Abstract
Mutations in the TET2 gene are frequent in myeloid disease, although their biologic and prognostic significance remains unclear. We analyzed 355 patients with myelodysplastic syndromes using “next-generation” sequencing for TET2 aberrations, 91 of whom were also subjected to single-nucleotide polymorphism 6.0 array karyotyping. Seventy-one TET2 mutations, with a relative mutation abundance (RMA) ≥ 10%, were identified in 39 of 320 (12%) myelodysplastic syndrome and 16 of 35 (46%) chroni myelomonocytic leukemia patients (P < .001). Interestingly, 4 patients had multiple mutations likely to exist as independent clones or on alternate alleles, suggestive of clonal evolution. “Deeper” sequencing of 96 patient samples identified 4 additional mutations (RMA, 3%-6.3%). Importantly, TET2 mutant clones were also found in T cells, in addition to CD34+ and total bone marrow cells (23.5%, 38.5%, and 43% RMA, respectively). Only 20% of the TET2-mutated patients showed loss of heterozygosity at the TET2 locus. There was no difference in the frequency of genome-wide aberrations, TET2 expression, or the JAK2V617F 46/1 haplotype between TET2-mutated and nonmutated patients. There was no significant prognostic association between TET2 mutations and World Health Organization subtypes, International Prognostic Scoring System score, cytogenetic status, or transformation to acute myeloid leukemia. On multivariate analysis, age (> 50 years) was associated with a higher incidence of TET2 mutation (P = .02).
Introduction
The myelodysplastic syndromes (MDSs) are clonal disorders of hematopoiesis characterized by dysplasia, ineffective hematopoiesis, and peripheral cytopenias. Evolution to acute myeloid leukemia (AML) occurs in approximately 25% of cases.1 Based on the International Prognostic Scoring System (IPSS), patients in low-risk, intermediate-1, intermediate-2 and high-risk subgroups have a median survival of 5.7, 3.5, 1.2, and 0.4 years, respectively.2 The early genetic lesions that lead to clonal hematopoiesis in MDS remain poorly understood,3 with a low frequency of point mutations in genes, such as TP53 (5%-10%), RUNX1 (2-10%), NRAS (10%-15%), and FLT3 (2%-5%).4
Several groups have used single-nucleotide polymorphism (SNP) genotyping to identify genomic aberrations, including uniparental disomy (UPD), in MDS patients, which were undetectable by conventional metaphase cytogenetic analysis.5 We identified a common region of UPD spanning chromosome 4q24 in 8% of MDS patients that has been confirmed by other groups and led to the discovery of mutations in the TET2 gene.6-13 The TET family consists of 3 proteins (TET1, TET2, and TET3) sharing 2 highly conserved domains. TET1 protein has been implicated in leukemia as a fusion partner of the MLL gene product in the t(10;11)(p12;q23) chromosomal translocation and has been shown to be capable of hydroxymethylation and thereby possibly contributing to cytosine methylation patterns and epigenetic gene regulation.14-18
Mutations in the TET2 gene have been identified in 19%-26% of MDS and 50% of chronic myelomonocytic leukemia (CMML) patients, making it one of the most frequently mutated genes in these myeloid neoplasms.9-11,13 Although, TET2 mutations have been shown to be an early-disease event and pre-JAK2 lesion11 in myeloproliferative neoplasms (MPNs), recent data from colony assays, in some MPN patients, show that TET2 and JAK2 mutations can occur independently of each other.19 This is highlighted in MPN cases in whom TET2 mutations were only detected at the time of myeloid leukemic transformation.20-22 The prognostic significance of TET2 aberrations in MDS remains unclear, with conflicting reports on the effect of mutant TET2 on overall survival.10,23 The majority of reported TET2 mutations are likely to cause inactivation of the TET2 protein, suggesting a tumor-suppressor role for this protein. Furthermore, TET2 mRNA is highly expressed in normal myeloid progenitor cells, granulocytes, and erythroid cells, whereas granulocytes isolated from MDS cases have reduced TET2 expression, highlighting the importance of wild-type levels of TET2 protein in myelopoeisis. Decreased TET2 expression is also noted in CD34+ cells, CD13+/33+ myelomonocytic cells, and CD71+ erythroid cells isolated from bone marrow, and mature granulocytes obtained from patients with no detectable mutations, suggesting alternative mechanisms of TET2 dysregulation.11
The advent of next-generation “deep” sequencing technology has enabled cost-effective, high-throughput sequencing, with the added advantage of increased sensitivity more than the conventional Sanger methodology. To examine the prevalence of TET2 mutations and, especially, lower abundance TET2 mutant clones, we used ultrasensitive “454-deep sequencing” to investigate the integrity of the TET2 gene in a well-characterized cohort of 355 de novo MDS and CMML patients. The presence of TET2 mutations existing as major/minor clones or single/multiple mutants was analyzed for association with conventional clinical and prognostic parameters as well as TET2 gene expression, JAK2V617F mutation, and 46/1 haplotype status. We also analyzed DNA from CD34+, CD3+ cells and constitutional DNA to determine mutant clone size and to ascertain the stage of acquisition of the TET2 aberration. In 91 patients, high-resolution Affymetrix SNP6 microarrays were used to detect cryptic genomic aberrations and their relationship to TET2 mutations.
Methods
Patients and sample preparation
A total of 355 patients with a median age of 64 years (range, 17-96 years) were identified between 2 hematology centers (Kings College Hospital, London, United Kingdom, and Haematologie, Onkologie und Klinische Immunologie, Heinrich-Heine-Universität, Düsseldorf, Germany). Diagnosis, relevant details, and sample preparation are detailed in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
TET2 mutation screen
TET2 (NM.001127208) mutations and polymorphisms were analyzed from bone marrow DNA using the Roche GS FLX sequencing platform (supplemental Table 1). Methodology has been described previously.12 We sequenced 31 amplicons for 355 patients across the entire TET2 coding region. Patient-specific “barcodes,” adapted to polymerase chain reaction (PCR) primers in the second round of amplification, were derived from Roche GS Titanium protocol. (supplemental Table 2A). Average sequencing coverage (X) across the entire TET2 coding region was 130× and > 95% of the sequenced amplicons had sequence coverage more than 40× (> 98% with more than 10× coverage). This reliably allowed detection of mutations down to 10% relative mutation abundance (RMA), defined as the proportion of sequence reads containing the mutation. Independent PCR and Genome Sequencer (GS) FLX sequencing experiments were performed for the confirmation of mutations in all cases (giving > 500× total coverage, on average). These independent experiments reduce sampling errors for RMA calculations owing to a higher combined sequencing coverage. Sanger sequencing confirmation was performed in all cases where the mutation had a RMA of ≥ 25%. To detect low-abundance mutant clones, we resequenced 96 patient samples, including TET2 mutant and nonmutant cases, at much greater depth (450× vs 130×), and for 13 of these patients at an average coverage of 950×. Mutations found at ≤ 10% RMA were validated in 2 additional independent PCR and GS FLX sequencing experiments, giving a combined coverage of > 800×. In 12 cases, where we had identified TET2 mutations in the total bone marrow cells, the mutation status was also assessed in paired bone marrow, CD34+, and CD3+ T cells, and in 8 of these cases, TET2 mutational status was also assessed in the skin biopsy.
Affymetrix SNP6 analysis
Genotyping was performed using the Affymetrix SNP6 platform and processed according to the manufacturer's instructions (Affymetrix). Further details are available in supplemental Methods. All microarray data are available at the Gene Expression Omnibus public database less than accession number GSE2330.
JAK2 genotyping
One informative SNP (RS12345895) within the 46/1 JAK2 haplotype was analyzed by pyrosequencing using primers and conditions described previously.24
Quantitative PCR for JAK2V617F and TET expression
JAK2 V617F allele burden was determined by real-time PCR using MGB-hydrolysis probes (Applied Biosystems).25 TET gene expression was analyzed using probes from the Universal Probe Library (Roche) and primers specific for TET1 (NM_030625.2), TET2 var 1 (NM_017628.3), TET2 var 2 (NM_001127208.1), and TET3 (NM_144993.1), as detailed in supplemental Table 2B.
Statistical analysis
Statistical analysis was performed using SPSS version 17.0 (SPSS Inc) as previously described and expanded in supplemental Methods.12 A P value of less than .05 was set as the threshold of clinical significance.
Results
Classifying and mapping mutations
Samples from 320 MDS and 35 CMML patients (Table 1) were screened for TET2 mutations at an average sequence coverage of 130× across the gene-coding region using the Roche GS FLX “next-generation sequencing” (NGS) platform. In total, 82 variants were initially identified in the TET2 gene in 65 (18.3%) patients from total bone marrow DNA, where the proportion of sequence reads containing the variant (RMA) was ≥ 10% (supplemental Table 1). Of these, 71 TET2 mutations predicted to alter protein structure/function were identified in 39 of 320 (12%) MDS and 16 of 35 (46%) CMML patients (P < .001; Table 2; Figure 1). These functionally relevant mutations were used as the primary dataset for all subsequent analyses and consisted of 43 nonsense mutations or Indel mutations, 3 splice-site mutations, and 25 nonsynonymous amino acid changes within the protein-conserved domains.9,11,12,23 Independent experiments confirmed mutations in all cases and allowed accurate estimates of RMA. Likewise, Sanger sequencing was used to confirm mutations where RMA was > 25%. For 2 common SNPs (rs34402524 and rs2454206), the respective heterozygous SNP sequence frequency (mean ± SD) was 47.8% ± 2.54% and 48.5% ± 2.18% within the first 96 patient samples, indicating the RMA estimates were accurate. Known SNPs and novel polymorphisms that were identified in more than 2 patients and confirmed in paired constitutional DNA, were excluded from the analysis. Significantly, 90% of the mutations identified here were previously uncharacterized.
Clinical and biological characteristics of the MDS cohort and relationship to TET2 mutations found at > 10% RMA
| Patient characteristics . | All n (%) . | TET2 mutated . | TET2 nonmutated . | P . |
|---|---|---|---|---|
| Number | 355 (100) | 55 (15.5) | 300 (84.5) | |
| Median age, y | 64.4 | 68.3 | 64.0 | < .01 |
| Range, y | (17.0-96.4) | (42.9-96.0) | (17.0-96.4) | |
| Sex, M/F | 214/141 | 34/21 | 180/120 | .06 |
| WHO classification | < .01 | |||
| RA/RCMD | 131 (36.9) | 17 (30.9) | 114 (38.0) | |
| RARS/RCMD-RS | 33 (9.3) | 6 (10.9) | 27 (9.0) | |
| RAEB-I/II | 105 (29.6) | 12 (21.8) | 93 (31.0) | |
| sAML | 35 (9.8) | 4 (7.3) | 31 (10.3) | |
| CMML-I/II | 35 (9.8) | 16 (29.1) | 19 (6.3) | |
| 5q- syndrome | 8 (2.3) | 0 (0) | 8 (2.7) | |
| MDS/MPN-U | 8 (2.3) | 0 (0) | 8 (2.7) | |
| Karyotype | .09 | |||
| Favorable | 192 (54.1) | 32 (58.2) | 160 (53.3) | |
| Intermediate | 60 (16.9) | 12 (21.9) | 48 (16.0) | |
| Poor | 71 (20.0) | 7 (12.7) | 64 (21.3) | |
| Missing | 32 (9.0) | 4 (7.2) | 28 (9.4) | |
| IPSS | .25 | |||
| Low/Int-1 | 192 (61.5) | 40 (72.7) | 152 (59.1) | |
| Int-2/High | 97 (31.1) | 13 (23.9) | 84 (32.7) | |
| Missing | 23 (7.4) | 2 (3.4) | 21 (8.2) | |
| Treatment | .08 | |||
| None/BSC | 211 (59.5) | 41 (74.6) | 170 (56.7) | |
| EPO/GCSF | 29 (8.2) | 1 (1.8) | 28 (9.3) | |
| 5-azacitidine | 14 (3.9) | 1 (1.8) | 13 (4.3) | |
| Intensive chemo ± transplant | 82 (23.1) | 11 (20.0) | 71 (23.7) | |
| Treatment data not available | 19 (5.3) | 1 (1.8) | 18 (6.0) |
| Patient characteristics . | All n (%) . | TET2 mutated . | TET2 nonmutated . | P . |
|---|---|---|---|---|
| Number | 355 (100) | 55 (15.5) | 300 (84.5) | |
| Median age, y | 64.4 | 68.3 | 64.0 | < .01 |
| Range, y | (17.0-96.4) | (42.9-96.0) | (17.0-96.4) | |
| Sex, M/F | 214/141 | 34/21 | 180/120 | .06 |
| WHO classification | < .01 | |||
| RA/RCMD | 131 (36.9) | 17 (30.9) | 114 (38.0) | |
| RARS/RCMD-RS | 33 (9.3) | 6 (10.9) | 27 (9.0) | |
| RAEB-I/II | 105 (29.6) | 12 (21.8) | 93 (31.0) | |
| sAML | 35 (9.8) | 4 (7.3) | 31 (10.3) | |
| CMML-I/II | 35 (9.8) | 16 (29.1) | 19 (6.3) | |
| 5q- syndrome | 8 (2.3) | 0 (0) | 8 (2.7) | |
| MDS/MPN-U | 8 (2.3) | 0 (0) | 8 (2.7) | |
| Karyotype | .09 | |||
| Favorable | 192 (54.1) | 32 (58.2) | 160 (53.3) | |
| Intermediate | 60 (16.9) | 12 (21.9) | 48 (16.0) | |
| Poor | 71 (20.0) | 7 (12.7) | 64 (21.3) | |
| Missing | 32 (9.0) | 4 (7.2) | 28 (9.4) | |
| IPSS | .25 | |||
| Low/Int-1 | 192 (61.5) | 40 (72.7) | 152 (59.1) | |
| Int-2/High | 97 (31.1) | 13 (23.9) | 84 (32.7) | |
| Missing | 23 (7.4) | 2 (3.4) | 21 (8.2) | |
| Treatment | .08 | |||
| None/BSC | 211 (59.5) | 41 (74.6) | 170 (56.7) | |
| EPO/GCSF | 29 (8.2) | 1 (1.8) | 28 (9.3) | |
| 5-azacitidine | 14 (3.9) | 1 (1.8) | 13 (4.3) | |
| Intensive chemo ± transplant | 82 (23.1) | 11 (20.0) | 71 (23.7) | |
| Treatment data not available | 19 (5.3) | 1 (1.8) | 18 (6.0) |
TET2 mutations and clinical details
| UPC . | WHO . | Residue change/ mutation position . | RMA (mean) . | SD of RMA . | JAK2 haplotype . | Disease progression . | IPSS . | Outcome . |
|---|---|---|---|---|---|---|---|---|
| 4 | RAEB-II | T1726fsX18 | 39.5 | 0.7 | No | No | 3 | Dead |
| 55 | sAML | P1937S | 15.3 | 1.2 | No | Yes | 3 | Dead |
| P1941S | 17.7 | 0.6 | ||||||
| 571 | CMML-I | A1344E | 46.0 | 7.1 | Yes | No | 1 | Dead |
| 682G | RAEB-II | G1275E [19] | 68.0 | 1.4 | No | Yes | 3 | Dead |
| G1282D | 68.0 | 1.4 | ||||||
| D1844G | 38.0 | 4.2 | ||||||
| 750 | RCMD-RS | R1216Q | 21.5 | 4.9 | No | No | 1 | Dead |
| 783 | RA | Q1529X | 25.0 | 1.0 | No | n/a | n/a | n/a |
| 798 | sAML | H1380P | 47.0 | 1.0 | No | No | 2 | Dead |
| 919 | RAEB-II | A619fsX19 | 86.7 | 1.5 | Yes | Yes | n/a | Dead |
| 957 | RCMD | W564X | 32.5 | 3.5 | Yes | No | 1 | Dead |
| 1074 | RCMD-RS | V872fsX44 | 38.5 | 0.7 | No | No | 0 | Dead |
| 1160 | CMML-I | Splice acceptor 5′ exon 9 | 42.3 | 0.6 | No | No | 1 | Dead |
| 1161 | RCMD | R1216X [9,11] | 18.0 | 2.6 | Yes | No | 1 | Dead |
| 1190 | CMML-I | R1216X [9, 11] | 31.5 | 2.1 | No | No | 1 | Dead |
| T1726fsX18 | 61.0 | 1.4 | ||||||
| 1217 | CMML-I | H1904Q | 45.0 | 4.2 | Yes | Yes | 1 | Dead |
| E1845fsX42 | 42.0 | 1.4 | ||||||
| 1288 | RARS | Q726X | 24.0 | 5.7 | No | No | 0 | Dead |
| 1326 | RCMD | C1193W | 49.0 | 3.0 | No | Yes | 0 | Dead |
| 1392 | CMML-I | D905fsX16 | 38.5 | 3.5 | Yes | No | 0 | Dead |
| 1444 | RCMD | L371X | 47.5 | 0.7 | No | No | 0 | Alive |
| Y1569fsX1 | 37.0 | 2.8 | ||||||
| 1490 | CMML-I | Q1699X | 49.3 | 1.5 | No | Yes | 0 | Dead |
| G1288V | 43.3 | 2.1 | ||||||
| 1586 | RCMD | R1216X [11] | 41.0 | 1.4 | No | No | 1 | Dead |
| 1619 | RAEB-I | Q888X | 44.7 | 0.6 | Yes | No | 1 | Alive |
| R1465X [23] | 35.0 | 2.8 | ||||||
| 1625 | RCMD | S716X | 44.7 | 0.6 | Yes | No | 0.5 | Alive |
| 1672 | RCMD-RS | F1377fsX22 | 84.5 | 0.7 | No | No | 1 | Dead |
| 1681 | RCMD | I1195V | 47.0 | 0.0 | No | No | 0 | Alive |
| 1684 | CMML-I | N1743fsX17 | 45.5 | 0.7 | No | No | 0 | Alive |
| 1702 | CMML-I | V1371fsX76 | 94.0 | 1.4 | No | No | 0 | Dead |
| 1740 | RCMD | Q1529X | 38.0 | 4.2 | Yes | No | 0.5 | Alive |
| 1757 | RAEB-II | Q644X | 36.3 | 3.2 | No | No | 3 | Dead |
| 1857 | CMML-I | R550X | 45.5 | 0.7 | No | Yes | 1 | Dead |
| 1869 | RCMD | Q526X | 41.0 | 1.4 | Yes | No | 1 | Dead |
| 1906 | RAEB-II | Splice acceptor 5′ exon 5 | 79.0 | 1.4 | No | No | 3 | Dead |
| 1932* | RCMD | V1862fsX24 | 97.5 | 0.7 | Homozygous | No | 0 | Alive |
| 1980 | CMML-I | A707fsX9 | 49.5 | 0.7 | No | No | 0 | Alive |
| 2007 | RCMD | P1962L [11] | 40.0 | 2.1 | Yes | No | 0 | Alive |
| L1360R | 36.0 | 4.2 | ||||||
| Q644X | 35.5 | 0.7 | ||||||
| 2222 | RAEB-II | A1355P | 26.0 | 2.0 | No | No | 2 | Dead |
| V160fsX22 | 19.0 | 2.6 | ||||||
| 2236 | CMML-I | Splice donor 3′ exon 10 | 42.0 | 2.6 | No | No | 0 | Dead |
| Y1631fsX28 | 50.5 | 0.7 | ||||||
| 2296 | RCMD-RS | E259X | 42.7 | 3.1 | No | n/a | 1 | Dead |
| Q888X | 47.5 | 0.7 | ||||||
| 2298 | RAEB-I | E576X | 43.5 | 0.7 | No | No | 1 | Alive |
| 2306* | RCMD | T1393P | 39.0 | 2.1 | No | No | 0 | Alive |
| 2392 | RCMD | P1962L [11] | 50.0 | 0.5 | No | No | 0 | Dead |
| 2483 | CMML-I | R1216X | 92.7 | 3.1 | Yes | No | 1 | Alive |
| 2545 | RAEB-II | F1287fsX75 | 20.3 | 7.1 | Yes | Yes | 3 | Alive |
| 2582 | sAML | R1261G | 42.0 | 2.8 | Yes | Yes | 3 | Dead |
| 2644 | RCMD-RS | A1876G | 46.7 | 1.5 | Yes | Yes | 0 | Alive |
| H1904R | 46.0 | 1.0 | ||||||
| 2705 | sAML | P1194A | 41.0 | 1.4 | No | No | 3 | Alive |
| 2719* | MDS/MPN-U | C1273F | 70.7 | 2.1 | No | Yes | 0 | Alive |
| R1452X | 20.0 | 3.0 | ||||||
| 2789 | RAEB-II | R1465Q | 40.0 | 0.0 | Yes | Yes | 2 | Dead |
| 2859 | RCMD | I1873N | 39.0 | 1.4 | Yes | No | 0.5 | Dead |
| Q1687X | 40.5 | 0.7 | ||||||
| 2874 | RCMD | P555fsX5 [11] | 49.5 | 0.7 | No | No | 1 | Alive |
| 2984 | CMML-II | H1380Y | 50.0 | 0.6 | Yes | No | 2 | Dead |
| 3181 | CMML-II | Q888X | 43.0 | 1.4 | Yes | No | . | Dead |
| 3198 | RAEB-II | P1629fsX31 | 89.5 | 0.7 | No | Yes | 2 | Alive |
| 3232 | CMML-I | S1303fsX58 | 44.0 | 1.4 | Yes | No | 0 | Alive |
| 3266 | CMML-I | Q1020fsX16 | 85.0 | 6.1 | No | Yes | 0.5 | Alive |
| 3338 | RCMD | Q910X | 37.0 | 1.4 | No | No | 0.5 | Alive |
| UPC . | WHO . | Residue change/ mutation position . | RMA (mean) . | SD of RMA . | JAK2 haplotype . | Disease progression . | IPSS . | Outcome . |
|---|---|---|---|---|---|---|---|---|
| 4 | RAEB-II | T1726fsX18 | 39.5 | 0.7 | No | No | 3 | Dead |
| 55 | sAML | P1937S | 15.3 | 1.2 | No | Yes | 3 | Dead |
| P1941S | 17.7 | 0.6 | ||||||
| 571 | CMML-I | A1344E | 46.0 | 7.1 | Yes | No | 1 | Dead |
| 682G | RAEB-II | G1275E [19] | 68.0 | 1.4 | No | Yes | 3 | Dead |
| G1282D | 68.0 | 1.4 | ||||||
| D1844G | 38.0 | 4.2 | ||||||
| 750 | RCMD-RS | R1216Q | 21.5 | 4.9 | No | No | 1 | Dead |
| 783 | RA | Q1529X | 25.0 | 1.0 | No | n/a | n/a | n/a |
| 798 | sAML | H1380P | 47.0 | 1.0 | No | No | 2 | Dead |
| 919 | RAEB-II | A619fsX19 | 86.7 | 1.5 | Yes | Yes | n/a | Dead |
| 957 | RCMD | W564X | 32.5 | 3.5 | Yes | No | 1 | Dead |
| 1074 | RCMD-RS | V872fsX44 | 38.5 | 0.7 | No | No | 0 | Dead |
| 1160 | CMML-I | Splice acceptor 5′ exon 9 | 42.3 | 0.6 | No | No | 1 | Dead |
| 1161 | RCMD | R1216X [9,11] | 18.0 | 2.6 | Yes | No | 1 | Dead |
| 1190 | CMML-I | R1216X [9, 11] | 31.5 | 2.1 | No | No | 1 | Dead |
| T1726fsX18 | 61.0 | 1.4 | ||||||
| 1217 | CMML-I | H1904Q | 45.0 | 4.2 | Yes | Yes | 1 | Dead |
| E1845fsX42 | 42.0 | 1.4 | ||||||
| 1288 | RARS | Q726X | 24.0 | 5.7 | No | No | 0 | Dead |
| 1326 | RCMD | C1193W | 49.0 | 3.0 | No | Yes | 0 | Dead |
| 1392 | CMML-I | D905fsX16 | 38.5 | 3.5 | Yes | No | 0 | Dead |
| 1444 | RCMD | L371X | 47.5 | 0.7 | No | No | 0 | Alive |
| Y1569fsX1 | 37.0 | 2.8 | ||||||
| 1490 | CMML-I | Q1699X | 49.3 | 1.5 | No | Yes | 0 | Dead |
| G1288V | 43.3 | 2.1 | ||||||
| 1586 | RCMD | R1216X [11] | 41.0 | 1.4 | No | No | 1 | Dead |
| 1619 | RAEB-I | Q888X | 44.7 | 0.6 | Yes | No | 1 | Alive |
| R1465X [23] | 35.0 | 2.8 | ||||||
| 1625 | RCMD | S716X | 44.7 | 0.6 | Yes | No | 0.5 | Alive |
| 1672 | RCMD-RS | F1377fsX22 | 84.5 | 0.7 | No | No | 1 | Dead |
| 1681 | RCMD | I1195V | 47.0 | 0.0 | No | No | 0 | Alive |
| 1684 | CMML-I | N1743fsX17 | 45.5 | 0.7 | No | No | 0 | Alive |
| 1702 | CMML-I | V1371fsX76 | 94.0 | 1.4 | No | No | 0 | Dead |
| 1740 | RCMD | Q1529X | 38.0 | 4.2 | Yes | No | 0.5 | Alive |
| 1757 | RAEB-II | Q644X | 36.3 | 3.2 | No | No | 3 | Dead |
| 1857 | CMML-I | R550X | 45.5 | 0.7 | No | Yes | 1 | Dead |
| 1869 | RCMD | Q526X | 41.0 | 1.4 | Yes | No | 1 | Dead |
| 1906 | RAEB-II | Splice acceptor 5′ exon 5 | 79.0 | 1.4 | No | No | 3 | Dead |
| 1932* | RCMD | V1862fsX24 | 97.5 | 0.7 | Homozygous | No | 0 | Alive |
| 1980 | CMML-I | A707fsX9 | 49.5 | 0.7 | No | No | 0 | Alive |
| 2007 | RCMD | P1962L [11] | 40.0 | 2.1 | Yes | No | 0 | Alive |
| L1360R | 36.0 | 4.2 | ||||||
| Q644X | 35.5 | 0.7 | ||||||
| 2222 | RAEB-II | A1355P | 26.0 | 2.0 | No | No | 2 | Dead |
| V160fsX22 | 19.0 | 2.6 | ||||||
| 2236 | CMML-I | Splice donor 3′ exon 10 | 42.0 | 2.6 | No | No | 0 | Dead |
| Y1631fsX28 | 50.5 | 0.7 | ||||||
| 2296 | RCMD-RS | E259X | 42.7 | 3.1 | No | n/a | 1 | Dead |
| Q888X | 47.5 | 0.7 | ||||||
| 2298 | RAEB-I | E576X | 43.5 | 0.7 | No | No | 1 | Alive |
| 2306* | RCMD | T1393P | 39.0 | 2.1 | No | No | 0 | Alive |
| 2392 | RCMD | P1962L [11] | 50.0 | 0.5 | No | No | 0 | Dead |
| 2483 | CMML-I | R1216X | 92.7 | 3.1 | Yes | No | 1 | Alive |
| 2545 | RAEB-II | F1287fsX75 | 20.3 | 7.1 | Yes | Yes | 3 | Alive |
| 2582 | sAML | R1261G | 42.0 | 2.8 | Yes | Yes | 3 | Dead |
| 2644 | RCMD-RS | A1876G | 46.7 | 1.5 | Yes | Yes | 0 | Alive |
| H1904R | 46.0 | 1.0 | ||||||
| 2705 | sAML | P1194A | 41.0 | 1.4 | No | No | 3 | Alive |
| 2719* | MDS/MPN-U | C1273F | 70.7 | 2.1 | No | Yes | 0 | Alive |
| R1452X | 20.0 | 3.0 | ||||||
| 2789 | RAEB-II | R1465Q | 40.0 | 0.0 | Yes | Yes | 2 | Dead |
| 2859 | RCMD | I1873N | 39.0 | 1.4 | Yes | No | 0.5 | Dead |
| Q1687X | 40.5 | 0.7 | ||||||
| 2874 | RCMD | P555fsX5 [11] | 49.5 | 0.7 | No | No | 1 | Alive |
| 2984 | CMML-II | H1380Y | 50.0 | 0.6 | Yes | No | 2 | Dead |
| 3181 | CMML-II | Q888X | 43.0 | 1.4 | Yes | No | . | Dead |
| 3198 | RAEB-II | P1629fsX31 | 89.5 | 0.7 | No | Yes | 2 | Alive |
| 3232 | CMML-I | S1303fsX58 | 44.0 | 1.4 | Yes | No | 0 | Alive |
| 3266 | CMML-I | Q1020fsX16 | 85.0 | 6.1 | No | Yes | 0.5 | Alive |
| 3338 | RCMD | Q910X | 37.0 | 1.4 | No | No | 0.5 | Alive |
TET2 mutations in the 355 MDS patient cohort. Details of all TET2 mutations and patient context are shown. The average (mean) and standard deviation (SD) for relative mutation abundance (RMA) are shown and are based on a combined sequence coverage of > 500×. Relevant clinical details are also shown for each patient, who are referred to by a unique patient code (UPC).
Genomic aberrations at the TET2 locus relating to SNP6 experiments are indicated. All frame-shift mutations (fs) are, by their nature, Indel mutations and are described by the number of amino acids corresponding to the length of the new frame after the Indel event, which is indicated at the end of the mutation description. Mutations previously referenced by the 4 largest studies, to date, are highlighted and referenced. The presence of the JAK2V617F 46/1 predisposition genotype (haplotype), indicated by a “yes,” corresponds to a heterozygous state.
Seventy-one mutations in 55 of 355 persons were identified by NGS and mapped to the TET2 coding region. Mutations were identified that existed at ≥ 10% relative mutation abundance (RMA). These were mapped against TET2 translated exons 3-11 (NM.001127208; 2002 amino acids). Classification of mutations is indicated in the figure key and includes nonsense or insertion/deletion mutations (Indels), nonsynonymous amino acid changes, and splice-site mutations, indicated by orange, blue, and pink bars, respectively. Mutation level is also defined as > or ≤ 25% RMA, indicated by the solid or stippled bars, respectively. Regions conserved across the TET protein family, and implicated in the conversion of 5′ methylcytosine to 5′ methylhydroxycytosine (5-hmC), are shown (cons'd regions 1 and 2) and correspond to amino acids 1104-1478 and 1845-2002, respectively ([11] [18]).
Seventy-one mutations in 55 of 355 persons were identified by NGS and mapped to the TET2 coding region. Mutations were identified that existed at ≥ 10% relative mutation abundance (RMA). These were mapped against TET2 translated exons 3-11 (NM.001127208; 2002 amino acids). Classification of mutations is indicated in the figure key and includes nonsense or insertion/deletion mutations (Indels), nonsynonymous amino acid changes, and splice-site mutations, indicated by orange, blue, and pink bars, respectively. Mutation level is also defined as > or ≤ 25% RMA, indicated by the solid or stippled bars, respectively. Regions conserved across the TET protein family, and implicated in the conversion of 5′ methylcytosine to 5′ methylhydroxycytosine (5-hmC), are shown (cons'd regions 1 and 2) and correspond to amino acids 1104-1478 and 1845-2002, respectively ([11] [18]).
Fourteen patients had multiple mutations (Table 2). Three patients had 2 mutations that were associated with the same amplicon being sequenced (unique patient codes [UPCs] 55, 682G, and 2644). Of these 3, UPC 682G codes had 2 mutations, which were found in the same DNA molecule and therefore resided in the same mutant clone, whereas the other 2 patients had mutations found in different DNA molecules and therefore resided in different clones or on alternate alleles, as illustrated for UPC 55 in Figure 2. Furthermore, 2 additional patients (UPCs 1190 and 2719) had multiple mutations where the higher abundance mutation was present at a frequency > 1.9-fold the lower abundance mutation, suggesting distinct mutation events in the evolution of the disease. In the remaining 9 patients, mutant clones were found at a similar RMA.
NGS clonal sequencing allows direct detection of independent disease clones. (A) Two C to T substitutions in TET2 exon 11 are depicted by the dominant red bars and are found in approximately 17% of sequence reads (17% variation [left axis] from the reference sequence [x-axis]). The sequence coverage (number of reads) across the region is shown on the right axis and is traced by the blue line. Experimental noise is shown underneath the dotted line with reads for bases A (green bars), C (blue bars), G (black bars), T (red bars), and base deletions (gray bars) at less than 4% of reads. (B) A selection of individual sequence reads lined up against each other is shown and demonstrates that the 2 mutations are found in separate molecules being sequenced and thus belong to independent clones or alternate alleles.
NGS clonal sequencing allows direct detection of independent disease clones. (A) Two C to T substitutions in TET2 exon 11 are depicted by the dominant red bars and are found in approximately 17% of sequence reads (17% variation [left axis] from the reference sequence [x-axis]). The sequence coverage (number of reads) across the region is shown on the right axis and is traced by the blue line. Experimental noise is shown underneath the dotted line with reads for bases A (green bars), C (blue bars), G (black bars), T (red bars), and base deletions (gray bars) at less than 4% of reads. (B) A selection of individual sequence reads lined up against each other is shown and demonstrates that the 2 mutations are found in separate molecules being sequenced and thus belong to independent clones or alternate alleles.
Nine mutations were found at ≤ 25% RMA in 8 patients (Table 2; Figure 1). One of these patients (UPC 55) had 2 low-level mutations (14% and 17% RMA) that resided in different sequenced molecules. Five of these patients (UPCs 750, 783, 1161, 1288, and 2545) had single mutations, and the 2 remaining patients (UPCs 2222 and 2719) had multiple mutations where 1 was above 25% RMA.
In 45 patients (82%), TET2 mutations ranged between 15% and 61% RMA (median, 41%). In the remaining 10 patients (18%) with mutational abundance of ≥ 65% RMA, the increased mutation burden was consistent, with a genomic aberration at the TET2 locus (see below).
Deep sequencing for low-abundance disease clones
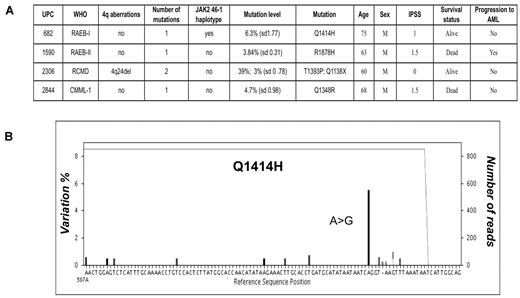
To assess the frequency of very low-abundance disease clones, we resequenced the first 96 patients at much greater depth (450× vs 130×). We identified and verified 4 additional mutations with an RMA of 3%-6.3% (Figure 3A). Nonsense mutation Q1138X was found in a patient (UPC 2306) with refractory cytopenia with multilineage dysplasia (RCMD), for whom an additional high-frequency (39% RMA), nonsynonymous mutation had already been identified, implicating distinct mutant clones (Figure 3B). The other 3 mutations were found in 1 case of refractory anemia with excess blasts (RAEB-II; UPC 1590), one case of CMML-I (UPC 2844) and one case of RAEB-I (UPC 682), in whom no previous mutations were detected. Importantly, these additional mutations were all located within the conserved domains of TET2, suggesting that these are not functionally silent events. To determine whether we were missing any frequent low-level mutation events, additional sequencing experiments of 13 TET2 mutation-negative patients within this subgroup were performed to increase the average sequence depth to > 900×. No additional mutations were discovered at this increased sensitivity. In addition to this, no mutations were found in a control group of bone marrow DNA from 12 normal healthy persons after sequencing at depth (average, 500×).
Deep sequencing reveals low-level mutant clones. (A) After deeper sequencing at 450× coverage for 96 patients, an additional 4 mutations were identified, which were present at between 3% and 6.3% RMA within total bone marrow DNA. One of these mutations was found in a patient with an additional mutation, which had previously been identified. Means and standard deviations for RMA corresponding to 3 independent experiments are shown. (B) Representative sequence readout for 1 of these mutations is exampled and shows an A>G substitution, denoted by the prominent black bar, in 6% of sequence reads (6% variation from the reference sequence, left axis), leading to a Q1414H nonsynonymous amino acid change. The sequence depth is 820× across this region traced by the gray line running across the top of the graph. Experimental noise accounts for ≤ 1% of variation.
Deep sequencing reveals low-level mutant clones. (A) After deeper sequencing at 450× coverage for 96 patients, an additional 4 mutations were identified, which were present at between 3% and 6.3% RMA within total bone marrow DNA. One of these mutations was found in a patient with an additional mutation, which had previously been identified. Means and standard deviations for RMA corresponding to 3 independent experiments are shown. (B) Representative sequence readout for 1 of these mutations is exampled and shows an A>G substitution, denoted by the prominent black bar, in 6% of sequence reads (6% variation from the reference sequence, left axis), leading to a Q1414H nonsynonymous amino acid change. The sequence depth is 820× across this region traced by the gray line running across the top of the graph. Experimental noise accounts for ≤ 1% of variation.
46/1 JAK2V617F status
The frequency of the 46/1 JAK2V617F mutation predisposition genotype was analyzed in 344 of 355 patients and identified in 148 (43%) patients with an allelic frequency of 24% (130 heterozygous, 18 homozygous). This was not significantly different to levels reported in the general population.24 Furthermore, there was no significant difference in the incidence of the 46/1 genotype between TET2-mutant versus nonmutant patients (20% vs 16% [P = .31] occurrence with an allelic frequency of 20% and 25% for mutant versus nonmutant patients, respectively). A screen for the JAK2V617F mutation in 200 patients from our cohort identified only one patient (MDS/MPN-unclassifiable [U]) carrying the V617F mutation with no coexistent TET2 mutation, suggesting that in MDS such mutations were not a downstream effect of a TET2 lesion.
TET gene expression
Expression of TET1, TET2, and TET3 was analyzed in a subset of 116 patients [RCMD = 48, RAEB-II = 26, RAEB-I = 13, AML with MDS-related changes (sAML) = 12, MDS/MPN-U = 8, CMML-I = 3, CMML-II = 3, and RCMD with ringed sideroblasts (RS) = 3], of which 18 had TET2 mutations. There was no significant difference in the mRNA levels of TET1 (P = .82), TET2 var1 (P = .26), TET2 var2 (P = .44), or TET3 (P = .67) between TET2-mutant and nonmutant patients (Figure 4A). Analysis of TET expression in different subtypes of myeloid neoplasms did not identify any significant differences (P > .4; Figure 4B).
Bar graphs showing TET1, TET2 var1, TET2 var2, and TET3 quantitative PCR gene-expression analysis in 116 MDS cases. (A) Gene-expression ratios in mutant (n = 18) and wild-type cases (n = 98) with standard error bars. (B) Gene-expression ratios of TET genes in different disease subtypes within our cohort.
Bar graphs showing TET1, TET2 var1, TET2 var2, and TET3 quantitative PCR gene-expression analysis in 116 MDS cases. (A) Gene-expression ratios in mutant (n = 18) and wild-type cases (n = 98) with standard error bars. (B) Gene-expression ratios of TET genes in different disease subtypes within our cohort.
Tracing TET2 mutations through differentiation
To investigate the origins of TET2 mutations and the selective advantage these lesions confer, we quantified the mutation load in CD34+ and CD3+ cells for 12 patients with TET2 mutations (Table 3). The RMA was similar between CD34+ cells (median 38.5%) and total bone marrow (median 43%). However, in UPC 1932, we detected an expansion of the TET2 mutation from 50% to 97% RMA for CD34+ and total bone marrow, respectively. In this patient, SNP array karyotyping identified a UPD at the TET2 locus in the total bone marrow DNA (see below). Importantly, in 9 of 11 patients, CD3+ cells had a reduced, but significant, mutation load, compared with total bone marrow and CD34+ cells, maintaining a mutant allele burden of ≥ 14% (median, 23.5% RMA) and indicating a very early event affecting a primitive stem cell. Significantly, relevant levels of mutant clones were not detected in paired skin samples, confirming the acquired nature of these mutations.
Abundance of patient-specific TET2 mutations across different tissues
| % relative mutation abundance . | ||||
|---|---|---|---|---|
| Patient . | BM . | CD34+ . | CD3+ . | Skin . |
| 2984 | 50 | 40 | 22 | 5 |
| 2236 | 51/42 | 37/35 | 36/36 | 8/4 |
| 1932* | 98 | 49 | 15 | 6 |
| 2859† | 39/41 | 46/42 | 30/34 | 2/5 |
| 1619 | 45/35 | 40/30 | 26/17 | 6/2 |
| 1684 | 46 | 42 | 4 | na |
| 2719 | 71/20 | 60/21 | 54/18 | na |
| 1490 | 49/43 | 25/27 | 19/15 | 5/7 |
| 1625 | 45 | 45 | 26 | 6 |
| 1326 | 49 | 42 | 4 | 0 |
| 1444 | 48/37 | 43/27 | 27/25 | na |
| 2222 | 26 | 32 | 5 | 1 |
| % relative mutation abundance . | ||||
|---|---|---|---|---|
| Patient . | BM . | CD34+ . | CD3+ . | Skin . |
| 2984 | 50 | 40 | 22 | 5 |
| 2236 | 51/42 | 37/35 | 36/36 | 8/4 |
| 1932* | 98 | 49 | 15 | 6 |
| 2859† | 39/41 | 46/42 | 30/34 | 2/5 |
| 1619 | 45/35 | 40/30 | 26/17 | 6/2 |
| 1684 | 46 | 42 | 4 | na |
| 2719 | 71/20 | 60/21 | 54/18 | na |
| 1490 | 49/43 | 25/27 | 19/15 | 5/7 |
| 1625 | 45 | 45 | 26 | 6 |
| 1326 | 49 | 42 | 4 | 0 |
| 1444 | 48/37 | 43/27 | 27/25 | na |
| 2222 | 26 | 32 | 5 | 1 |
TET2 mutations are an early event. Twelve patients who were found to have mutant TET2 clones within their bone marrow tissue were also screened for mutation status within paired germline (skin), CD34+, and CD3+ T-cell tissue, where available. Data for % relative mutation abundance (RMA) refers to the same mutation(s) across different tissues for any 1 patient. The median RMA for all patients is 43% for bone marrow, 38.5% CD34+ cells, 23.5% for CD3+ cells, and 5% for germline (skin) tissue. Data for individual RMA readings is an average based on at least 2 independent experiments. Patients with more than 1 mutation are denoted as n/n RMA.
Higher mutation abundance in total bone marrow than CD34+ cells, consistent with a genomic aberration rendering the TET2 locus hemizygous after differentiation.
High abundance of mutation within the T-cell population.
TET2 mutations and SNP karyotyping
We analyzed 91 cases with no previous history of hematologic malignancy, using SNP 6.0 arrays. We used a size cut-off of: > 270 kb for deletions, > 500 kb for gains, and > 6.9 Mb for UPD, which were based on detecting deletions (106 kb), gains (207 kb), and UPD (2.2 Mb) at the 95th percentile in normal tissue. DNA from bone marrow (n = 88) and peripheral blood samples (n = 3), as well as paired constitutional DNA from 12 patients, was analyzed. In addition, 155 somatic aberrations were identified in 41 patients (supplemental Figure 1). These consisted of 67 deletions (43%) of median size 2 Mb (range, 0.3-158 mb), 54 gains (35%) of median size 3.7 Mb (range, 0.5-171 Mb), and 34 regions (22%) of UPD of median size 14 Mb (range, 6.9-104 Mb). Aberrations were detected in 15 RA/RCMD, 1 RARS, 12 RAEB-I/II, 3 CMML-I/II, 6 sAML, and 4 MDS/MPN-U patients. There was no significant difference observed in detected aberrations between cytogenetic risk groups (good, intermediate, and poor) and IPSS scores (low and high risk).
Of the 41 patients with genomic aberrations, 8 (20%) had TET2 mutations. Of these, UPCs 1932 and 2306 had deletions spanning the TET2 locus at 4q24, and UPC 2719 had UPD on 4q21.21-q35.2. UPC 1932 and 2719 had no additional aberrations, whereas UPC 2306 had additional deletions at 4q21.22-q22.1 (7.3 Mb) and 4q34.3 (492 kb). The remaining 5 patients (UPC 1326, UPC 1625, UPC 2844, UPC 2984, and UPC 1740) had no genomic aberrations associated with chromosome 4q24, but had additional aberrations as follows: UPC 1326 had UPD6q14.1-q15 (8.8 Mb), UPC 1625 had del 7q22.1 (736 kb), and UPD had 7q22.1-q36.3 (57 Mb), UPC 2844 had del 12p13.2 (277 kb), UPC 1740 had del 6q12.1-11.2 (2.1 Mb), and UPC 2984 had del 17p132.1-p13.1 (1.6 Mb), gains on 13q34 (518 kb), and 17q25.3 (751 kb) and UPDs on chromosomes 13 q13.3-q14.2 (10.4 Mb), q21.1-q21.33 (13.2 Mb), and q33.1-q34 (10.2 Mb). Nine patients with a TET2 mutation had no detectable genomic aberrations. Of note, there was no significant difference (P < .462) in the total number of genomic aberrations between TET2-mutated and nonmutated patients.
Sequencing versus SNP array data
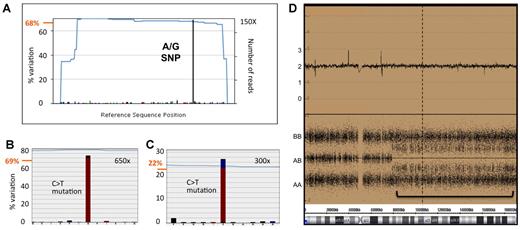
Patients 1932 and 2719 had TET2 mutations with an RMA of 98% and 71/20% (2 clones), respectively, consistent with loss oh heterozygosity (LOH) at the TET2 locus (see above). Interestingly, patient 2306, with LOH on the TET2 locus, had a mutation that was at 39% RMA, indicative of the mutation arising after the chromosomal abnormality. Consistent with these findings, patient 2306 was homozygous for SNPs within the TET2 coding region, according to the sequencing data (not shown). Interestingly, the sequencing data were also predictive of a further 8 LOH regions at the TET2 locus where SNP array analysis was not informative, having mutations that were found at greater than 65% RMA and consistent SNP frequencies (Table 1; example shown in Figure 5). Furthermore, the quantitative nature of the sequencing data allowed us to confirm cellular mixes, comprising clones with LOH and ones without. As an example, patient 2719 had an A/G SNP (nucleotide position 5284, rs2454206) that was found at 68% RMA, indicative of an LOH within a clonal mix (Figure 5A). This is consistent with the mutation status of this patient with 2 mutations, one at 69% and one at 22% RMA (shown in Figure 5B-C as a representative experiment and in Table 2 as an average of 3 such experiments [71% and 20%]), suggesting the existence of 2 disease clones and the occurrence of a second mutation event after recombination. Alternatively, this may also indicate multiple mutant clones existing entirely independently, perhaps as products of a single reciprocal recombination event occurring in an early clone containing both mutations. SNP data for this patient showed a UPD at this locus with a relatively high level of background noise, in line with expectations for a mixed population (Figure 5D). This could also be the case for other patients with multiple mutations that differ significantly in frequency from each other (see above). Conversely, patients with no genomic aberrations at the TET2 locus had a mutation frequency (RMA) consistent with heterozygosity.
The quantitative nature of NGS provides greater insight into LOH events. For UPC 2719, representative sequence read-out for one SNP (A) and 2 mutations (B-C) are displayed alongside SNP array data (D), showing a UPD over the TET2 locus. (A) An A to G SNP found in TET2 exon 11 is depicted by the dominant black bar and is found at 68% RMA (68% variation from the reference sequence indicated on the left axis), indicative of homozygosity and LOH in a population with mixed clonality. (B-C) Likewise, a C to T mutation in TET2 exon 7 is found at 69% RMA and another in exon 10 at 22% RMA, respectively, again indicative of multiple and distinct disease clones. The sequence coverage (number of reads) across the regions is indicated outside or inside the right axis in panel A or panels B and C, respectively, and traced by the blue lines in all cases. (D) Consistent with the sequence data, SNP6 array data show a region with LOH, indicative of UPD, highlighted by the green bar. The top panel represents a smooth copy number signal plot and a copy number of 2. The bottom panel shows individual genotypes for the SNP probes and a background of AB heterozygote calls consistent with mixed clonality.
The quantitative nature of NGS provides greater insight into LOH events. For UPC 2719, representative sequence read-out for one SNP (A) and 2 mutations (B-C) are displayed alongside SNP array data (D), showing a UPD over the TET2 locus. (A) An A to G SNP found in TET2 exon 11 is depicted by the dominant black bar and is found at 68% RMA (68% variation from the reference sequence indicated on the left axis), indicative of homozygosity and LOH in a population with mixed clonality. (B-C) Likewise, a C to T mutation in TET2 exon 7 is found at 69% RMA and another in exon 10 at 22% RMA, respectively, again indicative of multiple and distinct disease clones. The sequence coverage (number of reads) across the regions is indicated outside or inside the right axis in panel A or panels B and C, respectively, and traced by the blue lines in all cases. (D) Consistent with the sequence data, SNP6 array data show a region with LOH, indicative of UPD, highlighted by the green bar. The top panel represents a smooth copy number signal plot and a copy number of 2. The bottom panel shows individual genotypes for the SNP probes and a background of AB heterozygote calls consistent with mixed clonality.
Clinical associations
The association between TET2 mutation status and clinical variables is shown in Table 1. Previous studies have shown a high incidence of TET2 mutations in CMML patients.10 Likewise, a significantly higher proportion of CMML cases (46%) in our cohort carried the TET2 mutation (Table 1), compared with other World Health Organization (WHO) subtypes (12%; P < .001). Of note, none of the 8 cases with isolated 5q- abnormalities had TET2 mutations. Cytogenetic status, sex, WHO subtypes, transformation to AML, and IPSS score did not correlate with TET2 mutations. Similarly, no clinical association was attributed to the size of the mutant clone (P = .921) or the presence of single versus multiple TET2 mutations (P = .627). In addition, there was no significant difference in the treatment modalities received by TET2-mutated and nonmutated patients. Furthermore, exclusion of patients treated with 5-azacitidine or EPO/G-CSF (erythropoietin/granulocyte colony-stimulating factor) did not alter the significance (P = .68). However, increasing patient age was significantly associated with a higher incidence of TET2 mutations: 17% of patients above 50 years had a TET2 mutation, compared with 9% of patients aged less than 50 years (P = .02).
Effect of TET2 mutations on survival
At last follow-up, there were 170 deaths in the cohort, with 33 deaths being from the group of 55 patients with TET2 mutations. The median overall survival (OS) for the entire patient cohort was 35.0 months (95% confidence interval [CI]: 26.8-43.2). There was no significant difference in OS between patients with or without TET2 mutations (median OS: 30.0 months; 95% CI: 14.5-45.5 vs 36.0 months; 95% CI: 28.0-44.0.7; P = .37; Figure 6). When CMML (n = 35) and MDS (n = 320) patients were analyzed separately, no significant difference in OS was seen (P = .12 and P = .42, respectively). Among patients with a TET2 aberration, there was no difference in OS between patients with a high (> 25% RMA) or low (≤ 25%) level mutation (median OS: 26.0 months; 95% CI: 22.3-29.7 vs 35.0 months; 95% CI: 23.0-47.0; P = .45). In addition, homozygosity for TET2 mutations, defined as > 65% RMA, did not affect the OS. Inclusion of the 4 low-level mutations identified after sequencing the first 96 patients at greater depth did not alter the survival outcome in that subgroup. Likewise, there was no significant difference in OS between patients based on the presence of single or multiple mutations, even when all the variants initially identified (82 variants) were included in the analysis (65 patients; P = .15). Furthermore, when nonmutant patients with TET2 mRNA expression levels less than the 95th percentile (n = 15) were included together with cases having a TET2 mutation, OS remained nonsignificant (P = .77).
Survival curve depicting the impact of the presence or absence of TET2 mutations on the overall survival of the cohort.
Survival curve depicting the impact of the presence or absence of TET2 mutations on the overall survival of the cohort.
Within the cohort, 289 patients with MDS were evaluated based on the IPSS score at the time of diagnosis. Among the 192 patients with low/Int-1 IPSS scores, there was no difference in OS between patients with or without TET2 mutations (median OS: 52 months; 95% CI: 30.9-69.1 vs 65.0 months; 95% CI: 41.9.0-90.3; P = .33). Likewise, for the 97 patients with int-2/high-risk disease, the overall survival for TET2-mutated and nonmutated patients was equivalent (17.4 months; 95% CI: 3.3-31.4 vs 17.2 months; 95% CI: 11.3-23.0; P = .20). In the low/Int-1 subgroup of patients who were untreated (n = 162), there was no difference in OS between TET2-mutated and nonmutated patients (54.0 months; 95% CI: 40.1-68.5.0 vs 68.0 months; 95% CI: 16.6-119.4; P = .42).
Univariate analysis performed on the subgroup of patients with normal cytogenetics demonstrated that the presence of a TET2 mutation was associated with a significantly worse OS (32 vs 90 months; P = .03). However, this association could be confounded by the fact that patients with normal cytogenetics and a TET2 mutation were older, and more likely to have CMML.
Multivariate analysis showed that the presence of a TET2 mutation did not influence overall survival (odds ratio [OR]: 1.11; 95% CI: 0.98-1.25; P = .08). Advanced patient age (OR: 1.21; 95% CI: 1.05-1.38; P = .02), WHO classification (OR: 1.18; 95% CI: 1.07-1.38; P < .01), and cytogenetic subgroup (OR: 2.02; 95% CI: 1.66-2.46; P < .01) were the only variables significantly affecting OS (supplemental Table 3).
Discussion
TET2 mutations have been identified in a range of myeloid malignancies and are one of the most frequently acquired genetic aberrations associated with MDS.11,26 However, the prognostic significance of TET2 mutations remains unclear, with some studies suggesting favorable prognosis, whereas others show no significant contribution to survival in MDS.11,23,27 Previous studies of TET2 mutations have used conventional Sanger sequencing, which is constrained by a high cost and limitation in sensitivity, making this technology unsuitable for large-scale sequencing projects and inefficient in mutation screening. We have recently shown the utility of NGS technology for the detection of novel mutations across the TET2 gene, with a mutant-allele detection sensitivity of 1%-2% in total bone marrow DNA.12 The present study is the first to use NGS and high-resolution SNP array technology in 355 and 91 MDS/CMML patients, respectively, to simultaneously map and quantify mutant TET2 clones and additional genomic aberrations, and correlate the findings to the clinical status of these patients.
NGS technology has recently been used to sequence the complete genome of a person with AML, primarily confirming aberrations in known disease-associated genes. We used the massively parallel pyrosequencing NGS technique (Roche GS FLX), which, because of its relatively long sequence reads, is particularly suitable for mutation screening through exonic PCR amplicons.12 Furthermore, the increased sensitivity of NGS allows the identification of low-abundance mutant clones, enabling the study of disease evolution.28 We demonstrated an overall TET2 mutation rate of 15.5% in a cohort of 355 patients, with a TET2 mutation rate of 12%, specifically among MDS patients (vs 46% in CMML). As these mutations were predicted to significantly alter protein function, with a majority being nonsense or Indel mutations likely to abrogate TET2 protein function completely,9,11,12,23 the data strongly implicate TET2 mutations as having a distinct, yet unknown, biologic significance, rather than being a chance event, and is consistent with a tumor-suppressor role for this protein.
A prominent finding in our study was the infrequency of low-abundance TET2 mutant clones. Identification of such low-abundance mutant clones is important in understanding the relevance of TET2 protein function in disease and its contribution to clonal evolution. By increasing the sequence depth and sensitivity by 3-fold for 96 patients, we identified only 4 patients with low-abundance TET2-mutant clones (RMA of between 3% and 6.3%), of whom 3 had no previously detectable mutations. Moreover, by doubling sequence depth yet again for 13 of these patients, no additional TET2 mutations were discovered, suggesting that the moderate sequencing depth achieved in the primary mutation screen was adequate in identifying the majority of mutant cases. Of note is that all the low-abundance mutations seen here were predicted to abrogate protein function and therefore were unlikely to be background or neutral events. Moreover, the existence of low, but significant, numbers of minor clones is indicative of clonal disease evolution.29,30 This is highlighted by several multiple-mutant cases here, where we see one dominant and one low-abundance population, suggestive of clonal competition.
Previous reports have highlighted the occurrence of TET2 mutations in CD34+ cells, reinforcing the view that this aberration is an early disease lesion.9,11 To extend these studies, we screened CD34+ cells, CD3+ T cells, and skin tissue for TET2 mutations that were present in the bone marrow of affected patients. In line with previous findings,12 matched TET2 mutations were found in both CD34+ and bone marrow cells at similar levels of abundance (median, 38.5% and 43% RMA, respectively). Significantly, TET2 mutations were also found in CD3+ T cells (median, 23.5% RMA), confirming our previous findings.12 Background mutation levels in skin confirmed the acquired nature of these aberrations. This indicated that TET2 mutations were ubiquitous across myeloid and lymphoid lineages and suggests that such events occur early in hematopoiesis. Previous reports have suggested that TET2 mutations may precede the JAK2V617F mutation in MPN.27 However, after screening our samples for the JAK2V617F mutation and associated 46/1 predisposition genotype, no significant difference was observed between TET2 mutant versus nonmutant patients, showing that the 46/1 genotype was not associated with TET2 mutations in MDS and CMML.
We have previously identified a common region of UPD on chromosome 4q24 in MDS (8%) using 250-K SNP arrays that were subsequently shown to harbor TET2 mutations.6,12 In the present study, we used high-resolution SNP6 arrays in 91 patients and found that aberrations affecting the 4q24 region were identified in only 3 of 18 patients with TET2 mutations. These results support previous studies, which suggest that TET2 mutations and genomic aberrations affecting the 4q24 locus are independent events.7 using NGS sequencing to complement SNP6 arrays, we identified 8 additional cases with SNPs and mutations at > 65% RMA, indicative of LOH. Thus, 11 of 55 (20%) of TET2 mutant cases were associated with LOH at the TET2 locus. Interestingly, we identified 2 patients with 4q24 UPD and TET2 mutations at RMA 97% and 39%, respectively. The former mutation is consistent with homozygosity, whereas the latter would have had to have been acquired after the recombination event. Moreover, one patient (UPC 2719) who had UPD at TET2 also had 2 mutations that differed significantly in abundance from each other in bone marrow (69% vs 22% RMA), a relationship that was maintained in isolated CD34+ and T-cell populations, suggesting that LOH and TET2 mutations are independent events in some MDS patients.
An important feature of MDS is the increased frequency of UPD,5,6,8 which is an indirect measure of genomic instability.31,32 There was no significant difference between patients with mutant or wild-type TET2 in the frequency of UPD or copy-number aberrations, suggesting that TET2 defects do not contribute to general genomic instability. Furthermore, TET2 mutations are not linked to the 46/1 JAK2V617F mutation predisposition genotype, the only characterized genetic background associated with myeloid disease.24,33,34
We did not identify any correlation between the presence of TET2 mutations and overall survival for either MDS or CMML patients. This is in contrast to Kosmider et al,23 who have, in 2 separate studies, shown TET2 mutations as an independent favorable prognostic factor in MDS and associated with poor outcome in CMML patients. Any contradiction between the Kosmider MDS study and our own may, in part, be due to a relatively frequent transformation to AML in the non–TET2-mutated cases for the aforementioned study. Given that both Kosmider studies were retrospective in nature, it is possible that patient selection bias in either study could also have contributed to the disparity in observations. Our study benefited from the uniformity of data collected from a large cohort of patients from only 2 centers. Although the incidence of TET2 mutations in this study was slightly lower (15.5% overall; 12% MDS and 46% CMML) than that reported by other centers,9,11 this could be accounted for by the observation that advanced age correlates with an increased incidence of TET2 mutations, and that the median age in our study (64 years) was lower, compared with other studies.
The size of the mutant TET2 clone, presence of multiple versus single mutations and TET2 mRNA expression levels did not affect the overall survival. In addition, there is no evidence from our study that cases with homo- or hemizygous TET2 mutations show reduced survival relative to heterozygous mutations, as might be expected if aberrant TET2 were driving the disease independently.
The lack of correlation between the presence of TET2 mutations and prognosis in MDS suggests that several other factors affect the MDS phenotype. Nonetheless, the acquisition of TET2 mutations is an important event, both in the pathogenesis as well as the transformation of MDS. The latter is supported by 2 studies showing that, in MPN, the acquisition of TET2 mutations is associated with transformation to AML, and additional mutations in genes, such as ASXL1 or IDH1, are present in either a TET2 mutant or wild-type background bone marrow culture.20,21 Whether mutations in TET2 create a permissive environment or provide a “mutator phenotype”35 for subsequent mutations, perhaps via alterations in the hydroxymethylome, remains to be seen.18 Studies that interrogate both TET2 function and clonal evolution in parallel are likely to clarify the role of this gene in disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We acknowledge the Leukemia Lymphoma Research Fund (UK)/British Society for Hematology for supporting A.K. and King's College London for funding the Kings College Hemato-Oncology Tissue Bank, from which all local samples were processed. We acknowledge Rajani Chelliah for assisting with sample processing and tissue separation. We also thank the British Society for Hematology for funding travel for A.S. to present the results of this study.
Authorship
Contribution: A.E.S. and A.M.M. contributed equally and were involved with all aspects of the study's design, execution, analysis, and manuscript preparation; G.J.M. contributed to design, analysis, and manuscript preparation as well as providing project leadership; J.G. and N.L. contributed to experimental design and analysis; G.J.M., A.K., Z.L., and N.W. provided clinical details and analysis; N.W. provided Tissue Bank support and contributed to manuscript preparation; S.M. and both C.S. and E.N. contributed to sequencing and SNP array experiments, respectively; C.S., N.G., and U.G. provided clinical samples and details; and B.P. and J.M. were involved with SNP array analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ghulam J. Mufti, Department of Hematological Medicine, King's College London, The Rayne Institute, 123 Coldharbour Lane, London, SE5 9NU, United Kingdom; e-mail: ghulam.mufti@kcl.ac.uk.
References
Author notes
A.E.S. and A.M.M. contributed equally to this work.

![Figure 1. Seventy-one mutations in 55 of 355 persons were identified by NGS and mapped to the TET2 coding region. Mutations were identified that existed at ≥ 10% relative mutation abundance (RMA). These were mapped against TET2 translated exons 3-11 (NM.001127208; 2002 amino acids). Classification of mutations is indicated in the figure key and includes nonsense or insertion/deletion mutations (Indels), nonsynonymous amino acid changes, and splice-site mutations, indicated by orange, blue, and pink bars, respectively. Mutation level is also defined as > or ≤ 25% RMA, indicated by the solid or stippled bars, respectively. Regions conserved across the TET protein family, and implicated in the conversion of 5′ methylcytosine to 5′ methylhydroxycytosine (5-hmC), are shown (cons'd regions 1 and 2) and correspond to amino acids 1104-1478 and 1845-2002, respectively ([11] [18]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/19/10.1182_blood-2010-03-274704/5/m_zh89991060180001.jpeg?Expires=1769123893&Signature=4P7uKSniMJ95AbxJQDrxy3Znmck0gBl9soP5rLBuO5m6wJ-tZQutdhdu33jKd-u9riPxWwL1cdqy5ynfrZm~6m6BW2~kjmkEmKNhyBbXYd7RTL2eqeQx-bJ28E5lGAM4-4N4UH-RZ3oYNg78rx3FsRBK9kh6DmYJ0oI0XEZBkkT7gKMO4PeRTzBk-ZajS3PxsNw83hGy7E16EZzCgfEph4LQlron1DhVHRzJikPnX9~r7qElqZPpFDhKBdjibqpqzIFuTxztobgHE83DJaQhhChCj~6HXNdKSaOju9y~VkhtnDXwY9mEH35rZngoAvc1MlkfeDGvpXqYkpoYCPTIvA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. NGS clonal sequencing allows direct detection of independent disease clones. (A) Two C to T substitutions in TET2 exon 11 are depicted by the dominant red bars and are found in approximately 17% of sequence reads (17% variation [left axis] from the reference sequence [x-axis]). The sequence coverage (number of reads) across the region is shown on the right axis and is traced by the blue line. Experimental noise is shown underneath the dotted line with reads for bases A (green bars), C (blue bars), G (black bars), T (red bars), and base deletions (gray bars) at less than 4% of reads. (B) A selection of individual sequence reads lined up against each other is shown and demonstrates that the 2 mutations are found in separate molecules being sequenced and thus belong to independent clones or alternate alleles.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/19/10.1182_blood-2010-03-274704/5/m_zh89991060180002.jpeg?Expires=1769123893&Signature=bP~NuIosHvRag4wF46KvnaYlmlHTVUye8aQ~lifEZKC-yGYyNcVWBfmdT-5sU4a2QuW~utdQLigTD39Zdy~py3tLqK9acpO9MA6nDtIpetir9hpaoOX~FAt2Upf4BxUlNRu~Jcv7GBMzq6zGUhhpGqxSyWU~coER7S19cfMEXfoW5lXa3iEO3sPPGlfD3nEORjhpdFJFO9OdEjJNGQYDqOd0g8kw6WVyYUMqJLuHFDRSurMNxZTqp~1JVZR8Fuga-8YgT5-AzMOcmS1fru~02ZcY-8WU6YNZ8bTwZjtdAWZaXLZ7MwcorydOWV1tvYtgNP5ytBXpp3rRrEAeLtMcEw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)