Abstract
The ability of dendritic cells (DCs) to cross-present protein tumor antigens to cytotoxic T lymphocytes (CTLs) underpins the success of therapeutic cancer vaccines. We studied cross-presentation of the cancer/testis antigen, NY-ESO-1, and the melanoma differentiation antigen, Melan-A by human DC subsets. Monocyte-derived DCs (MoDCs) efficiently cross-presented human leukocyte associated (HLA)–A2-restricted epitopes from either a formulated NY-ESO-1/ISCOMATRIX vaccine or when either antigen was mixed with ISCOMATRIX adjuvant. HLA-A2 epitope generation required endosomal acidification and was proteasome-independent for NY-ESO-1 and proteasome-dependent for Melan-A. Both MoDCs and CD1c+ blood DCs cross-presented NY-ESO-1–specific HLA-A2157-165–, HLA-B760-72–, and HLA-Cw392-100–restricted epitopes when formulated as an NY-ESO-1/ISCOMATRIX vaccine, but this was limited when NY-ESO-1 and ISCOMATRIX adjuvant were added separately to the DC cultures. Finally, cross-presentation of NY-ESO-1157-165/HLA-A2, NY-ESO-160-72/HLA-B7, and NY-ESO-192-100/HLA-Cw3 epitopes was proteasome-dependent when formulated as immune complexes (ICs) but only proteasome-dependent for NY-ESO-160-72/HLA-B7–restricted cross-presentation facilitated by ISCOMATRIX adjuvant. We demonstrate, for the first time, proteasome-dependent and independent cross-presentation of HLA-A–, B–, and C–restricted epitopes within the same full-length tumor antigen by human DCs. Our findings identify important differences in the capacities of human DC subsets to cross-present clinically relevant, full-length tumor antigens and how vaccine formulation impacts CTL responses in vivo.
Introduction
A major goal of tumor antigen vaccination is the generation and expansion of cytotoxic T lymphocytes (CTLs) that can recognize and kill tumor cells. To prime a naive or to reactivate memory tumor antigen-specific CD8+ T cells, exogenous tumor cell-derived or vaccine-associated antigens must first be cross-presented on class I major histocompatibility (MHC) molecules.1-4 Dendritic cells (DCs) are considered the principal cross-presenting antigen-presenting cell that specializes in the priming of antigen-specific CD8+ T cells and, as a consequence, represent targets for tumor antigen vaccination strategies.5-9 DCs take up exogenous tumor antigens into endosomes and/or phagolysosomes and translocate these into the cytosol so as to enter the class I MHC processing pathway for presentation to tumor-specific CD8+ T cells.6,10,11 However, not all DCs are equal with respect to cross-presenting capacity.12,13
The cross-presenting capacity of various DC subsets has been most thoroughly studied in the mouse system where CD8α+ DCs resident in lymphoid tissues are highly specialized and can constitutively cross-present exogenous antigen.14 Although recent reports suggest that the rare BDCA3+ blood circulating human DC subset most closely resembles the cross-presenting murine CD8α+ DCs,15,16 a direct human equivalent has not been conclusively identified. Thus, drawing functional comparisons between mouse and human DC subsets has limitations and highlights the need to understand the mechanisms by which human DC subsets generate tumor antigen- or virus-specific CTL epitopes.
The majority of human studies and clinical trials have used in vitro-generated monocyte-derived DCs (MoDCs),16-18 which most closely represent inflammatory-type DCs (rather than lymphoid tissue-resident, steady-state DCs) in vivo.19 In human blood, there are at least 3 distinct DC populations: (1) CD1c+ DCs, (2) BDCA3+ DCs, and (3) plasmacytoid DCs (pDCs).20-22 However, there are limited reports on the cross-presenting efficiency of these human blood DC subsets compared with MoDCs.4 Thus, understanding the mechanisms governing epitope-specific tumor antigen cross-presentation by human DC subsets is critical to the rational approach used to generate tumor antigen vaccine-specific CTLs.
The choice of tumor antigen and adjuvant will likely play a critical role in the success of any given cancer vaccine. The cancer/testis antigen, NY-ESO-1, and the melanoma differentiation antigen, Melan-A, are clinical targets that have generated considerable interest. NY-ESO-1 is expressed in a wide range of solid cancer types,23-25 is highly immunogenic resulting in spontaneous antibody and T-cell responses in NY-ESO-1+ cancer patients,25 has been used as a therapeutic vaccine target in several clinical trials, and generates broad CD4+ and CD8+ T-cell responses in humans.26 In contrast, Melan-A is unusual. First, a large proportion of healthy human leukocyte associated-A2 (HLA-A2)–restricted individuals possess naive CD8+ T cells specific for the naturally presented Melan-A26-35/HLA-A2 epitope.27 Second, the Melan-A26-35/HLA-A2 epitope is strikingly immunodominant with few other CD8+ T-cell epitopes described.28 Third, the Melan-A26-35/HLA-A2 epitope is generated and presented by HLA-A2+ melanoma cells, and these are efficiently lysed by Melan-A26-35/HLA-A2-specific CTLs.29 Finally, the Melan-A26-35/HLA-A2 epitope is generated by conventional proteasomes but may be destroyed by immunoproteasomes expressed by DCs or by interferon-γ (IFN-γ)–treated tumor cells.30 This final point makes the development of a Melan-A protein vaccine problematic as immunoproteasome-expressing DCs will fail to generate tumor-relevant CTLs in vivo, thus restricting the development of a Melan-A vaccine to peptide-based strategies, which have shown limited clinical efficacy because of multiple tumor escape mechanisms.31
Soluble protein antigens are typically poorly cross-presented by DCs, and adjuvants have been used in vitro and in vivo to enhance this process. ISCOMATRIX adjuvant is based on the immune-stimulating complexes (ISCOMS) technology combining an efficient antigen delivery system with the immune-stimulatory activity of saponin.32 ISCOMATRIX-formulated vaccines are effective in promoting both humoral and cellular immune responses, and the NY-ESO-1/ISCOMATRIX vaccine induces tumor-specific CTLs in humans and tumor protection in mice.26,33
In our current report, we describe the DC subset and epitope-specific cross-presentation of NY-ESO-1 and Melan-A facilitated by ISCOMATRIX adjuvant or by immune complexes (ICs). We find that tumor antigen and adjuvant formulation significantly impact the ability of human DCs to cross-present multiple tumor-specific CTL epitopes and that DCs use diverse processing mechanisms to generate distinct epitopes from individual protein antigens.
Methods
Cell culture
Peripheral blood mononuclear cells (PBMCs) from buffy coats of healthy donors (Red Cross Blood Bank) were prepared by Ficoll-Paque density gradient centrifugation (GE Healthcare). Monocytes or CD1c+ peripheral blood (PB) DCs were isolated by positive selection using magnetic beads specific for CD14 or CD1c, respectively (Miltenyi Biotec). Cell purity was greater than 96% as determined by flow cytometry.
MoDCs were generated by culturing CD14+ monocytes with granulocyte-macrophage colony-stimulating factor (GM-CSF; 20 ng/mL) and interleukin-4 (IL-4; 10 ng/mL) for 6 to 7 days. Cell cultures were maintained in RPMI 1640 supplemented with 20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 60 mg/L penicillin, 12.5 mg/L streptomycin, 2mM l-glutamine, 1% nonessential amino acids, and 10% heat-inactivated fetal calf serum (culture media).
Generation of recombinant NY-ESO-1 and Melan-A formulations
Full-length NY-ESO-1 and Melan-A protein were produced in Escherichia coli by the Ludwig Institute for Cancer Research (LICR) and purified as described.34 Endotoxin levels ranged between 3 and 31 EU/0.1 mg of protein (limit < 175 EU/0.1 mg protein). Formulated NY-ESO-1/ISCOMATRIX vaccine was generated whereby NY-ESO-1 antigen was physically incorporated into ISCOMATRIX particles as described,33 and the protein concentration was determined by amino acid analysis. ISCOMATRIX is a registered trademark of ISCOTEC AB, a CSL Limited company. NY-ESO-1/anti-NY-ESO-1 ICs were generated by mixing NY-ESO-1 protein with anti-NY-ESO-1 monoclonal antibody ES121 at a 2:1 molar ratio in serum-free RPMI at 37°C for 30 minutes.
Generation of NY-ESO-1 and Melan-A-specific human CD8+ T-cell lines and clones
CD8+ T-cell lines specific for NY-ESO-1157-165/HLA-A2, NY-ESO-160-72/HLA-B7, NY-ESO-192-100/HLA-Cw3, or the Melan-A26-35/HLA-A2 epitope were established from melanoma patient PBMCs. Cells were collected under protocols approved by the Human Research Ethics Committee of the Austin Hospital (Melbourne, Australia) and the LICR. T-cell lines were maintained in medium containing 20 IU/mL IL-2 (Cetus) and restimulated with irradiated feeder cells and irradiated peptide-pulsed antigen-presenting cells every 12 to 14 days. The percentage of NY-ESO-1 or Melan-A-specific T cells ranged between 10% to 90%. NY-ESO-1157-165/HLA-A2 and NY-ESO-160-72/HLA-B7 T-cell clones were generated from specific CTL lines by tetramer-guided flow cytometry. Single cells were expanded on irradiated pooled feeder PBMCs with 1 μg/mL phytohemagglutinin and 600 IU/mL IL-2 as described.35
Tumor antigen cross-presentation assays
MoDCs or freshly isolated CD1c+ DCs were plated at 1 × 105 cells in 200 μL in 96-well, round-bottom culture plates in their own day 6 or 7 conditioned media or fresh culture media supplemented with GM-CSF, respectively. DCs plated in media were then pulsed with antigens in the absence or presence of ISCOMATRIX adjuvant or as ICs for 0.5 to 18 hours and, in some cases (see Figures 2 and 5), simultaneously stimulated with combinations of 2 μg/mL CD40L-trimer (a kind gift from Amgen), 100 ng/mL IFN-γ (PeproTech), 1800 U/mL IFN-α (supplied through the LICR clinical program), 2.5 μg/mL CpG-A (Invivogen), 1 μg/mL R848 (Sapphire Bioscience), 12.5 μg/mL polyinosinic acid–polycytidylic acid (Poly I:C), or 100 ng/mL E coli–derived lipopolysaccharide (LPS; Sigma-Aldrich).
In some assays, DCs were incubated for 45 minutes before and throughout the antigen pulse period with lactacystin, Ala-Ala-Phe-chloromethylketone (AAF-CMK; Sigma-Aldrich) or concanamycin B (ConB) (a kind gift from Dr J. Villadangos, Walter and Eliza Hall Institute). To inhibit tripeptidyl peptidase II (TPPII) activity, DCs were washed 3 times to remove serum and then plated in specific serum-free media (AIM-V; Invitrogen) supplemented with 10 ng/mL GM-CSF before addition of butabindide oxalate (Tocris Bioscience) 45 minutes before and throughout the antigen pulse period. DCs pulsed with relevant NY-ESO-1 or Melan-A–restricted peptides (Auspep) served as positive controls. After washing, DCs and NY-ESO-1 or Melan-A peptide-specific CTL lines or clones were cocultured in 96-well, round-bottom plates in the presence of 10 μg/mL brefeldin A (BFA; Sigma-Aldrich) for 4 hours. Cells were then washed and stained with anti-CD4 and anti-CD8 monoclonal antibodies for 20 minutes at 4°C, fixed with 1% paraformaldehyde, washed, and stained with anti-IFN-γ monoclonal antibody (BD Biosciences) in a 0.25% saponin buffer. Samples were analyzed by flow cytometry, and data were analyzed using FlowJo software (Version 3.4; TreeStar).
Statistics
Unless indicated, results are expressed as the mean plus or minus 1 SD of 3 or more donors or the mean plus or minus 1 SD of triplicate cultures, and P values were calculated using an unpaired 2-tailed Student t test with P less than .02 considered significant.
Results
ISCOMATRIX adjuvant facilitates cross-presentation of NY-ESO-1157-165 and Melan-A26-35 on HLA-A2
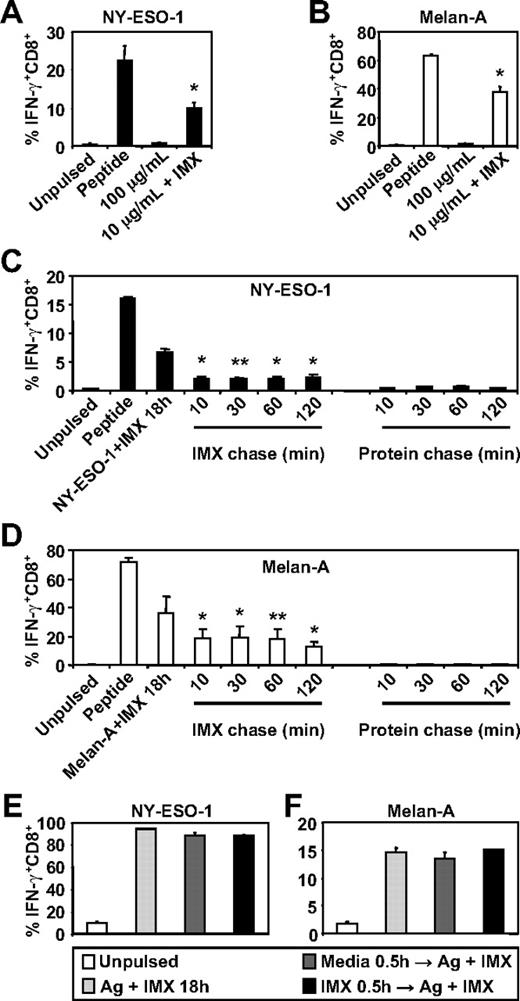
To investigate whether ISCOMATRIX adjuvant influences the generation of tumor antigen-specific CTL epitopes by human DC subsets, we first compared the ability of MoDCs to cross-present either NY-ESO-1157-165 or Melan-A26-35 onto HLA-A2 molecules either as soluble protein antigen alone or when mixed with ISCOMATRIX adjuvant. We found that MoDCs were ineffective in cross-presenting, even at relatively high concentrations of soluble NY-ESO-1 or Melan-A protein, but efficiently cross-presented either antigen (even at a 10-fold lower antigen dose) when added to the cultures with ISCOMATRIX adjuvant (Figure 1A-B). Surprisingly, pulse-chase experiments revealed that ISCOMATRIX adjuvant could facilitate cross-presentation of both NY-ESO-1 and Melan-A proteins by MoDCs, even when added 2 hours after the antigen pulse (Figure 1C-D). In contrast, this did not occur if MoDCs were first pulsed with ISCOMATRIX adjuvant and then exposed to antigen (Figure 1C-D). To exclude the possibility that pre-exposure to ISCOMATRIX adjuvant shuts down the capacity of MoDCs to cross-present, we plated MoDCs with or without ISCOMATRIX adjuvant and, after washing, copulsed these DCs with antigen and ISCOMATRIX adjuvant. The results demonstrate that ISCOMATRIX adjuvant does not negatively affect the subsequent capacity of MoDCs to cross-present either NY-ESO-1 or Melan-A (Figure 1E-F), and taken together, indicate that ISCOMATRIX adjuvant did not alter the functional capacity of MoDCs to cross-present per se, but rather influenced intracellular antigen trafficking after antigen uptake.
MoDCs cross-present the HLA-A2-restricted epitopes from NY-ESO-1 and Melan-A in the presence of ISCOMATRIX adjuvant. Immature MoDCs (1 × 105) were either unpulsed or pulsed with 100 or 10 μg/mL of (A) NY-ESO-1 or (B) Melan-A protein alone or together with 10 μg/mL ISCOMATRIX adjuvant for 18 hours in 96-well, round-bottom plates. After being washed, MoDCs were cocultured with HLA-A2-restricted NY-ESO-1 or Melan-A-specific CTL lines for 4 hours in the presence of 10 μg/mL BFA. A standard intracellular cytokine stain (ICS) was then performed, and IFN-γ levels were assessed by flow cytometry. MoDCs pulsed with cognate peptide for 30 minutes before the 4-hour co-culture were also included to assess maximal peptide-specific IFN-γ responses. (C-F) Immature MoDCs were pulsed with 10 μg/mL of NY-ESO-1 or Melan-A protein together with 10 μg/mL ISCOMATRIX adjuvant for 18 hours or pulsed with antigen or ISCOMATRIX for 30 minutes before being washed 3 times, and then chased with antigen or ISCOMATRIX at the times indicated (C-D) or with antigen together with ISCOMATRIX (E-F). Pulsed MoDCs were cultured for a further 18 hours before being cocultured with specific CTL lines as in panels A and B. Data are mean ± SD of 3 separate donors. **P < .02, *P < .01 vs no addition of ISCOMATRIX (A-B) and vs the protein chase (C-D).
MoDCs cross-present the HLA-A2-restricted epitopes from NY-ESO-1 and Melan-A in the presence of ISCOMATRIX adjuvant. Immature MoDCs (1 × 105) were either unpulsed or pulsed with 100 or 10 μg/mL of (A) NY-ESO-1 or (B) Melan-A protein alone or together with 10 μg/mL ISCOMATRIX adjuvant for 18 hours in 96-well, round-bottom plates. After being washed, MoDCs were cocultured with HLA-A2-restricted NY-ESO-1 or Melan-A-specific CTL lines for 4 hours in the presence of 10 μg/mL BFA. A standard intracellular cytokine stain (ICS) was then performed, and IFN-γ levels were assessed by flow cytometry. MoDCs pulsed with cognate peptide for 30 minutes before the 4-hour co-culture were also included to assess maximal peptide-specific IFN-γ responses. (C-F) Immature MoDCs were pulsed with 10 μg/mL of NY-ESO-1 or Melan-A protein together with 10 μg/mL ISCOMATRIX adjuvant for 18 hours or pulsed with antigen or ISCOMATRIX for 30 minutes before being washed 3 times, and then chased with antigen or ISCOMATRIX at the times indicated (C-D) or with antigen together with ISCOMATRIX (E-F). Pulsed MoDCs were cultured for a further 18 hours before being cocultured with specific CTL lines as in panels A and B. Data are mean ± SD of 3 separate donors. **P < .02, *P < .01 vs no addition of ISCOMATRIX (A-B) and vs the protein chase (C-D).
DC activation neither enhances nor inhibits cross-presentation of NY-ESO-1157-165 or Melan-A26-35 on HLA-A2
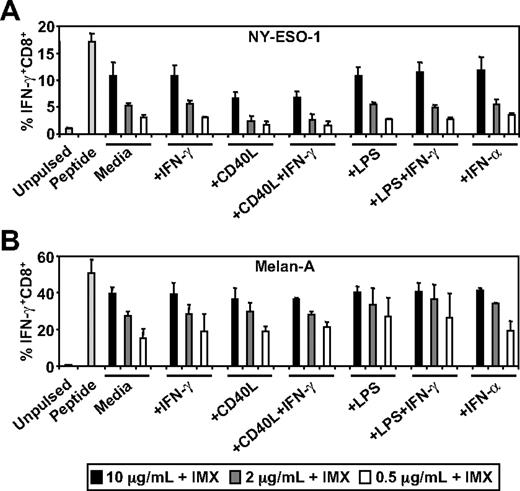
Stimuli, such as CD40L and type I interferon, can induce the conversion of the somatic proteasome to the immunoproteasome in DCs which, in turn, can enhance or inhibit antigen cross-presentation.30,36,37 This is particularly relevant in the case of Melan-A where it has been demonstrated that conversion of the somatic proteasome to the immunoproteasome results in the inability of DCs or tumor cells to generate the immunodominant Melan-A26-35/HLA-A2 CTL epitope.30 To test the effects of DC activation on ISCOMATRIX-mediated cross-presentation, we pulsed MoDCs with NY-ESO-1 or Melan-A together with ISCOMATRIX adjuvant, in either the absence or presence of potent stimulators of DC activation. Surprisingly, we found that additional DC activation had neither an enhancing nor inhibitory effect on the cross-presentation of NY-ESO-1157-165/HLA-A2 or Melan-A26-35/HLA-A2 epitopes (Figure 2).
Effects of DC activation on ISCOMATRIX facilitated HLA-A2-restricted cross-presentation of NY-ESO-1 or Melan-A. Immature MoDCs (1 × 105) were either unpulsed or pulsed with 0.5 to 10 μg/mL of (A) NY-ESO-1 or (B) Melan-A protein together with 10 μg/mL ISCOMATRIX adjuvant in the absence or presence of 100 ng/mL IFN-γ, 2 μg/mL CD40L, 100 ng/mL LPS, or 1800 U/mL IFN-α in 96-well, round-bottom plates for 18 hours. After being washed, MoDCs were cocultured with HLA-A2-restricted NY-ESO-1 or Melan-A-specific CTL lines for 4 hours in the presence of 10 μg/mL BFA. A standard ICS was then performed, and IFN-γ levels were assessed by flow cytometry. Data are mean ± SD of 3 separate donors.
Effects of DC activation on ISCOMATRIX facilitated HLA-A2-restricted cross-presentation of NY-ESO-1 or Melan-A. Immature MoDCs (1 × 105) were either unpulsed or pulsed with 0.5 to 10 μg/mL of (A) NY-ESO-1 or (B) Melan-A protein together with 10 μg/mL ISCOMATRIX adjuvant in the absence or presence of 100 ng/mL IFN-γ, 2 μg/mL CD40L, 100 ng/mL LPS, or 1800 U/mL IFN-α in 96-well, round-bottom plates for 18 hours. After being washed, MoDCs were cocultured with HLA-A2-restricted NY-ESO-1 or Melan-A-specific CTL lines for 4 hours in the presence of 10 μg/mL BFA. A standard ICS was then performed, and IFN-γ levels were assessed by flow cytometry. Data are mean ± SD of 3 separate donors.
Cross-presentation of Melan-A26-35/HLA-2 by MoDCs is proteasome dependent, whereas NY-ESO-1157-165/HLA-A2 is proteasome independent
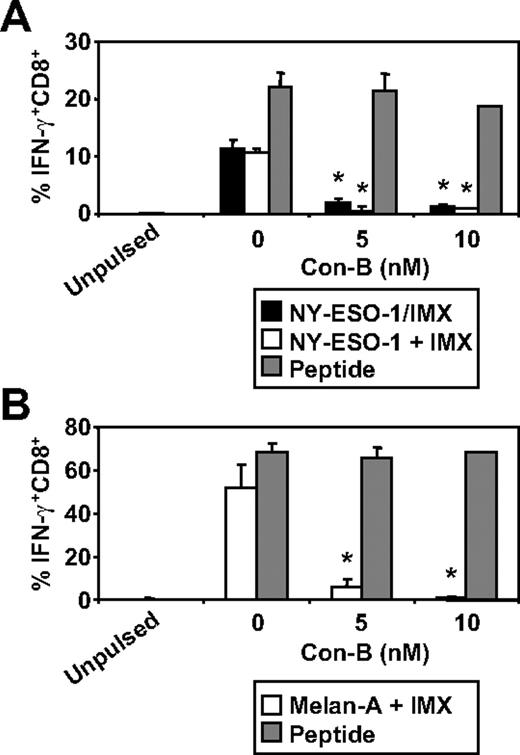
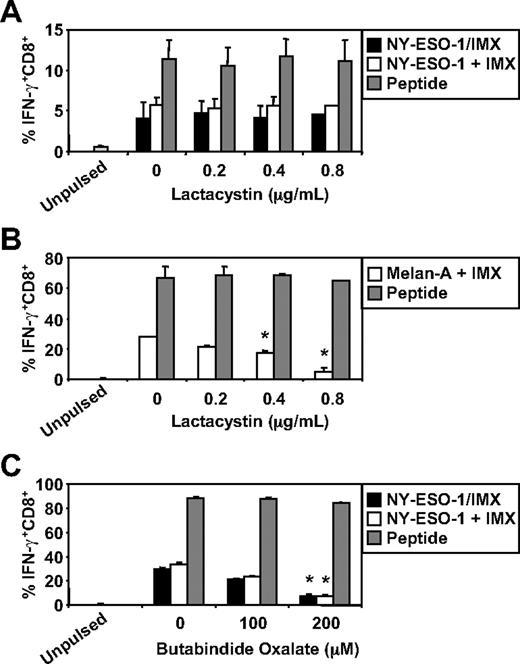
We have recently documented some of the processing mechanisms used by DCs for the HLA-A2-restricted cross-presentation of a formulated NY-ESO-1/ISCOMATRIX vaccine (associated antigen/adjuvant complexes) and NY-ESO-1/ICs (antigen/antibody complex). These studies demonstrated that the HLA-A2-restricted cross-presentation of each of these NY-ESO-1 antigen formulations required endosomal acidification but were either proteasome independent or proteasome dependent, respectively.4,38 To more fully understand the mechanisms used by DCs for the efficient cross-presentation of NY-ESO-1 and Melan-A, we again inhibited components of the antigen-processing machinery. As shown in Figure 3, inhibition of endosomal acidification via the vacuolar type H[+]-ATPase protein pump inhibitor, ConB, abolished NY-ESO-1157-165/HLA-A2-restricted cross-presentation of the NY-ESO-1/ISCOMATRIX vaccine as well as inhibited both the cross-presentation of NY-ESO-1157-165/HLA-A2 and Melan-A26-35/HLA-A2 epitopes when antigen and adjuvant were added separately to the MoDC cultures. Furthermore, cytosolic proteasomal processing was differentially required for these 2 tumor antigens. Generation of the NY-ESO-1157-165/HLA-A2 epitope was unaffected by the proteasome inhibitor, lactacystin, irrespective of whether NY-ESO-1 was formulated as a vaccine or when each component was added separately to the cultures (Figure 4A). Conversely, the generation of the Melan-A26-35/HLA-A2 epitope was significantly inhibited by lactacystin (Figure 4B) but was insensitive to the serine protease inhibitor, AAF-CMK (not shown). This suggests that, unlike the NY-ESO-1157-165/HLA-A2 epitope, the generation of the Melan-A26-35/HLA-A2 epitope is proteasome-dependent. Finally, as an extension to our previous findings, we found that generation of the NY-ESO-1157-165/HLA-A2 epitope, irrespective of antigen and adjuvant formulation, required processing by TPPII as in each case cross-presentation was significantly inhibited by addition of butabindide oxalate (Figure 4C).
Effects of inhibiting endosomal acidification on ISCOMATRIX facilitated HLA-A2-restricted cross-presentation of NY-ESO-1 or Melan-A by MoDCs. Immature MoDCs (1 × 105) were either untreated or treated with 5 or 10nM Con-B for 45 minutes before and throughout the 18-hour pulse with 10 μg/mL NY-ESO-1/ISCOMATRIX vaccine or 10 μg/mL NY-ESO-1 (A) or Melan-A protein (B) together with 10 μg/mL ISCOMATRIX adjuvant in 96-well, round-bottom plates. In parallel, untreated or inhibitor-treated MoDCs were pulsed with cognate peptide for 30 minutes and then washed thoroughly before the 4-hour coculture with CTL lines to control for nonspecific effects of the inhibitor. A standard ICS was then performed, and IFN-γ levels were assessed by flow cytometry. Data are mean ± SD of 3 separate donors. *P < .01 vs no Con-B treatment.
Effects of inhibiting endosomal acidification on ISCOMATRIX facilitated HLA-A2-restricted cross-presentation of NY-ESO-1 or Melan-A by MoDCs. Immature MoDCs (1 × 105) were either untreated or treated with 5 or 10nM Con-B for 45 minutes before and throughout the 18-hour pulse with 10 μg/mL NY-ESO-1/ISCOMATRIX vaccine or 10 μg/mL NY-ESO-1 (A) or Melan-A protein (B) together with 10 μg/mL ISCOMATRIX adjuvant in 96-well, round-bottom plates. In parallel, untreated or inhibitor-treated MoDCs were pulsed with cognate peptide for 30 minutes and then washed thoroughly before the 4-hour coculture with CTL lines to control for nonspecific effects of the inhibitor. A standard ICS was then performed, and IFN-γ levels were assessed by flow cytometry. Data are mean ± SD of 3 separate donors. *P < .01 vs no Con-B treatment.
Effects of proteasome inhibition on ISCOMATRIX facilitated HLA-A2-restricted cross-presentation of NY-ESO-1 or Melan-A by MoDCs. Immature MoDCs (1 × 105) were either untreated or treated with 0.2 to 0.8μM lactacystin for 45 minutes before and throughout the 18-hour pulse with 10 μg/mL NY-ESO-1/ISCOMATRIX vaccine or 10 μg/mL NY-ESO-1 (A) or Melan-A protein (B) together with 10 μg/mL ISCOMATRIX adjuvant in 96-well, round-bottom plates. (C) MoDCs were first washed 3 times to remove serum before being treated with butabindide oxalate for 45 minutes before and throughout the 18-hour antigen pulse. In parallel, untreated or inhibitor-treated MoDCs were pulsed with cognate peptide for 30 minutes and then washed thoroughly before the 4-hour coculture with CTL lines to control for nonspecific effects of the inhibitors. A standard ICS was then performed, and IFN-γ levels assessed by flow cytometry. Data are mean ± SD of 3 separate donors. *P < .01 vs no addition of lactacystin or butabindide oxalate.
Effects of proteasome inhibition on ISCOMATRIX facilitated HLA-A2-restricted cross-presentation of NY-ESO-1 or Melan-A by MoDCs. Immature MoDCs (1 × 105) were either untreated or treated with 0.2 to 0.8μM lactacystin for 45 minutes before and throughout the 18-hour pulse with 10 μg/mL NY-ESO-1/ISCOMATRIX vaccine or 10 μg/mL NY-ESO-1 (A) or Melan-A protein (B) together with 10 μg/mL ISCOMATRIX adjuvant in 96-well, round-bottom plates. (C) MoDCs were first washed 3 times to remove serum before being treated with butabindide oxalate for 45 minutes before and throughout the 18-hour antigen pulse. In parallel, untreated or inhibitor-treated MoDCs were pulsed with cognate peptide for 30 minutes and then washed thoroughly before the 4-hour coculture with CTL lines to control for nonspecific effects of the inhibitors. A standard ICS was then performed, and IFN-γ levels assessed by flow cytometry. Data are mean ± SD of 3 separate donors. *P < .01 vs no addition of lactacystin or butabindide oxalate.
CD1c+ blood DCs are inefficient at cross-presentation of either NY-ESO-1157-165/HLA-A2 or Melan-A26-35/HLA-A2 epitopes when mixed with ISCOMATRIX adjuvant
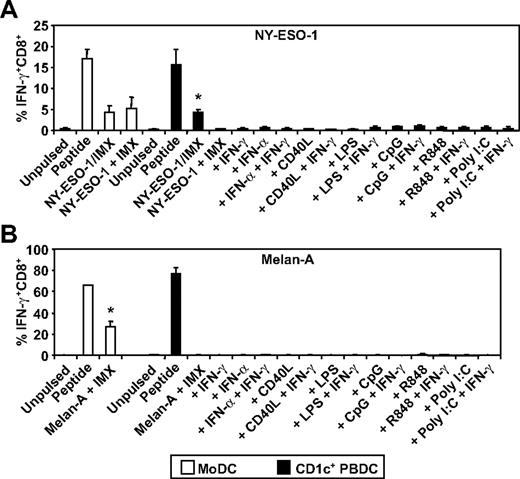
To extend our analysis, we compared 2 human DC subsets for their ability to cross-present the NY-ESO-1157-165/HLA-A2 or the Melan-A26-35/HLA-A2 epitope. To do this, we purified HLA-A2+ CD1c+ DCs from blood (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and compared these with in vitro generated MoDCs in their ability to cross-present the NY-ESO-1/ISCOMATRIX vaccine or each full-length tumor antigen when added separately to the cultures with ISCOMATRIX adjuvant. In each case, the tumor antigens were efficiently cross-presented by MoDCs; but surprisingly, CD1c+ DCs only cross-presented the formulated NY-ESO-1/ISCOMATRIX vaccine and were largely incapable of cross-presentation when antigen and adjuvant were added separately to the cultures (Figure 5). Furthermore, this poor capacity to cross-present could not be enhanced after DC activation with an array of stimuli (Figure 5A-B). Importantly, each DC subset was equally capable of cross-presenting the NY-ESO-1/ISCOMATRIX vaccine and of presenting exogenously loaded NY-ESO-1157-165/HLA-A2 or Melan-A26-35/HLA-A2 peptides (Figure 5A-B).
ISCOMATRIX facilitated HLA-A2-restricted cross-presentation of NY-ESO-1 or Melan-A by MoDCs and CD1c+ DCs. Immature MoDCs (1 × 105) or freshly isolated CD1c+ DCs (1 × 105) were either unpulsed or pulsed with 10 μg/mL NY-ESO-1/ISCOMATRIX vaccine or 10 μg/mL NY-ESO-1 (A) or Melan-A protein (B) together with 10 μg/mL ISCOMATRIX adjuvant in the absence or presence of 100 ng/mL IFN-γ, 1800 U/mL IFN-α, 2 μg/mL CD40L, 100 ng/mL LPS, 2.5 μg/mL CpG-A (2216), 1 μg/mL R848, or 12.5 μg/mL Poly I:C in 96-well, round-bottom plates. After 18 hours, DCs were washed and cocultured with HLA-A2-restricted NY-ESO-1 or Melan-A-specific CTL lines for 4 hours in the presence of 10 μg/mL BFA. A standard ICS was then performed, and IFN-γ levels were assessed by flow cytometry. DCs pulsed with cognate peptide for 30 minutes before the 4-hour coculture with CTL lines served as positive controls. Data are mean ± SD of 3 separate donors. (A) *P < .01 vs unpulsed CD1c+ DCs and vs CD1c+ DCs pulsed with NY-ESO-1 and ISCOMATRIX adjuvant separately with or without additional stimulation. (B) *P < .01 vs unpulsed MoDCs.
ISCOMATRIX facilitated HLA-A2-restricted cross-presentation of NY-ESO-1 or Melan-A by MoDCs and CD1c+ DCs. Immature MoDCs (1 × 105) or freshly isolated CD1c+ DCs (1 × 105) were either unpulsed or pulsed with 10 μg/mL NY-ESO-1/ISCOMATRIX vaccine or 10 μg/mL NY-ESO-1 (A) or Melan-A protein (B) together with 10 μg/mL ISCOMATRIX adjuvant in the absence or presence of 100 ng/mL IFN-γ, 1800 U/mL IFN-α, 2 μg/mL CD40L, 100 ng/mL LPS, 2.5 μg/mL CpG-A (2216), 1 μg/mL R848, or 12.5 μg/mL Poly I:C in 96-well, round-bottom plates. After 18 hours, DCs were washed and cocultured with HLA-A2-restricted NY-ESO-1 or Melan-A-specific CTL lines for 4 hours in the presence of 10 μg/mL BFA. A standard ICS was then performed, and IFN-γ levels were assessed by flow cytometry. DCs pulsed with cognate peptide for 30 minutes before the 4-hour coculture with CTL lines served as positive controls. Data are mean ± SD of 3 separate donors. (A) *P < .01 vs unpulsed CD1c+ DCs and vs CD1c+ DCs pulsed with NY-ESO-1 and ISCOMATRIX adjuvant separately with or without additional stimulation. (B) *P < .01 vs unpulsed MoDCs.
Association of NY-ESO-1 antigen with ISCOMATRIX adjuvant enables cross-presentation of multiple CTL epitopes by CD1c+ blood DCs
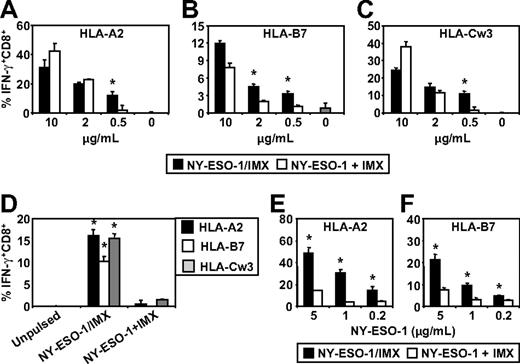
We next investigated whether antigen and adjuvant formulation influenced the capacity of MoDCs or CD1c+ DCs to cross-present tumor antigen-specific CTL epitopes on class I MHC molecules other than HLA-A2. For these studies, we specifically focused on the NY-ESO-1 antigen system as we could directly compare NY-ESO-1 mixed with ISCOMATRIX adjuvant with the NY-ESO-1/ISCOMATRIX-formulated vaccine. The NY-ESO-1/ISCOMATRIX vaccine has been previously tested in human clinical trials on melanoma patients and shown to be capable of generating CTL responses against a broad array of novel epitopes as well as enhancing preexisting CTL responses of diverse HLA haplotypes.26 Epitopes of particular interest were those that were frequently boosted by vaccination and are restricted by HLA-A2, HLA-B7, and HLA-Cw3. To allow us to expand our analysis to NY-ESO-1 epitope generation other than the NY-ESO-1157-165/HLA-A2, we also generated individual NY-ESO-1-specific CTL lines restricted by HLA-B7 and HLA-Cw3 and identified a healthy donor who was positive for all 3 alleles as a source of HLA-matched DCs. Using this approach, it was possible to pulse HLA-A2, HLA-B7, and HLA-Cw3 triple-positive DCs either with NY-ESO-1 protein mixed with ISCOMATRIX adjuvant or the formulated NY-ESO-1/ISCOMATRIX vaccine and assess the cross-presentation of all 3 CTL epitopes by the same DC populations using individual epitope-specific T-cell lines.
We found that MoDCs were capable of cross-presenting the NY-ESO-1157-165/HLA-A2, NY-ESO-160-72/HLA-B7, and NY-ESO-192-100/HLA-Cw3 epitopes irrespective of whether NY-ESO-1 was formulated as an NY-ESO-1/ISCOMATRIX vaccine or when NY-ESO-1 and ISCOMATRIX adjuvant were added separately to the MoDCs (Figure 6A-C). However, these results also show that, under conditions where antigen was limiting, the NY-ESO-1/ISCOMATRIX vaccine continued to generate efficient cross-presentation of all 3 epitopes, whereas this was not seen when NY-ESO-1 and ISCOMATRIX adjuvant were added separately to the MoDCs. This indicates that the antigen and adjuvant formulation significantly affect the efficiency with which cross-presentation of these epitopes can occur. To extend these findings, we compared the cross-presentation capacity of HLA-A2, HLA-B7, and HLA-Cw3 triple-positive CD1c+ blood DCs. In contrast to MoDCs, CD1c+ DCs were largely ineffective in cross-presenting NY-ESO-1157-165/HLA-A2, NY-ESO-160-72/HLA-B7, and NY-ESO-192-100/HLA-Cw3 epitopes when NY-ESO-1 and ISCOMATRIX adjuvant were added separately into the CD1c+ DC cultures (Figure 6D). However, formulation of NY-ESO-1 as an associated NY-ESO-1/ISCOMATRIX vaccine enabled CD1c+ DCs to cross-present all 3 NY-ESO-1-derived epitopes (Figure 6D). To understand the mechanisms behind these differences in more detail, we enhanced the sensitivity of our assays by generating a T-cell clone for the HLA-A2 epitope and an enriched T-cell line for the HLA-B7 epitope (> 95% peptide-specific CD8+ cells; data not shown). Using these enriched CTLs as reporters, we found that, as with MoDCs, CD1c+ DCs can cross-present NY-ESO-1 when mixed with ISCOMATRIX adjuvant but that this is much less efficient than cross-presentation of the associated NY-ESO-1/ISCOMATRIX vaccine, highlighting that antigen and adjuvant formulation significantly alters the cross-presentation efficiency (Figure 6E-F).
ISCOMATRIX facilitated cross-presentation of HLA-A2, HLA-B7, and HLA-Cw3-restricted epitopes from NY-ESO-1 by MoDCs and CD1c+ DCs. Immature HLA-A2, HLA-B7, and HLA-Cw3 triple-positive MoDCs (A-C) or triple-positive CD1c+ DCs (D) or singly positive CD1c+ DCs (E-F) were plated in triplicate in 96-well, round-bottom plates and either unpulsed or pulsed with 0.2 to 10 μg/mL of NY-ESO-1 protein added separately with ISCOMATRIX adjuvant or with NY-ESO-1/ISCOMATRIX vaccine for 18 hours. After being washed, the DCs were divided and cocultured with HLA-A2 or HLA-B7 or HLA-Cw3-restricted NY-ESO-1-specific CTL lines (A-D) or HLA-A2 or HLA-B7 clones (E-F) for 4 hours in the presence of 10 μg/mL BFA. A standard ICS was then performed, and IFN-γ levels were assessed by flow cytometry. Data are mean ± SD of triplicate wells, and one of 3 experiments is shown. *P < .01 vs NY-ESO-1 added separately with ISCOMATRIX adjuvant to the cultures.
ISCOMATRIX facilitated cross-presentation of HLA-A2, HLA-B7, and HLA-Cw3-restricted epitopes from NY-ESO-1 by MoDCs and CD1c+ DCs. Immature HLA-A2, HLA-B7, and HLA-Cw3 triple-positive MoDCs (A-C) or triple-positive CD1c+ DCs (D) or singly positive CD1c+ DCs (E-F) were plated in triplicate in 96-well, round-bottom plates and either unpulsed or pulsed with 0.2 to 10 μg/mL of NY-ESO-1 protein added separately with ISCOMATRIX adjuvant or with NY-ESO-1/ISCOMATRIX vaccine for 18 hours. After being washed, the DCs were divided and cocultured with HLA-A2 or HLA-B7 or HLA-Cw3-restricted NY-ESO-1-specific CTL lines (A-D) or HLA-A2 or HLA-B7 clones (E-F) for 4 hours in the presence of 10 μg/mL BFA. A standard ICS was then performed, and IFN-γ levels were assessed by flow cytometry. Data are mean ± SD of triplicate wells, and one of 3 experiments is shown. *P < .01 vs NY-ESO-1 added separately with ISCOMATRIX adjuvant to the cultures.
DCs use proteasome-dependent and -independent mechanisms for tumor antigen processing
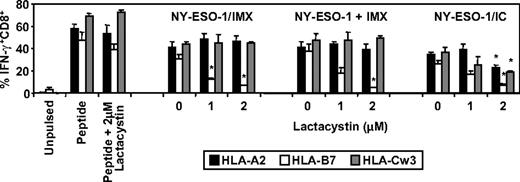
Finally, we investigated the requirement for proteasomal processing in the generation of the NY-ESO-1157-165/HLA-A2, NY-ESO-160-72/HLA-B7, and NY-ESO-192-100/HLA-Cw3 epitopes by MoDCs for the aforementioned NY-ESO-1 formulations and included NY-ESO-1/anti-NY-ESO-1 ICs as a third formulation previously shown to be efficiently cross-presented by MoDCs.4,38,39 We found that proteasome inhibition significantly reduced NY-ESO-1157-165/HLA-A2, NY-ESO-160-72/HLA-B7, and NY-ESO-192-100/HLA-Cw3-restricted cross-presentation when NY-ESO-1 was presented as an IC (Figure 7). In contrast, proteasome inhibition had no effect on the generation of either the NY-ESO-1157-165/HLA-A2 or NY-ESO-192-100/HLA-Cw3 epitopes but dramatically inhibited NY-ESO-160-72/HLA-B7 epitope generation, either when DCs were exposed to formulated NY-ESO-1/ISCOMATRIX vaccine or when NY-ESO-1 and ISCOMATRIX adjuvant were added separately to the DCs (Figure 7). Taken together, these results suggest that there is differential processing of these 3 epitopes.
Effects of proteasome inhibition on ISCOMATRIX facilitated cross-presentation of HLA-A2, HLA-B7, and HLA-Cw3-restricted epitopes from NY-ESO-1 by MoDCs. Immature HLA-A2, HLA-B7, and HLA-Cw3 triple-positive MoDCs (1 × 105) were plated in triplicate in 96-well, round-bottom plates and untreated or treated with 1 or 2μM lactacystin for 45 minutes before and throughout the 18-hour pulse with 10 μg/mL of the NY-ESO-1/ISCOMATRIX vaccine or NY-ESO-1 protein added separately with ISCOMATRIX adjuvant or with NY-ESO-1/ICs. After being washed, MoDCs were divided and cocultured with HLA-A2 or HLA-B7 or HLA-Cw3-restricted NY-ESO-1-specific CTL lines for 4 hours in the presence of 10 μg/mL BFA. A standard ICS was then performed, and IFN-γ levels were assessed by flow cytometry. Data represent the mean ± SD of triplicate wells, and one of 3 experiments is shown. *P < .01 vs no addition of lactacystin.
Effects of proteasome inhibition on ISCOMATRIX facilitated cross-presentation of HLA-A2, HLA-B7, and HLA-Cw3-restricted epitopes from NY-ESO-1 by MoDCs. Immature HLA-A2, HLA-B7, and HLA-Cw3 triple-positive MoDCs (1 × 105) were plated in triplicate in 96-well, round-bottom plates and untreated or treated with 1 or 2μM lactacystin for 45 minutes before and throughout the 18-hour pulse with 10 μg/mL of the NY-ESO-1/ISCOMATRIX vaccine or NY-ESO-1 protein added separately with ISCOMATRIX adjuvant or with NY-ESO-1/ICs. After being washed, MoDCs were divided and cocultured with HLA-A2 or HLA-B7 or HLA-Cw3-restricted NY-ESO-1-specific CTL lines for 4 hours in the presence of 10 μg/mL BFA. A standard ICS was then performed, and IFN-γ levels were assessed by flow cytometry. Data represent the mean ± SD of triplicate wells, and one of 3 experiments is shown. *P < .01 vs no addition of lactacystin.
Discussion
Full-length tumor proteins are attractive antigens for vaccine development because they contain multiple epitopes recognized by both CD4+ and CD8+ T lymphocytes. However, unlike peptide-based vaccines, full-length tumor antigen-based vaccines critically require enzymatic processing and cross-presentation by DCs to generate cellular immune responses. Our studies show that the mode of antigen delivery to DCs can influence the processing mechanisms that are used and thereby which peptides are cross-presented. This in turn may affect the fine specificity and quality of the ensuing immune responses. Understanding these mechanisms is critical for optimizing vaccine development. We have therefore evaluated the processing and cross-presentation of 2 human tumor antigens by different DC populations, delivered in conjunction with ISCOMATRIX adjuvant, which is known to facilitate cross-presentation of protein antigens.4,40,41
ISCOMATRIX adjuvant composes 40nM cage-like particles, which can be mixed with antigen or into which hydrophobic antigens can be physically incorporated (“formulated”). We have previously shown that ISCOMATRIX adjuvant dramatically affects antigen processing and presentation,4,38 and this study was undertaken to more deeply assess the capacity of ISCOMATRIX adjuvant to facilitate the cross-presentation of multiple tumor antigen-derived CTL epitopes. Comparisons have also been made with antigen delivered as ICs. We focused on antigens that are representative of 2 tumor antigen classes: NY-ESO-1, a cancer/testis antigen, and the melanoma differentiation antigen, Melan-A. We found that individual epitopes within these proteins presented on different HLA molecules undergo differential processing via different intracellular pathways, a phenomenon dependent on the mode of antigen delivery and the DC subset.
We first compared cross-presentation of soluble antigen and antigen mixed with ISCOMATRIX adjuvant using MoDCs generated in vitro. MoDCs failed to cross-present HLA-A2-restricted epitopes from either soluble NY-ESO-1 or Melan-A, which was in contrast to the highly efficient cross-presentation that occurred when antigens were mixed with ISCOMATRIX adjuvant (even at 10-fold lower antigen dose). This did not require the simultaneous addition of ISCOMATRIX because addition of the adjuvant 2 hours after antigen exposure still facilitated cross-presentation, albeit to a lesser extent than when antigen and ISCOMATRIX adjuvant were coadministered to the DCs (Figure 1C-D). Although the mechanism for this remains unclear, cross-presentation was not enhanced if the order was reversed and ISCOMATRIX adjuvant administered first. It is possible under these conditions that ISCOMATRIX adjuvant induced some level of DC activation that subsequently impaired antigen cross-presentation.42 Although we have not found ISCOMATRIX adjuvant to induce activation of human MoDCs in vitro, we formally excluded this as an unlikely explanation for these differences by demonstrating that preadministration of ISCOMATRIX adjuvant to MoDCs did not negatively impact their subsequent capacity to cross-present either NY-ESO-1 or Melan-A. Taken together, this suggests that ISCOMATRIX adjuvant facilitates processing of preexisting antigen stores. For NY-ESO-1, this may have been in association with cell surface calreticulin, as previously described,43 which had been internalized into endosome/phagolysosomal compartments.38 Similar mechanisms may apply for Melan-A, even though cell surface binding with calreticulin has not been described. As previously shown, the addition of ISCOMATRIX likely facilitates translocation into the cytosol, thereby providing tumor antigen access to the class I MHC pathway.38
We next evaluated the effect of DC activation on cross-presentation. Class I MHC antigen processing is largely dependent on the proteasome, which has alternative forms depending on immune activation. Stimulation of DCs via CD40 or Toll-like receptor ligation or exposure to cytokines, such as IFN-γ or IFN-α, has been shown to induce conversion of the somatic proteasome to the immunoproteasome by replacement of the β1, β2, and β5 subunits with their immunoproteasome subunit counterparts LMP2, MECL-1, and LMP7, respectively.36,37 Because the immunoproteasome cleaves protein substrates at alternative residues, it can alter the peptide repertoire that is subsequently generated. Of particular relevance here, it has been shown that the Melan-A26-35/HLA-A2 epitope is generated in tumors by the somatic proteasome but not the immunoproteasomes of DCs.30 This potentially renders vaccination with Melan-A protein ineffective because the one critical dominant epitope would not be generated by any vaccination strategy that requires processing by immunoproteasome-expressing DCs. We were therefore particularly interested in evaluating the effect of ISCOMATRIX adjuvant on the cross-presentation of Melan-A26-35 on HLA-A2 molecules. Interestingly, and in agreement with others,37 we have found by Western blot analysis that MoDCs express both immunoproteasome and somatic proteasome subunits (not shown). Furthermore, we found that ISCOMATRIX adjuvant facilitated efficient cross-presentation of the Melan-A26-35/HLA-A2 epitope by MoDCs through a proteasome-dependent mechanism (Figure 4). Taken together, these findings show that immature MoDCs can generate the Melan-A26-35/HLA-A2 epitope if facilitated by ISCOMATRIX adjuvant. This shows, for the first time, that manipulating the delivery system can modify the proteolytic cleavage preference for processing of Melan-A protein, thereby generating the critical immunodominant epitope that would otherwise be lost or generated at insufficient levels.
The present study also shows differential effects of ISCOMATRIX adjuvant when different DC subsets cross-present the HLA-A2-restricted epitopes from NY-ESO-1 and Melan-A. In this regard, MoDCs cross-presented the NY-ESO-1157-165/HLA-A2 and the Melan-A26-35/HLA-A2 epitopes irrespective of antigen and adjuvant association. In contrast, CD1c+ DCs were inefficient under these conditions. Furthermore, MoDCs also cross-presented the NY-ESO-1157-165/HLA-A2, NY-ESO-160-72/HLA-B7, and NY-ESO-192-100/HLA-Cw3 epitopes, without the need for further activation, irrespective of formulation as an NY-ESO-1/ISCOMATRIX vaccine or when NY-ESO-1 and ISCOMATRIX adjuvant were added separately to the DCs. However, when antigen was limiting, the formulated NY-ESO-1/ISCOMATRIX vaccine was superior to separately adding the antigen and adjuvant to the DC cultures. This highlights important differences in the efficiency of tumor antigen cross-presentation between associated and nonassociated formulations that may critically impact vaccine efficacy. Furthermore, CD1c+ DCs were even more limited and could only cross-present all 3 epitopes efficiently when NY-ESO-1 was physically formulated with ISCOMATRIX adjuvant and were inefficient at generating these epitopes when NY-ESO-1 and ISCOMATRIX adjuvant were added separately. The reason for this important difference is not clear. Possibilities include: (1) differences in the efficiency of antigen uptake by MoDCs and CD1c+ DCs, (2) differences in their intracellular organization of antigen-processing pathways, which may affect the cytosolic translocation of antigen that in turn affect the efficiency of cross-presentation, or (3) differences in rates of functional maturation and their effects on antigen processing functions. In this regard, CD1c+ DCs rapidly and spontaneously mature in vitro.44 As a result, CD1c+ DCs may lose the capacity to internalize and process antigen compared with MoDCs. The combined effects of reduced antigen uptake capacity and limiting intracellular colocalization of antigen and adjuvant may explain the differing efficiencies of cross-presentation between these 2 DC subsets. Taken together, we demonstrate that preformed antigen and adjuvant association is an important parameter for consideration as it results in more efficient CTL activation.
Finally, we have previously demonstrated that NY-ESO-1 formulation, either as ICs or as ISCOMATRIX vaccine, determines whether human DCs use the classical proteasome-dependent processing pathway or a TPPII-dependent processing pathway for NY-ESO-1157-165/HLA-A2-restricted cross-presentation, respectively.4,38 In agreement with these findings, we show here that, when NY-ESO-1 is delivered as an IC, all 3 epitopes have some dependence on the proteasome to be generated (ie, cross-presentation was partly inhibited by lactacystin). In contrast, the NY-ESO-1157-165/HLA-A2 and NY-ESO-192-100/HLA-Cw3 epitopes were generated in a proteasome-independent manner irrespective of whether NY-ESO-1 and ISCOMATRIX adjuvant were added separately or formulated as an NY-ESO-1/ISCOMATRIX vaccine. Furthermore, the NY-ESO-160-72/HLA-B7 epitope, which is an unusually long (13 amino acid) “bulged” epitope,45 was dependent on the proteasome for processing irrespective of the mode of delivery (ie, formulated with ISCOMATRIX adjuvant, added separately, or formulated as an IC) because its generation and cross-presentation on HLA-B7 were inhibited by lactacystin in each instance. Thus, different epitopes from the same protein can be processed by different mechanisms; and although the mode of antigen delivery can redirect processing in some instances (eg, NY-ESO-1 on HLA-A2157-165 and NY-ESO-192-100 on HLA-Cw3), this was not the case for NY-ESO-160-72 on HLA-B7.
These findings offer novel insights into the diverse mechanisms used by human DC subsets for the processing and cross-presentation of an array of tumor antigen epitopes, even when derived from a single protein. They show important differences in the capacities of human DC subsets to cross-present clinically relevant, full-length tumor antigens and how formulation can potentially impact CD8+ T-cell responses in vivo. The implication of this is that the fine specificity of an antitumor immune response can depend on how antigen is formulated and delivered. Understanding these details should assist in the design of vaccination strategies that can generate dominant immune responses against critical target epitopes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Lisa Ebert, Oliver Klein, and Suzanne Graham for helpful discussion and technical support and Amgen for provision of soluble CD40L trimer.
N.C.R. was supported by the Australian National Health and Medical Research Council (Industry Fellowship Grant) and the Ludwig Institute for Cancer Research and is currently a Wellcome Trust Fellow. M.S. was supported by the Deutsche Krebshilfe (Max Eder Research Grant) and the Deutsche Forschungsgemeinschaft (GK 1202). E.M. is an employee of CSL Limited and an Honorary Senior Research Fellow of the Ludwig Institute for Cancer Research. J.C. is a National Health and Medical Research Council Practitioner Fellow.
Authorship
Contribution: N.C.R. designed and performed research, collected and analyzed data, and wrote the paper; T.M., A.J.K., and A.S. performed research and collected and analyzed data; and M.S., W.C., E.M., and J.C. designed research, analyzed data, wrote the paper.
Conflict-of-interest disclosure: E.M. is employed by CSL Limited, whose potential product was studied in the present work. The remaining authors declare no competing financial interests.
Correspondence: Neil C. Robson, Ludwig Institute for Cancer Research, Centre for Clinical Sciences, Austin Health, Melbourne, VIC, 3084, Australia; e-mail: neil.robson@ed.ac.uk.
References
Author notes
W.C., E.M., and J.C. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal