of CD9031 ). The CD34+ cell fraction consistently engrafted in immunocompromised mice as AML exhibiting the same characteristics as the patient's primary leukemic cells (mutated/cytoplasmic NPM1 and CD34 negativity), suggesting it contains the LICs (or LSCs), which characteristically recapitulate human AML in mice.39 Moreover, as recently reported by Taussig et al,21 we found that also CD34− cells from a few NPM1-mutated AML patients had significant LIC potential in immunocompromised mice.
Definitive evidence that CD34+ cells belong to the leukemic clone and contain the LICs mainly apply to NPM1-mutated AML cases containing more than 1% of CD34+ cells. In contrast, the nature of CD34+ cells from our NPM1-mutated AML patients, when they represented less than 1% of MNCs (supplemental Table 1), remains controversial. Indeed, the CD34+/CD38− cell subpopulation from these cases contained a variable percentage of CD34+ cells with normal immunophenotypic features (supplemental Table 2), a finding that is in keeping with previous reports.32,40 In 3 of such patients (not shown), we detected NPM1-mutated protein by WB analysis of purified CD34+ cells, suggesting that the majority of cells may have belonged to the leukemic clone. However, definitive conclusions on this issue can only be drawn from the study of additional cases, possibly searching for the NPM1 mutant by single-cell polymerase chain reaction analysis.
Our findings that in most cases CD34+, but not CD34−, cells generated NPM1-mutated AML in immunocompromised mice are consistent with previous observations in other AML subtypes where the capability to engraft was shown to be mainly related to the CD34+/CD38− fraction.14 Moreover, engraftment of NPM1-mutated AML cells occurred preferentially within the bone marrow endosteal region, where primitive CD34+/CD38− AML cells have been described to home in NOG mice.41 Interestingly, the bone marrow endosteal region was identified as the microenvironmental niche for human primary LSCs42 which, during engraftment, appear to compete with normal hemopoietic stem cells for this area.41 Finally, if the small pool of CD34+ cells in NPM1-mutated AML were of leukemic origin, a clonal outgrowth of them might be expected at relapse. Notably, in 3 NPM1-mutated AML patients studied at diagnosis and relapse, we found a 6.8-, 15.8-, and 16.7-fold increase in CD34+ cell percentage at relapse, respectively (Table 1; supplemental Table 1, patients 2, 8, and 22). Increases in CD34+ cells at relapse have been reported in other AML subtypes.43
In immunocompromised mice, CD34+ cells from NPM1-mutated AML usually generated a leukemic outgrowth similar to the original patient's disease (ie, mainly CD34− cells with few CD34+ leukemic cells). It appears improbable that this small CD34+ cell pool derived from contaminating CD34− leukemic cells, as described for normal hemopoietic stem cells.44 In fact, flow cytometric studies at different time points showed progressive CD34 down-regulation and retention of a small pool of CD34+ leukemic cells that were consistent with a derivation of the leukemic population from CD34+ engrafted cells. Moreover, immunohistochemistry frequently showed dot-like CD34 positivity in cytoplasm, which could reflect CD34 internalization and down-regulation. Further studies are required to clarify the molecular mechanisms of CD34 down-regulation in NPM1-mutated AML.
In normal hemopoiesis, a cell compartment devoid of lineage markers and CD34 antigen with SCID-repopulating ability was isolated,13 where leukemic transformation could conceivably occur.37,38 Recently, Taussig et al21 reported that, in NPM1-mutated AML, the LICs were found only in the CD34− fraction or in both the CD34+ and CD34− subpopulations, suggesting that the phenotype of LICs is more heterogeneous than previously realized.
Our results that CD34− cells from a few NPM1-mutated AML patients showed significant LIC potential in immunocompromised mice is in keeping with the findings by Taussig et al.21 Moreover, use in combination of flow cytometry, histology, and immunohistochemistry showed that leukemic engraftment of CD34− fraction was more heterogeneous than that observed with the CD34+ fraction, giving rise to different patterns. Rarely, CD34− cells generated an AML with the same morphologic and immunohistologic features (cytoplasmic NPM1) as in patients' bone marrow trephines and in mice injected with NPM1-mutated CD34+ cells. More frequently, CD34− cells engrafted as leukemia showing more differentiated morphologic and immunohistologic features. We hypothesize that the latter pattern might be ascribed to a limited proliferative ability of CD34− cells, which allows them to expand in mice, although with a more limited engraftment potential than CD34+ cells. Further studies are needed to address this issue.
In conclusion, in our series of patients, both CD34+ and, at lower extent, CD34− cells exhibited LIC activity. Whether LSCs in NPM1-mutated AML originate from very early progenitors or committed myeloid precursors45 remains to be elucidated. Our studies have biologic and potential clinical implications. The finding that CD34+/CD38− cells from NPM1-mutated AML may harbor the same genetic lesion as the CD34− tumor bulk population adds to the evidence that the NPM1 mutation is a founder genetic lesion defining a new leukemia entity. This evidence includes: (1) specificity of NPM1 mutation for AML among human tumors1 ; (2) mutual exclusion of NPM1 mutation with other AML recurrent cytogenetic abnormalities46 ; (3) secondary nature of chromosomal aberrations in 15% of NPM1-mutated AML47 ; (4) association of NPM1-mutated AML with distinctive gene expression7,8 and microRNA profiles4 ; and (5) results of whole genomic sequencing in AML with normal karyotype.48,49
As LSCs from NPM1-mutated AML strongly express CD33 and CD123, immunotherapy with CD33 and/or CD12350 targeting drugs combined with chemotherapy is an attractive strategy. Development of novel therapeutic approaches is important because, although NPM1-mutated without FLT3-ITD is usually characterized by a favorable prognosis,5 a significant number of patients with NPM1-mutated AML still die of their disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Geraldine A. Boyd for editing the paper, Claudia Tibidò for secretarial assistance, Luca De Carolis, Chiara Balucani, Tiziana Zei, Roberta Iacucci, and Federica Cecchetti for their invaluable technical help, and the personnel of the Animal Facility of University of Perugia for their assistance.
This work was supported by the Associazione Italiana Ricerca Cancro, Fondazione Cassa di Risparmio di Perugia (grants 2007.0099.020 and 2008.020.058), and Fondazione Cassa di Risparmio di Spoleto.
Authorship
Contribution: M.P.M. and B.F. had the original idea for the study, designed experiments, and wrote the paper; V.P. designed and performed experiments, analyzed data, and wrote the paper; C.T. and U.O. performed FACS experiments, flow cytometry, and molecular analyses, analyzed data, and contributed to the writing of the manuscript; F.M. performed mice experiments; E.B., I.G., D.C., and L.B. performed immunophenotypic analysis; M.G. and L.D.V. performed FACS experiments; F.F., M.D.I., and R.C. were responsible for molecular diagnostics and analysis; N.M., R.R., and L.G. processed patient samples and performed WB studies; A.L., R.P., and A.T. performed immunohistochemical studies; and M.D., G.S., F.D.R., and M.F.M. provided patient samples and clinical information and contributed to discussion of data.
Conflict-of-interest disclosure: B.F. applied for a patent on clinical use of NPM1 mutants. The remaining authors declare no competing financial interests.
Correspondence: Brunangelo Falini, Institute of Hematology, University of Perugia, Strada Sant'Andrea della Fratte, Perugia, 06132, Italy; e-mail: faliniem@unipg.it; and Maria Paola Martelli, Institute of Hematology, University of Perugia, Strada Sant'Andrea della Fratte, Perugia, 06132, Italy; e-mail: mpmartelli@libero.it.
References
Author notes
M.P.M. and V.P. contributed equally to this study.

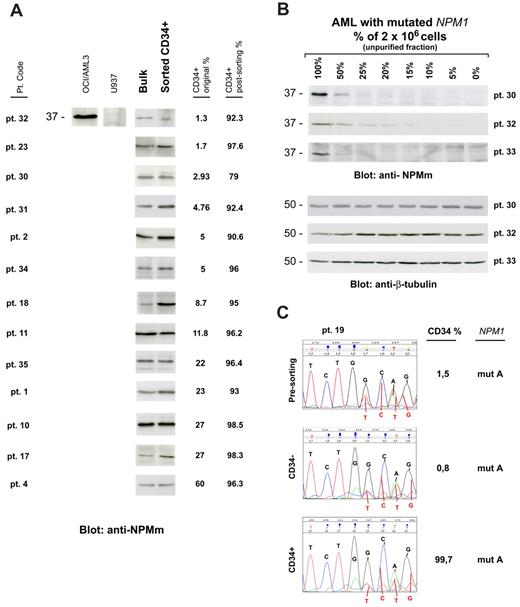
![Figure 2. CD34+/CD38− cells from AML with mutated NPM1 express mutated NPM1 gene and protein and display immunophenotypic features of LSCs. (A) CD34/CD38 flow cytometry staining patterns of peripheral blood MNCs (patients 2, 18, and 22) or whole bone marrow (patients 25 and 26) from the 5 NPM1-mutated AML patients studied for involvement of CD34+/CD38− cells by NPM1 mutation. Flow cytometric analysis was performed on either FACSAria (BD Biosciences; patients 22, 25, and 26) or Cytomics FC500 cytometer (Beckman Coulter; patients 2 and 18), as indicated in “Flow cytometric immunophenotyping.” Concomitant expression of CD123 and CD33 on the CD34+/CD38− cell population is shown in a representative example (patient 18, bottom right panels). Here, within the CD34+/CD38− cell population, only a very small cell fraction (gate Q: ∼ 0.8%) was negative for both CD123 and CD33 that could represent normal residual HSCs. (B) WB showing expression of NPM1 mutant protein in MACS-sorted CD34+ cells from 2 selected cases from Figure 1 (patients 2 and 18) where CD34+/CD38− cells represented almost the whole CD34+ cell population. Here, percentages of CD34+/CD38− and CD34+/CD38+ cells are shown. (C-D) CD34+/CD38− and CD34+/CD38+ cells were FACS-sorted from bone marrow of a patient (patient 25) with AML with cytoplasmic NPM1 with 0.3% CD34+ cells. Expression of CD123 and CD33 antigens in the 2 cell subpopulations is shown (C bottom panels). Molecular analysis for NPM1 and FLT3-ITD mutations was performed on genomic DNA by high-resolution fragment analysis (D). Results obtained on sorted CD34+/CD38− (purity of 99%; i) and CD34+/CD38+ (purity of 99%; ii) cells were compared with the total bone marrow mononuclear cells (BM-MNC; iii). As shown in green, the typical double peak indicating the NPM1 mutation (green arrows) was detectable at similar levels in both populations as well as in the MNCs. Peaks indicating FLT3 gene status (wild-type [wt] and FLT3-ITD [ITD] mutation) are in blue. FLT3-ITD mutation/FLT3-wt ratio (mut/wt) is shown on the right. (E) Flow cytometric analysis of a representative NPM1-mutated AML patient sample (patient 30) showing an evident double-cell population with distinct immunophenotypic features within CD34+/CD38− progenitor cells: (i) CD34+ cell percentage (2.93%) in the leukemic bulk from the patient's peripheral blood (MNCs); (ii) expression of CD38 on the purified CD34+ cells (CD34+ after sorting); and detection of 2 distinct cell subpopulations within CD34+/CD38− cells: CD34bright/CD38−/CD123−/CD33− (iii) versus CD34low/CD38−/CD123+/CD33+ (iv) cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/19/10.1182_blood-2009-08-238899/5/m_zh89991058840002.jpeg?Expires=1769164851&Signature=EdiOolQvpsLcQXuSFFIzrLuJ9LjcG1SjIV1oq8agGCFkQpqPMl3lOF6e~32VtbsuheoX0Iwo1UO-8Xv2IlsMEq2pfqBFpvlCw7NBzNOIrzRuiwgwK~lW2qIUfKxPab3ueu6YenAdj2n4Pa97H7uCsrbtDXWeMM7DeKW4UDy6CQrGiILy5dlCHe6pkZbuM6K9OZcq~wCku0gQId9zlXOaAaa-J~l51SxIbNw~lvCJ3m7sa1SgOajDRcR4qHaV3eRZ7SNTYi6mGeXtrOlX2bKgYsY18p0qZI3F0bPfDV2zqvYfRRIVz0savl5XQNlAebBk39czKvhrN0-6vWrE52Y7EA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

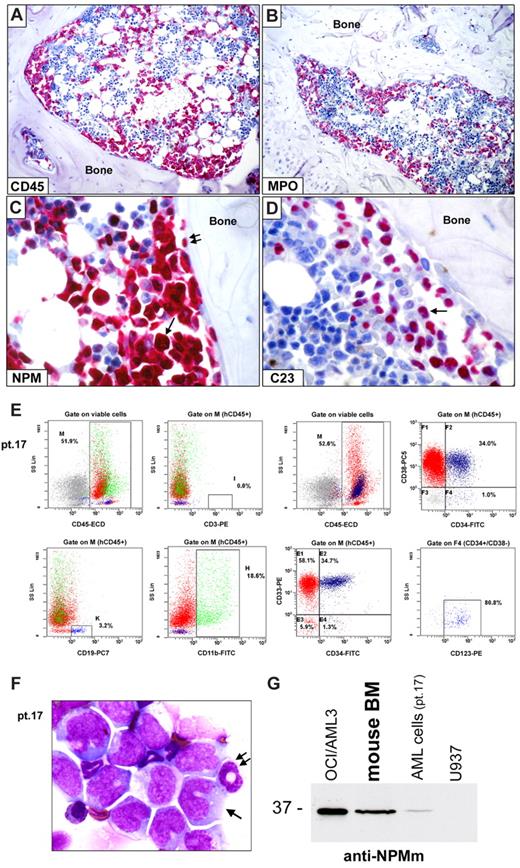
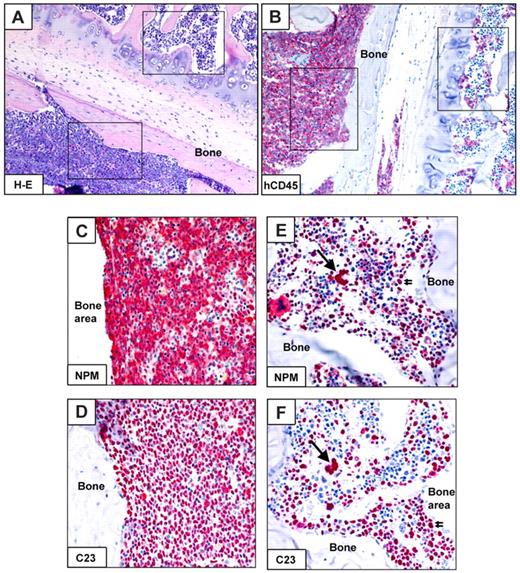
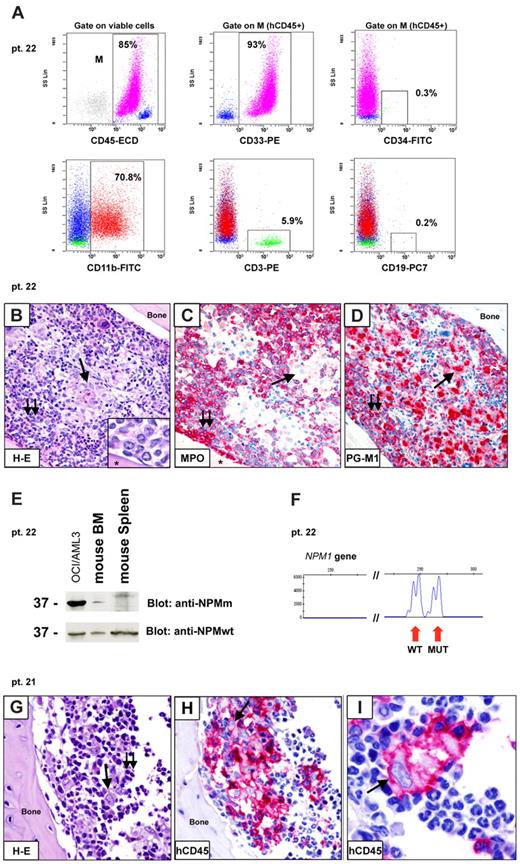
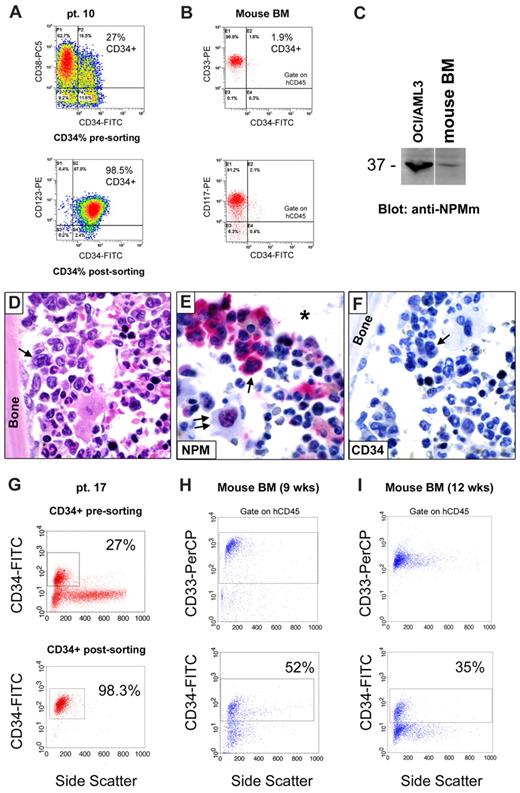
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal