SCD (or “HbSS”) can vary markedly in its clinical manifestations1 : in HbSS cells, regulatory factors that skew in association with disease severity may present new prognostic and/or therapeutic opportunities. In this issue of Blood, Sangokoya et al have applied unsupervised miRNA profiling to reveal elevated microRNA-144 levels in a severe anemia subset of SCD patients (despite an essential lack of mRNA transcripts, erythrocytes can retain miRNAs).2,3
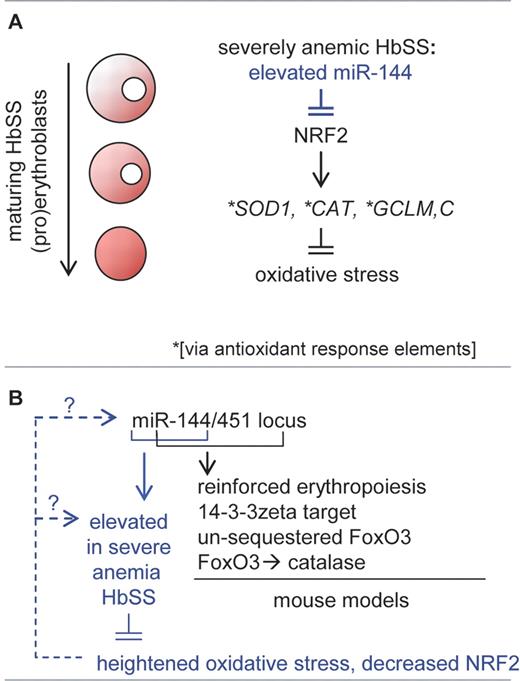
Evidence further is provided that the CNC-bZip transcription factor NRF2 is a target for decay by miR-144. NRF2 is known to activate the expression of several antioxidant encoding genes (eg, SOD1, CAT, GCL2) in part via antioxidant response elements. Increased miR-144 levels (and consequentially decreased NRF2 levels) therefore may elevate oxidative species (see figure, panel A). Oxidative events can compromise erythrocyte integrity, oxygen-carrying capacity, and red cell half-life—and erythrocytes from sickle cell disease (SCD) patients are known to display heightened sensitivity to oxidative stress.
Investigations by Sangokoya et al provide evidence that elevated miR-144 in HbSS erythroid cells from SCD patients with severe anemia can compromise antioxidizing capacities.2 Mechanistically, this may involve miR-144 decay of transcripts for NRF2, a CNC-bZip transcription factor known to regulate several antioxidant genes (A). Panel B outlines (in black font) roles for miR-144/451 as discovered this year via novel knockout mouse models. For erythroid HbSS cells (blue font), interesting parallel questions concerning mechanisms of miR-144/451 dysregulation are raised within a SCD context.
Investigations by Sangokoya et al provide evidence that elevated miR-144 in HbSS erythroid cells from SCD patients with severe anemia can compromise antioxidizing capacities.2 Mechanistically, this may involve miR-144 decay of transcripts for NRF2, a CNC-bZip transcription factor known to regulate several antioxidant genes (A). Panel B outlines (in black font) roles for miR-144/451 as discovered this year via novel knockout mouse models. For erythroid HbSS cells (blue font), interesting parallel questions concerning mechanisms of miR-144/451 dysregulation are raised within a SCD context.
In 2009, Pase et al used morpholino knockdown as well as Meunier mutant approaches to provide novel evidence in zebrafish for miR-144/451 roles in erythroid maturation (miR-144 and miR-451 are expressed from a conserved single locus).4 This year, 3 laboratories reported on miR-144/451 gene disruption investigations in mice.5-7 Rasmussen et al reported on mild anemia among miR-144/451 null animals, and described miR-144/451 tuning of the expression of a diverse gene set.5 Patrick et al and Yu et al further characterized miR-144/451 protection of erythroid cells from oxidative stress, in part via miR-451 targeting of 14-3-3 zeta.6,7 As a phospho-S/T–binding protein, 14-3-3 zeta can sequester phospho-FoxO3 and restrict nuclear entry of this group-O forkhead box transcription factor. In this system, one target of FoxO3 proved to be the antioxidant enzyme, catalase.7 Thus, strong genetic underpinnings exist to implicate miR-144/451 in late erythropoiesis and oxidative stress contexts. Interestingly, recent investigations by Godlewski et al in glioma cells similarly have related elevations in miR-451 levels to worsened stress consequences, but in a context of sensitization to glucose deprivation.8
In the present contexts of erythropoiesis, SCD, NRF2, and oxidative stress, the study by Sangokoya et al initially employs a K562 cell line model to map a 3′ untranslated region target site in NRF2 for degradation by miR-144.2 Ectopic expression of miR-144 then is demonstrated to decrease NRF2 levels in both K562 and primary erythroblasts, and to also potentiate peroxide-induced cell death. In K562 cells, enforced expression of miR-144 decreased superoxide dismutase (SOD1) levels, as well as glutathione synthesis enzyme subunits GCL-M plus GCL-C. Among HbSS cells, erythrocytes with high-level miR-144 similarly exhibited decreased SOD activity, and decreased GCL-M and GCL-C. In addition, NRF2 levels varied inversely with miR-144 levels, and in primary (pro)erythroblasts NRF2 was observed to decay at a developmental stage when miR-144 levels increased. Because erythrocytes have no nuclei, the demonstrated skewing of miR-144 plus NRF2 in primary HbSS (pro)erythroblasts fills a mechanistic gap.
Observed miR-144 increases in HbSS (pro)erythroblasts also suggest that microRNAs become dysregulated in progenitor pools. This raises a basic question as to what SCD factors might elevate miR-144. One potentially insightful feature involves HbSS erythrocyte elevation of miR-144, but not miR-451. In part, this could implicate SCD-specific stabilization of miR-144, and miRNA half-lives can be sharply regulated.9 In mouse models, and in a miR-144 vs miR-451 context, it is also interesting to note that miR-451 per se can phenocopy erythropoietic effects of miR-144/451.5-7 In addition, miR-451 processing has been shown to depend on nucleolytic activity of an Ago-2 Argonaute factor, and to be atypically Dicer-independent. In future studies, the extent to which miR-144 might modulate 14-3-3 zeta within HbSS cells also should be interesting to assess.
Sangokoya et al further raise the clinical prospect of using erythrocyte miRNA profiles as co-correlates for predicting SCD progression and/or susceptibility to stroke.2 Extended studies with larger sets of variant SCD samples will be needed to establish such possible uses as related to SCD's variable severity. Additional attention also may need to be paid to the potential younger ontogenic age of HbSS erythrocytes (and possibly, HbSS erythroid progenitors) due to accelerated turnover of the SCD erythron. For SCD, one established clinical correlate of severity relates to HbF levels. In closing, it is therefore noteworthy to at least cite recent advances toward understanding of gamma-globin gene regulators. These include the discoveries of BCL11A as a gamma-globin gene repressor1 and Klf1 as a regulator of Bcl11a.10 By speculation, potential roles of these factors (together with GATA1)7 in modulating miR-144/451 expression may also prove productive for consideration.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal