Abstract
Our previous in vitro studies led to proposals that the atypical chemokine receptor CCX-CKR is a scavenger of CCR7 ligand homeostatic chemokines. In the present study, we generated CCX-CKR−/− mice and confirm this scavenger function in vivo. Compared with wild-type mice, CCX-CKR−/− have a 5-fold increase in the level of CCL21 protein in blood, and 2- to 3-fold increases in CCL19 and CCL21 in peripheral lymph nodes. The effect of these protein increases on immunity was investigated after immunization with MOG35-55 peptide emulsified in complete Freund adjuvant (CFA). The subsequent characteristic paralysis develops with enhanced kinetics and severity in CCX-CKR−/− versus wild-type mice. Despite this effect, antigen-specific immune responses in the draining lymph nodes are diminished in CCX-CKR−/− mice. Instead, the earlier onset of disease is associated with enhanced T-cell priming in the CCX-CKR−/− spleen and a skewing of CD4+ T-cell responses toward Th17 rather than Th1. This observation correlates with increased expression of IL-23 in the CCX-CKR−/− spleen and increased CCL21 levels in the central nervous system postimmunization. The early onset of disease in CCX-CKR−/− mice is reversed by systemic administration of neutralizing anti-CCL21 antibodies. Thus, by regulating homeostatic chemokine bioavailability, CCX-CKR influences the localization, kinetics, and nature of adaptive immune responses in vivo.
Introduction
Chemokines comprise a superfamily of mainly secreted and pleiotropic proteins that play critical roles in regulating leukocyte homing and recirculation in vivo.1 They are subdivided into inflammatory and homeostatic subsets with inflammatory chemokines directing leukocytes to inflamed peripheral tissues while constitutive chemokines control basal trafficking of leukocytes during homeostasis. The homeostatic chemokines CCL19 and CCL21, acting via the chemokine receptor CCR7, play key roles in the development of immune responses.2,3 They have well-characterized roles in trafficking naive and central-memory T cells through secondary lymphoid organs,1 directing thymocytes during T-cell differentiation,4 and controlling migration of mature dendritic cells (DCs) to T-cell areas of lymphoid tissues.5 Studies using CCR7−/− and plt mice (a strain of mice lacking CCL19 and lymphoid tissue CCL21 expression) have revealed profound roles for this chemokine axis in immune system homeostasis and the generation of immune response.6-9 Despite this knowledge, less is known in regard to how this process is regulated—particularly as to how CCL19 and CCL21 bioavailability are controlled at a posttranslational level.
Recently, it has become evident that chemokines are regulated by a family of atypical chemokine receptors.10-12 These chemokine receptors differ from classical models as they are uncoupled from signal transduction cascades normally activated after chemokine receptor ligation and do not mediate cell migration. Instead, they have been implicated in regulation of chemokine bioavailability either by scavenging or by transporting chemokines across endothelial barriers. Two atypical chemokine receptors, D6 and DARC, have been the focus of much study, but a third member, CCX-CKR, is less well characterized. It is a high-affinity—but apparently silent—receptor for CCL19, CCL21, and CCL25,13,14 that is capable of continuous chemokine internalization in transfected cells in vitro.15 This property enables it to progressively scavenge large quantities of its ligands. A recent study16 using a knockin/knockout approach indicated that CCX-CKR expression was restricted to lymph node (LN) stromal cells, epidermal cells, and thymic epithelial cells, and that deletion of this receptor in vivo led to subtle defects in constitutive DC trafficking to draining LNs. However, to our knowledge, the role of CCX-CKR in the regulation of chemokines in vivo or in adaptive immune responses has not been reported. Therefore, in the present study, we generated and characterized CCX-CKR−/− mice and focused our investigations on the impact of CCX-CKR deletion on chemokine bioavailability and adaptive immune responses after immunization.
Methods
Mice
C57BL/6 mice were obtained from the University of Adelaide animal house. CCX-CKR−/− mice were generated at the Beatson Institute for Cancer Research (Glasgow, United Kingdom) as described in supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article), then backcrossed for 10 generations to the C57BL/6 background. Genetic compatibility of the F10 CCX-CKR−/− mice to the C57BL/6 strain at the University of Adelaide animal house was confirmed by microsatellite genotyping performed by Professor Alan Baxter (Comparative Genomics Center, James Cook University, Townsville, Australia) and the sole congeneic segment identified was at the locus of the targeted allele. Mice were maintained behind a specific pathogen–free barrier under standard temperature and light conditions and afforded food and water ad libitum.
Experimental autoimmune encephalomyelitis
Female mice aged 6-12 weeks of age were anesthetized with isofluorane and immunized in the hind flanks and scruff of the neck with a total of 25 μg MOG35-55 (MEVGWYRSPFSRVVHLYRNGK) peptide (Australian National University, Canberra) emulsified in 120 μL of CFA per mouse as previously described.17 On days 0 and 2, 300 ng of pertussis toxin (List Biological Laboratories) in 250 μL of endotoxin-free phosphate-buffered saline (PBS) was injected intravenously. Animals were monitored daily and clinical disease was measured by assessment of paralysis using the following criteria: .5 = slight tail weakness, 1 = tail weakness, 2 = full tail paralysis, 2.5 = tail and some hind limb paralysis, 3 = full hind limb paralysis, 3.5 = hind limb and some forelimb paralysis, 4 = full hind and forelimb paralysis, 5 = moribund. Day of disease onset was defined as the first day an animal achieved a score ≥ 1 or the second consecutive day that an animal was scored as 0.5. All experiments were carried out in accordance with approvals obtained from the University of Adelaide's institutional animal ethics committee.
Collection of peripheral blood, lymphoid tissues, and CNS tissue
Mice were euthanized by CO2 asphyxiation and peripheral blood collected by cardiac puncture. Harvested peripheral blood was incubated overnight at 4°C, centrifuged at 13 000g, and serum collected for enzyme-linked immunosorbent assay (ELISA) analysis. PBS perfusion through the left ventricle was performed to remove circulating leukocytes before spinal cords were dissected out. Organs were then either weighed and snap frozen for subsequent ELISA analysis or cell suspensions prepared. For cell suspensions, LNs and spleens were homogenized in cold PBS + 1% bovine serum albumin (BSA) + 0.04% sodium azide (PBA) and passed through a 70-μm mesh strainer. Splenocytes were then subjected to red cell lysis. Spinal cords were strained through 70-μm mesh and cells collected in 30 mL of RPMI + 1% fetal calf serum (FCS) and 20 mL of 90% isotonic Percoll (Amersham Pharmacia Biotech) was added before centrifugation at 1000g for 25 minutes, 4°C, with no brake. The myelin cake was then discarded and cells subjected to red blood cell lysis, washed twice, and resuspended in PBA.
Antibodies and flow cytometric analysis
Harvested cells (2 × 105) were resuspended in PBA while Fc receptors were blocked by incubating for 30 minutes at room temperature with 50 μg of murine γ-globulin (Rockland Immunochemicals)/106 cells. Cells were mixed with anti-mouse CCR6 monoclonal antibodies (R+D Systems) for 30 minutes at room temperature. Isotype-matched negative control antibodies were included as appropriate. Cells were then washed in PBA and anti-rat AlexFluor 488 (Molecular Probes) used to detect. Free binding sites on secondary antibodies were blocked using 50 μg of rat γ-globulin/106 cells before the addition of directly conjugated anti–mouse CD4-PE (phycoerythrin)–Cy7 (BD Biosciences). Intracellular FoxP3 staining was performed using a commercially available kit (eBioscience) according to manufacturer instructions and using a PE-conjugated anti–mouse FoxP3 antibody (clone FJK-16s; eBioscience). Flow cytometric analysis was then performed as previously described.18,19
Intracellular cytokine staining
Cells were harvested and cultured for 4 hours in complete RPMI-1640 containing 20 ng/mL phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich), 1nM ionomycin (Invitrogen) and GolgiStop (BD biosciences) at 1 × 106 cells/mL. Cells were then washed and stained for cell-surface CD4 as described above. After this procedure, cells were fixed and permeabilized using a cytofix/cytoperm kit (BD Biosciences) according to manufacturer instructions. Cells were then stained using anti–mouse IL-17A–PE (BD Biosciences) and IFNγ-FITC (fluorescein isothiocyanate; BD Biosciences).
ELISA
Chemokine ELISAs were carried out essentially as previously described.20 Briefly, serum and tissues were harvested from mice, weighed, snap frozen, and stored at −80°C. Costar high-binding 96-well trays (Corning) were coated with appropriate capture Ab (anti-mouse CCL19 or CCL21; R&D Systems), diluted in 0.1 M NaHCO3, and incubated at 4°C overnight. Plates were washed twice with PBS/Tween, blocked with 200 μL of PBS/3% BSA for 2 hours at 37°C, and washed twice before chemokine standards or samples added (100 μL per well), and incubated for 90 minutes at 37°C. Detection antibodies were biotin-conjugated anti-CCL19 or anti-CCL21 (R&D Systems), diluted in PBS/1% BSA, incubated for 45 minutes at room temperature, then washed 4 times with PBS/Tween. Streptavidin-HRP (Rockland, 100 μL/well) was then added to each well. The plates were incubated for 30 minutes at room temperature then washed 5 times with PBS/Tween. The reaction was developed after the addition of 200 μL of o-phenylenediamine dihydrochloride, OPD, substrate prepared as per manufacturer instructions (Sigma-Aldrich). It was developed for up to 15 minutes in the dark then stopped by the addition of 50 μL of 3M HCl. The intensity of signal at 490 nm was then determined with a Dynatech plate reader. Chemokine concentration in each sample was then determined from the standard curve using GraphPad Prism software. For lysates prepared from solid organs, this process was then normalized to the weight of tissue homogenized. For assessment of anti-MOG35-55 IgG levels, Costar high-binding 96-well trays (Corning) were coated with 200 ng of MOG35-55 peptide diluted in PBS overnight at 4°C. Plates were washed twice with PBS/Tween and blocked with 200 μL of PBS/3% BSA for 2 hours at 37°C. The plates were washed twice, as before, and 100 μL of serum diluted 1/5 or 1/10 in PBS added to each well and incubated for 90 minutes at room temperature, then extensively washed in PBS/Tween and anti–murine IgG conjugated to HRP added. Signal was developed and detected as described above. IL-6 and TGFβ were assessed using commercially available ELISA kits (eBioscience) according to manufacturer instructions. Before the addition to the plate, samples for the TGFβ ELISA were activated from latent into the immunoreactive form by acidification in 1 N HCI and neutralization in 1 N NaOH. IL-23 ELISAs used anti-IL-23p19 (clone G23-8, eBioscience) as a capture antibody and biotinylated anti-IL23/IL-12p70 (clone C17.8; eBioscience) as a detection antibody. Recombinant mIL-23 (eBioscience) was used as standard. After the addition of avidin-HRP (eBioscience), 200 μL of TMB substrate was added. The reaction was allowed to develop for 15 minutes in the dark and stopped by the addition of 50 μL of 1M H3PO4 to each well. The intensity of signal at 450 nm was then determined with a Dynatech plate reader.
In vitro Ag-specific proliferation assay
Proliferation of inguinal LN cells or red blood cell−lysed splenocytes from immunized mice was assessed using a modification of previously published protocols.17,21 Briefly, cells were labeled with CFSE (Molecular Probes) and cultured in complete RPMI-1640 at 2.5 × 106 cells/mL with MOG35-55 at a concentration between 0 and 50 μg/mL, or with 1.5 μg/mL Con A. After 4 days culture, the cells were harvested, labeled with anti–CD4-PE, and analyzed by flow cytometry using a BD LSRII and FACSDiva software (BD Biosciences). Cell division (proliferation) was determined as a progressive halving in CFSE fluorescence intensity.
Bromodeoxyuridine incorporation proliferation assay
Bromodeoxyuridine (BrdU; Sigma-Aldrich) was dissolved to 0.8 mg/mL in drinking water starting at day 6 postimmunization for experimental autoimmune encephalomyelitis (EAE). On day 9 postimmunization, spleens and LNs were harvested and cell-surface CD4 stained as normal. Cells were then washed, resuspended in 500 μL of 0.15M ice-cold NaCl, and then 1.2 mL of ice-cold 95% ethanol added as the samples were gently vortexed. After 30 minutes at 4°C, samples were washed in PBS and fixed in 1% PFA containing 0.01% Tween 20 for 30 minutes at room temperature. Cells were then treated with DNAase at 37°C and labeled with FITC-conjugated anti-BrdU monoclonal antibodies (BD Biosciences). As a negative control, cells from mice untreated with BrdU were included. Cells were then analyzed by flow cytometry as described.
Generation and administration of CCL21-neutralizing antibodies
Anti-mCCL21 antibodies were raised in New Zealand white rabbits by immunization with synthetically produced full-length mCCL21 that was active in chemotaxis and calcium mobilization assays.22-24 Serum IgG was purified using Protein A columns (Millipore) and mice were treated with neutralizing antibodies 24 hours prior to and 5 and/or 10 days after EAE induction. Intraperitoneal injection of 500 μg of affinity-purified rabbit anti-muCCL21 antibodies or normal rabbit IgG was used, as previously described.25
Reverse transcriptase polymerase chain reaction
Tissues were dissected, snap frozen in liquid nitrogen, and total RNA was extracted from homogenized tissue using TRIzol Reagent (Invitrogen) according to manufacturer instructions, as previously described.19 RNA was treated with TURBO DNA-free (Ambion) and 2 μg of RNA reverse-transcribed using Random Primers (Promega) and reverse-transcriptase Bioscript (Bioline). cDNA obtained was treated with DNase-free RNase, diluted and used as template in reactions using LightCycler 480 SYBR Green I master mix (Roche) according to manufacturer instructions. Primers for CCX-CKR were 5′-AAAACAAGAGTTGCCTGACTCAGA-3′ (forward) and 5′-AGCCATGGTACCGAGCTGTTTA-3′ (reverse). Primers for RPLP0 as reference gene were 5′-AGATGCAGCAGATCCGCAT-3′ (forward) 5′-GGATGGCCTTGCGCA-3′ (reverse). Relative level of CCX-CKR mRNA was calculated as 1/2ΔCT (ΔCT=CT of CCX-CKR − CT of RPLP0).
Statistical analysis
All statistical tests were performed using GraphPad Prism 5 software. Unpaired Student t tests were used for all statistical analysis except clinical data from EAE experiments where 2-way ANOVAs with Bonferroni posttests were performed. For all analyses, P < .05 was considered significant.
Results
Generation of CCX-CKR−/− mice
To study the function of CCX-CKR in vivo, we deleted the open reading frame of mouse CCX-CKR by gene targeting. Southern blotting of genomic DNA from the targeted embryonic stem clone used to generate these mice revealed a single homologous integration of the targeting construct. In addition, reverse transcriptase polymerase chain reaction (RT-PCR) of thymic and cardiac tissue revealed that targeting CCX-CKR successfully abolished expression of this gene (supplemental Figure 1). As previously reported,16 CCX-CKR deficiency did not impact mouse fertility or lead to obvious anatomical or developmental defects (data not shown).
CCX-CKR regulates the abundance of CCL19 and CCL21 in vivo
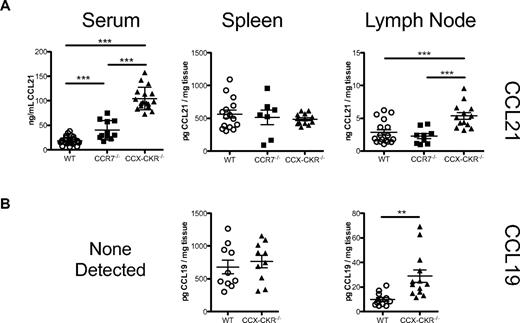
Our previous in vitro studies led to the proposal that CCX-CKR functions as a chemokine scavenger.15 Thus, initial investigations focused on the impact of CCX-CKR deletion on the abundance of protein levels of 2 of its chemokine ligands. Consistent with a scavenging function, a large increase in serum CCL21 was evident in CCX-CKR−/− compared with wild-type mice (Figure 1A). In addition, significant increases were apparent in CCL21 and CCL19 in lysates from LNs but not spleen (Figure 1A and B). Deletion of CCX-CKR therefore leads to dysregulation of homoeostatic chemokine abundance in the peripheral blood and LNs. To test if this result was a specific scavenging effect of CCX-CKR, and not solely due to the lack of a CCL19 and CCL21 binding receptor, CCL21 levels were assessed in CCR7−/− mice. CCL21 levels were normal in secondary lymphoid organs of CCR7−/− mice, and, although a modest but statistically significantly increase in CCL21 was measured in serum of CCR7−/− mice compared with wild-type mice, CCL21 levels were approximately 2.5-fold higher in CCX-CKR−/− serum than CCR7−/− serum, indicating that CCX-CKR plays a more important role than CCR7 in regulating CCL21 levels in vivo.
CCX-CKR regulates circulating and peripheral LN chemokine abundance. (A) Diluted serum, or protein lysates from LNs (pooled inguinal, axillary, and brachial LNs) or spleens, from resting wild-type, CCR7−/−, or CCX-CKR−/− mice (8- to 16-week-old males) were analyzed using a 2-site ELISA for CCL21. Concentrations were calculated from a standard curve of serial dilutions of recombinant CCL21. Each data point shown is the concentration of CCL21 (per milligram or per milliliter) in that tissue from each individual mouse. The mean ± SD is indicated (***P < .001, **P < .01). (B) Diluted serum, or protein lysates from LNs (pooled inguinal, axillary, and brachial LNs) or spleens, from resting wild-type or CCX-CKR−/− mice (8- to 16-week-old males) were analyzed using a 2-site ELISA for CCL19. Concentrations were calculated from a standard curve of serial dilutions of recombinant CCL19. Each data point shown is the concentration of CCL19 (per milligram or per milliliter) in that tissue from each individual mouse. The mean ± SD is indicated (***P < .001, **P < .01). As indicated, serum CCL19 levels were below the limit of detection (< 100 pg/mL). In all cases, data are pooled from at least 2 individual experiments.
CCX-CKR regulates circulating and peripheral LN chemokine abundance. (A) Diluted serum, or protein lysates from LNs (pooled inguinal, axillary, and brachial LNs) or spleens, from resting wild-type, CCR7−/−, or CCX-CKR−/− mice (8- to 16-week-old males) were analyzed using a 2-site ELISA for CCL21. Concentrations were calculated from a standard curve of serial dilutions of recombinant CCL21. Each data point shown is the concentration of CCL21 (per milligram or per milliliter) in that tissue from each individual mouse. The mean ± SD is indicated (***P < .001, **P < .01). (B) Diluted serum, or protein lysates from LNs (pooled inguinal, axillary, and brachial LNs) or spleens, from resting wild-type or CCX-CKR−/− mice (8- to 16-week-old males) were analyzed using a 2-site ELISA for CCL19. Concentrations were calculated from a standard curve of serial dilutions of recombinant CCL19. Each data point shown is the concentration of CCL19 (per milligram or per milliliter) in that tissue from each individual mouse. The mean ± SD is indicated (***P < .001, **P < .01). As indicated, serum CCL19 levels were below the limit of detection (< 100 pg/mL). In all cases, data are pooled from at least 2 individual experiments.
CCX-CKR delays the onset of EAE and suppresses disease severity
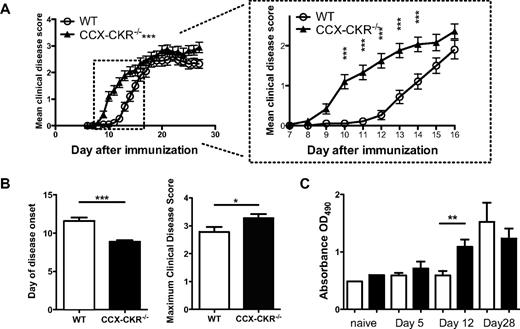
The observed alterations in CCL19 and CCL21 abundance led us to explore the impact of CCX-CKR deletion on the development of a model immune response. We compared the response of wild-type and CCX-CKR−/− mice to MOG35-55-induced chronic EAE in which animals are immunized subcutaneously on a single occasion causing the development of clinical symptoms of disease within 10-14 days in WT mice. After immunization, CCX-CKR−/− mice developed pathology an average of 3 days earlier than their wild-type counterparts (Figure 2A and B), and the disease was more severe with significantly higher peak disease scores (Figure 2B). Moreover, compared with wild-type animals, anti-MOG35-55-specific IgG appeared earlier in the serum of CCX-CKR−/− mice and was evident by day 12 after immunization though it was not detected in wild-type animals until day 21 (Figure 2C). Thus, absence of CCX-CKR accelerates the development of immune responses leading to EAE.
CCX-CKR−/− mice develop EAE with enhanced kinetics and severity. Female CCX-CKR−/− and wild-type C57BL/6 mice between 6-12 weeks of age were immunized for EAE and the clinical disease monitored daily. (A) Main graph: Clinical disease scores over a 28-day time course after immunization. Data are pooled from 3 separate experiments with 23 mice per group in total and are shown as the mean clinical disease score ± SEM (***P < .001 comparing CCX-CKR−/− with wild-type using 2-way ANOVA). (Inset) Clinical disease scores between days 6 and 16 postimmunization (***P < .001 between CCX-CKR−/− and wild-type mice using Bonferroni posttests on indicated days). (B) Day of onset and maximum scores of clinical disease symptoms from these experiments. Data shown are the mean day of onset ± SEM (***P < .001) and the mean maximum score ± SEM (***P < .001). (C) Mice were immunized for EAE and serum harvested from 5 or 6 mice per group per time point on days 0, 5, 12, and 21. Serum was then diluted 1/5 and analyzed for the presence of MOG35-55-specific IgG by ELISA. Plotted is the mean optical density from each group ± SEM (**P < .01).
CCX-CKR−/− mice develop EAE with enhanced kinetics and severity. Female CCX-CKR−/− and wild-type C57BL/6 mice between 6-12 weeks of age were immunized for EAE and the clinical disease monitored daily. (A) Main graph: Clinical disease scores over a 28-day time course after immunization. Data are pooled from 3 separate experiments with 23 mice per group in total and are shown as the mean clinical disease score ± SEM (***P < .001 comparing CCX-CKR−/− with wild-type using 2-way ANOVA). (Inset) Clinical disease scores between days 6 and 16 postimmunization (***P < .001 between CCX-CKR−/− and wild-type mice using Bonferroni posttests on indicated days). (B) Day of onset and maximum scores of clinical disease symptoms from these experiments. Data shown are the mean day of onset ± SEM (***P < .001) and the mean maximum score ± SEM (***P < .001). (C) Mice were immunized for EAE and serum harvested from 5 or 6 mice per group per time point on days 0, 5, 12, and 21. Serum was then diluted 1/5 and analyzed for the presence of MOG35-55-specific IgG by ELISA. Plotted is the mean optical density from each group ± SEM (**P < .01).
Encephalitogenic immune responses reduced in the draining LNs
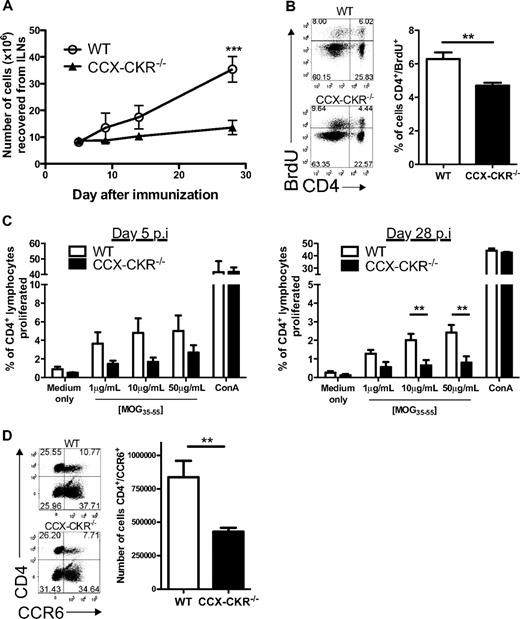
Because immunopathology developed with enhanced kinetics in CCX-CKR−/− mice, we hypothesized that CD4+ T-cell priming was occurring more rapidly in these animals. Thus, we examined encephalitogenic immune responses in skin-draining LNs throughout the course of disease. Remarkably, while the draining LNs of wild-type mice became progressively enlarged, those from CCX-CKR−/− mice barely contained any more cells than resting animals (Figure 3A). Furthermore, contrary to our expectations, BrdU-feeding experiments demonstrated that CCX-CKR−/− mice had a smaller proportion of CD4+ T cells in their draining LNs that had incorporated BrdU into their DNA during cell division between days 6 and 9 postimmunization compared with wild-type mice (Figure 3B). In addition, ex vivo antigen-specific proliferation assays using cells from draining LNs of immunized mice revealed a trend for reduced MOG35-55-specific CD4+ T-cell proliferative responses in CCX-CKR−/− mice at day 5 postimmunization that was statistically significant on day 28 postimmunization (Figure 3C). Moreover, flow cytometric analysis of the cells in the draining LNs of CCX-CKR−/− mice revealed that fewer CD4+ cells expressed the chemokine receptor CCR6 than in wild-type LNs (Figure 3D). Collectively, these data indicate that the enhanced disease kinetics observed in CCX-CKR−/− mice was not due to more efficient priming in the draining LN.
Immune priming in the draining LNs is reduced in CCX-CKR−/− mice. (A) Viable cell counts from inguinal LNs during the time course of EAE. Six mice per time point per group were analyzed. Data shown are the mean cell number ± SEM (**P < .01). (B) Six mice per group were immunized for EAE and fed BrdU at 0.8 mg/mL in their drinking water from day 6. On day 9, draining LNs were harvested, stained for CD4+ and BrdU, and analyzed by flow cytometry. Representative dot plots are shown. Data shown are the mean proportion of CD4+/BrdU+ cells ± SEM (**P < .01). (C) Ex vivo proliferation assay to MOG35-55 using cells from mice taken on days 5 and 28 postimmunization for EAE. Cells taken from 6 immunized mice per time point per group were analyzed. Data shown are the mean percentage of CD4+ cells that proliferated during the 4-day incubation ± the SEM (* P < .05). (D) On day 12 postimmunization for EAE, draining LNs were harvested, viable cells counted, and cells cultured overnight at 4 × 106 /mL in complete RPMI at 37°C, 5% CO2. Cells were then harvested and stained for flow cytometric analyses. Data shown are from 11 mice per group in total from 2 independent experiments pooled and represent the mean ± SEM (**P < .01). Representative dot plots showing the gating for CD4 and CCR6 are shown.
Immune priming in the draining LNs is reduced in CCX-CKR−/− mice. (A) Viable cell counts from inguinal LNs during the time course of EAE. Six mice per time point per group were analyzed. Data shown are the mean cell number ± SEM (**P < .01). (B) Six mice per group were immunized for EAE and fed BrdU at 0.8 mg/mL in their drinking water from day 6. On day 9, draining LNs were harvested, stained for CD4+ and BrdU, and analyzed by flow cytometry. Representative dot plots are shown. Data shown are the mean proportion of CD4+/BrdU+ cells ± SEM (**P < .01). (C) Ex vivo proliferation assay to MOG35-55 using cells from mice taken on days 5 and 28 postimmunization for EAE. Cells taken from 6 immunized mice per time point per group were analyzed. Data shown are the mean percentage of CD4+ cells that proliferated during the 4-day incubation ± the SEM (* P < .05). (D) On day 12 postimmunization for EAE, draining LNs were harvested, viable cells counted, and cells cultured overnight at 4 × 106 /mL in complete RPMI at 37°C, 5% CO2. Cells were then harvested and stained for flow cytometric analyses. Data shown are from 11 mice per group in total from 2 independent experiments pooled and represent the mean ± SEM (**P < .01). Representative dot plots showing the gating for CD4 and CCR6 are shown.
EAE immune activation occurs more efficiently in CCX-CKR−/− spleen
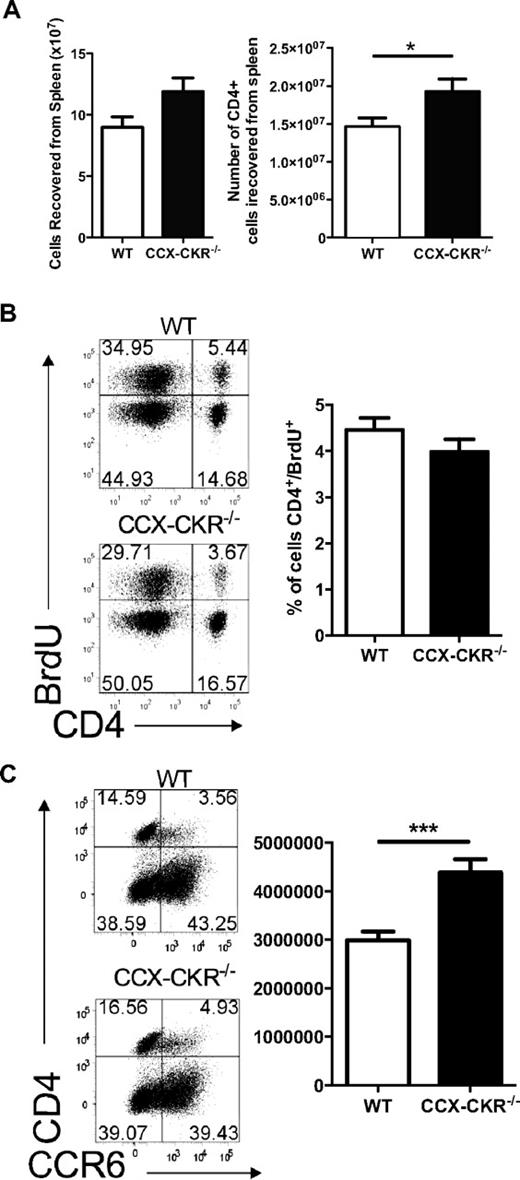
The observations from the draining LNs seemed counterintuitive given the enhanced disease severity and kinetics that were observed in CCX-CKR−/− mice. Therefore, we turned our attention to the spleen, an alternative site of immune priming in this model.26 We measured a trend for increased total cell numbers in the spleen of CCX-CKR−/− mice and significantly increased numbers of CD4+ cells on day 12 postimmunization (Figure 4A). Despite this result, BrdU-feeding experiments demonstrated that the proportion of CD4+ cells in the spleen that had proliferated on day 9 postimmunization was equivalent in both mice types (Figure 4B). However, when the phenotype of splenic CD4+ T cells during immune priming was analyzed in terms of chemokine receptor expression, a greater number of CD4+/CCR6+ cells were detected on day 12 postimmunization in the spleen of CCX-CKR−/− than in wild-type (Figure 4C). Together, these data suggest that, while overall T-cell priming in the spleen is not compromised by deletion of CCX-CKR, in the absence of this receptor, the dividing T cells more rapidly acquire surface expression of chemokine receptors indicative of effector T cells.
Immune priming in the spleen of wild-type and CCX-CKR−/− mice. (A) Total cell numbers and CD4+ cell numbers from wild-type and CCX-CKR−/− spleens on day 12 postimmunization for EAE. Data shown are the mean ± SEM (*P < .05). (B) Six mice per group were immunized for EAE and fed BrdU at 0.8 mg/mL in their drinking water from day 6. On day 9, draining LNs and spleens were harvested, stained for CD4+ and BrdU, and analyzed by flow cytometry. Representative dot plots are shown. Data are the mean proportion of CD4+/BrdU+ cells ± SEM. (C) On day 12 postimmunization for EAE, spleens were harvested, red blood cells lysed, viable cells counted, and cells cultured overnight at 4 × 106/mL in complete RPMI at 37°C, 5% CO2. Cells were then harvested and stained for flow cytometric analysis. Data shown are from 11 mice per group in total from 2 independent experiments pooled and represent the mean ± SEM (**P < .01). Representative dot plots showing the gating for CD4 and CCR6 are shown.
Immune priming in the spleen of wild-type and CCX-CKR−/− mice. (A) Total cell numbers and CD4+ cell numbers from wild-type and CCX-CKR−/− spleens on day 12 postimmunization for EAE. Data shown are the mean ± SEM (*P < .05). (B) Six mice per group were immunized for EAE and fed BrdU at 0.8 mg/mL in their drinking water from day 6. On day 9, draining LNs and spleens were harvested, stained for CD4+ and BrdU, and analyzed by flow cytometry. Representative dot plots are shown. Data are the mean proportion of CD4+/BrdU+ cells ± SEM. (C) On day 12 postimmunization for EAE, spleens were harvested, red blood cells lysed, viable cells counted, and cells cultured overnight at 4 × 106/mL in complete RPMI at 37°C, 5% CO2. Cells were then harvested and stained for flow cytometric analysis. Data shown are from 11 mice per group in total from 2 independent experiments pooled and represent the mean ± SEM (**P < .01). Representative dot plots showing the gating for CD4 and CCR6 are shown.
Th17 responses are enhanced in CCX-CKR−/− mice
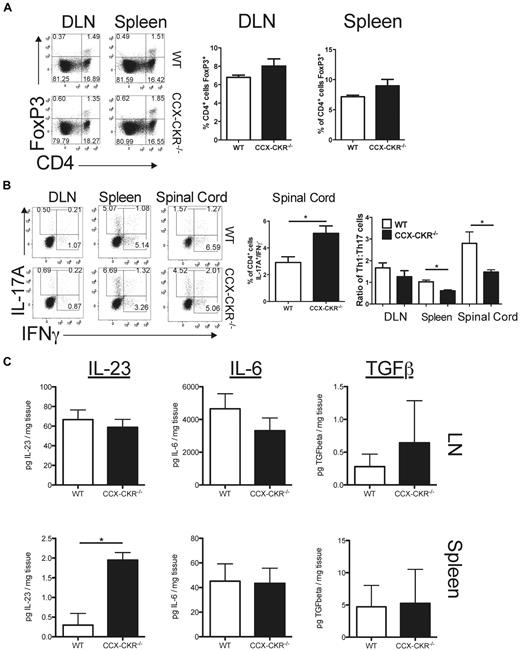
IL-17A-secreting Th17 cells and IFNγ-secreting Th1 cells have been reported to drive EAE pathology, while FoxP3+/CD4+ regulatory T cells suppress this disease.27-29 Therefore, the nature of the CD4+ T-cell responses occurring in wild-type and CCX-CKR−/− mice after immunization with MOG35-55 was investigated. No evidence of alterations in the frequency of CD4+/FoxP3+ regulatory T cells was apparent in either the draining LN or spleen of CCX-CKR−/− mice compared with wild-type on day 12 postimmunization for EAE (Figure 5A). Furthermore, intracellular cytokine staining of the CD4+ T cells recovered from the lymph nodes, spleen, and spinal cord at day 12 postimmunization revealed that a similar proportion of these cells in the central nervous system (CNS) had a typical Th1 (IFNγ+/IL-17A−) profile in both wild-type mice and CCX-CKR−/− mice (Figure 5B). By contrast, the proportion of Th17 cells (IL-17A+/IFNγ−) in the CNS was significantly increased in CCX-CKR−/− mice (Figure 5B). This trend was also apparent in the LNs and spleen although it did not reach statistical significance (data not shown). However, when these data are expressed as the number of Th1 cells per Th17 cell recovered from each of these tissues, there were significant differences between CCX-CKR−/− mice and wild-type mice, with fewer Th1 cells per Th17 cell in the CCX-CKR−/− spleen and spinal cord (Figure 5B). These data indicate that, in CCX-CKR−/− mice, Th17 responses predominate over Th1 responses both at a site of immune priming and at the effector site of EAE, which is likely to contribute to the enhanced disease evident in these animals. To rule out the possibility that the skewing toward Th17 responses measured in mice with EAE was due to intrinsic defects in differentiation of naive CCX-CKR−/− T cells, we determined the ability of these cells to differentiate into Th1 or Th17 cells in vitro after stimulation with αCD3 and αCD28 antibodies. In these cultures, IL-12–driven Th1 differentiation and Th17 differentiation driven by IL-23, IL-6, and TGFβ were normal when CCX-CKR−/− T cells were compared with wild-type (data not shown). Therefore, the possibility that the enhanced Th17 response during EAE in CCX-CKR−/− mice was due to in vivo alterations in key cytokines driving the differentiation and expansion of Th17 cells was tested. Th17 differentiation is driven essentially by IL-6 and TGFβ30,31 and these cells are expanded by IL-23.31-33 Therefore, levels of these cytokines in secondary lymphoid tissue during EAE were quantified by ELISA. On day 12 postimmunization, no differences in the levels of IL-6 or TGFβ in either LN or spleen were apparent between wild-type and CCX-CKR−/− mice, indicating that the enhanced Th17 response is unlikely to be due to alterations in production of these cytokines. However, IL-23 levels in the spleen, but not the LN, of CCX-CKR−/− mice were significantly increased compared with wild-type mice (Figure 5C). Together, these data suggest that the enhanced Th17 response may be due to alterations in the level of expression of IL-23 in CCX-CKR−/− mice at the site of immune priming.
Deletion of CCX-CKR does not affect regulatory T cells but leads to skewing of CD4+ T-cell responses from Th1 to Th17 and increased IL-23 levels. Mice were immunized for EAE and euthanized on day 12 postimmunization. (A) LN cells and splenocytes were harvested, cell-surface CD4 stained and then fixed/permeabilized overnight and stained for intracellular FoxP3. Cells were then analyzed by flow cytometry. Representative dot plots gating on lymphocytes by forward and side scatter are shown. Five mice per group were analyzed and a repeat experiment produced a similar dataset. Bar graphs showing the percentage (%) of CD4+ lymphocytes in draining lymph nodes and spleen-expressing FoxP3 are shown. (B) Leukocytes were harvested from spleen, draining LNs, and spinal cord and activated for 4 hours with PMA/ionomycin in the presence of GolgiStop. Cell-surface CD4 and intracellular IL-17 and IFNγ were then detected and cells analyzed by flow cytometry. Five mice per group were analyzed. Representative dot plots gated on CD4+ lymphocytes are shown. The proportion of CD4+ cells from spinal cord that have a Th17 phenotype is shown. Data are representative of the mean ± SEM (*P < .05). The ratio of Th1 to Th17 cells in draining LNs, spleen, and spinal cord is also shown. Data are representative of the mean ± SEM (*P < .05). (C) Spleen and draining lymph node protein lysates were prepared from female wild-type and CCX-CKR−/− mice on day 12 postimmunization for MOG35-55 EAE and IL-23, IL-6 and TGFβ protein levels assessed by ELISA. Data points show the mean amount of each of these proteins per milligram of tissue homogenized ± SEM (n = 9 mice per group per time point; **P < .01).
Deletion of CCX-CKR does not affect regulatory T cells but leads to skewing of CD4+ T-cell responses from Th1 to Th17 and increased IL-23 levels. Mice were immunized for EAE and euthanized on day 12 postimmunization. (A) LN cells and splenocytes were harvested, cell-surface CD4 stained and then fixed/permeabilized overnight and stained for intracellular FoxP3. Cells were then analyzed by flow cytometry. Representative dot plots gating on lymphocytes by forward and side scatter are shown. Five mice per group were analyzed and a repeat experiment produced a similar dataset. Bar graphs showing the percentage (%) of CD4+ lymphocytes in draining lymph nodes and spleen-expressing FoxP3 are shown. (B) Leukocytes were harvested from spleen, draining LNs, and spinal cord and activated for 4 hours with PMA/ionomycin in the presence of GolgiStop. Cell-surface CD4 and intracellular IL-17 and IFNγ were then detected and cells analyzed by flow cytometry. Five mice per group were analyzed. Representative dot plots gated on CD4+ lymphocytes are shown. The proportion of CD4+ cells from spinal cord that have a Th17 phenotype is shown. Data are representative of the mean ± SEM (*P < .05). The ratio of Th1 to Th17 cells in draining LNs, spleen, and spinal cord is also shown. Data are representative of the mean ± SEM (*P < .05). (C) Spleen and draining lymph node protein lysates were prepared from female wild-type and CCX-CKR−/− mice on day 12 postimmunization for MOG35-55 EAE and IL-23, IL-6 and TGFβ protein levels assessed by ELISA. Data points show the mean amount of each of these proteins per milligram of tissue homogenized ± SEM (n = 9 mice per group per time point; **P < .01).
EAE CCL21 levels in the CNS are elevated and neutralization of CCL21 delays disease onset
As deleting CCX-CKR leads to alterations in chemokine abundance in secondary lymphoid tissues and serum at homeostasis, we assessed levels of CCL21 in the spinal cord during EAE in CCX-CKR−/− mice.
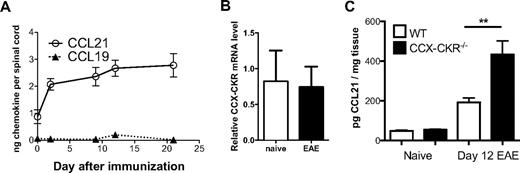
First, to provide background data to address this question, we assayed levels of CCR7 ligands in the spinal cord during EAE in wild-type mice and determined whether CCX-CKR expression in the CNS is regulated during EAE. We found that CCL21 was detectable constitutively in spinal cord lysates, and that this was increased approximately 3-fold by the peak of disease pathogenesis (Figure 6A). In contrast, CCL19 was only detectable at very low levels constitutively and we could not detect substantial increases in the level of this chemokine in the CNS during EAE (Figure 6A). Quantitative PCR analysis revealed that CCX-CKR is expressed in spinal cord tissue normally and that this is not changed by induction of EAE (Figure 6B). Therefore, CCL21 levels in wild-type and CCX-CKR−/− mice were analyzed at homeostasis and after the induction of EAE. No differences were observed in the levels of CCL21 in the CNS between wild-type and CCX-CKR−/− mice before immunization for EAE. However in CCX-CKR−/− mice, an approximately 2-fold greater level of CCL21 was evident in spinal cord lysates on day 12 postimmunization than in lysates from wild-type mice (Figure 6C), indicating that CCX-CKR regulates CCL21 levels in the CNS during EAE.
CCL21 abundance in the CNS is increased after immunization in CCX-CKR−/− mice. Female SJL/J mice were immunized for EAE. At days 0, 2, 4, 9, 12, and 21 mice were killed and spinal cords were collected with the protein fraction extracted. CCL19 and CCL21 protein levels were determined in the spinal cord using a 2-site ELISA assay. All data points represent the mean amount of chemokine recovered from the whole spinal cord ± SEM (n = 3 mice per time point). (B) Spinal cords from either naive wild-type or day 15 EAE mice were harvested, RNA extracted and cDNA reverse-transcribed. Real time PCR for CCX-CKR and acidic ribosomal phosphoprotein P0 (RPLP0) was then performed (n = 3 mice per group). (C) Spinal cord protein lysates were prepared from female wild-type and CCX-CKR−/− mice on days 0 and 12 postimmunization for MOG35-55 EAE and CCL21 protein levels assessed by ELISA. Data points show the mean amount of CCL21 per mg of spinal cord homogenized ± SEM (n = 9 mice per group per time point; **P < .01).
CCL21 abundance in the CNS is increased after immunization in CCX-CKR−/− mice. Female SJL/J mice were immunized for EAE. At days 0, 2, 4, 9, 12, and 21 mice were killed and spinal cords were collected with the protein fraction extracted. CCL19 and CCL21 protein levels were determined in the spinal cord using a 2-site ELISA assay. All data points represent the mean amount of chemokine recovered from the whole spinal cord ± SEM (n = 3 mice per time point). (B) Spinal cords from either naive wild-type or day 15 EAE mice were harvested, RNA extracted and cDNA reverse-transcribed. Real time PCR for CCX-CKR and acidic ribosomal phosphoprotein P0 (RPLP0) was then performed (n = 3 mice per group). (C) Spinal cord protein lysates were prepared from female wild-type and CCX-CKR−/− mice on days 0 and 12 postimmunization for MOG35-55 EAE and CCL21 protein levels assessed by ELISA. Data points show the mean amount of CCL21 per mg of spinal cord homogenized ± SEM (n = 9 mice per group per time point; **P < .01).
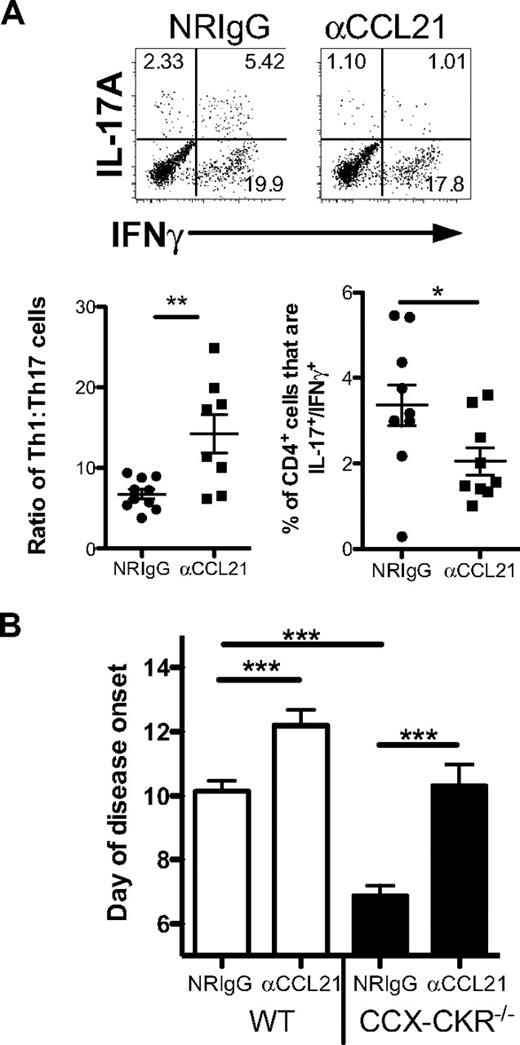
Next, to investigate whether the enhanced EAE and Th17 response of CCX-CKR−/− mice was linked to the increase in chemokine abundance seen in these animals, we used protein A-purified rabbit anti-mouse CCL21 polyclonal antibodies to neutralize CCL21 in vivo during EAE. A precedent for this approach is provided by previously published work that adopted a similar means to demonstrate chemokine-dependence of a phenotype observed in mice in which the inflammatory chemokine scavenger D6 was deleted.34 The anti-CCL21 antibodies were shown to neutralize CCL21 by virtue of their ability to inhibit the migration of CCR7-expressing cells toward CCL21 (data not shown). Administration of these antibodies to mice during immune priming for EAE lead to a skewing of the T-helper response from Th17 to Th1 in the spleen, increasing the relative proportion of Th1 cells to Th17 cells by approximately 2-fold (Figure 7A). In addition, treatment with anti-CCL21 antibodies also lead to a reduction in the proportion of CD4+/IL-17A+/IFNγ+ in the spleen, a cell population shown to be highly pathogenic in EAE. Next, the antibodies were administered to wild-type and CCX-CKR−/− mice before and during the induction of disease. The day of onset was compared with animals receiving normal rabbit IgG. Treatment with the anti-CCL21 antibodies led to a 2-day delay in the day of disease onset in wild-type mice (Figure 7B). However, CCL21 neutralization had a more dramatic effect on CCX-CKR−/− mice, delaying disease by approximately 3.5 days compared with animals treated with normal rabbit IgG, such that it was not significantly different from wild-type animals treated with normal rabbit IgG (Figure 7B).
Neutralization of CCL21 in vivo reduces Th17 responses and delays EAE onset. (A) Mice were immunized for EAE and injected intraperitoneally with 500 μg of protein A–purified rabbit anti-CCL21 antisera or 500 μg of protein A–purified normal rabbit IgG (NRIgG) on days 1 and 6 postimmunization. On day 9 postimmunization, spleens were harvested and intracellular IL-17A and IFNγ detected in CD4+ lymphocytes after 4 hours activation in the presence of PMA/ionomycin in the presence of GolgiStop. Representative dot plots showing intracellular cytokines gated on CD4+ lymphocytes are presented along with data for Th1:Th17 from each mouse. The proportion of CD4+ cells that express both IL-17A+ and IFNγ+ was also quantified. Data shown are the mean ± SEM (**P < .01; *P < .05). (B) Mice were administered with either 500 μg of protein A–purified polyclonal anti-CCL21 antibodies or protein A–purified normal rabbit IgG via intraperitoneal injection the day before immunization, and on days 5 and 10 postimmunization. Mice were then monitored until disease onset. Data are pooled from 2 independent experiments with 5-8 mice in each group in each experiment. The mean day of disease onset of each mouse in each group is shown ± SEM (***P < .001; **P < .01).
Neutralization of CCL21 in vivo reduces Th17 responses and delays EAE onset. (A) Mice were immunized for EAE and injected intraperitoneally with 500 μg of protein A–purified rabbit anti-CCL21 antisera or 500 μg of protein A–purified normal rabbit IgG (NRIgG) on days 1 and 6 postimmunization. On day 9 postimmunization, spleens were harvested and intracellular IL-17A and IFNγ detected in CD4+ lymphocytes after 4 hours activation in the presence of PMA/ionomycin in the presence of GolgiStop. Representative dot plots showing intracellular cytokines gated on CD4+ lymphocytes are presented along with data for Th1:Th17 from each mouse. The proportion of CD4+ cells that express both IL-17A+ and IFNγ+ was also quantified. Data shown are the mean ± SEM (**P < .01; *P < .05). (B) Mice were administered with either 500 μg of protein A–purified polyclonal anti-CCL21 antibodies or protein A–purified normal rabbit IgG via intraperitoneal injection the day before immunization, and on days 5 and 10 postimmunization. Mice were then monitored until disease onset. Data are pooled from 2 independent experiments with 5-8 mice in each group in each experiment. The mean day of disease onset of each mouse in each group is shown ± SEM (***P < .001; **P < .01).
Discussion
The importance of homeostatic chemokine regulation in controlling adaptive immune response is not well understood. In this study, we have shown for the first time that the atypical chemokine receptor CCX-CKR functions in vivo to control the abundance of homeostatic chemokines, influences CD4+ T-cell differentiation and regulates immune response kinetics.
The data presented in this study provide unequivocal confirmation in vivo of a role for CXC-CKR in regulating the levels of CCL19 and CCL21 in several anatomical locations. In the absence of CCX-CKR, levels of CCL19 and CCL21 were increased in peripheral LNs, and CCL21 was overabundant in serum. While CCL19 was not detectable in serum of either wild-type or CCX-CKR−/− mice, it remains possible that it is increased in the serum of CCX-CKR−/− mice although more sensitive means of CCL19 detection will be required to determine this. Nonetheless, these results are consistent with our previous in vitro data showing that CCX-CKR targets homeostatic chemokines for degradation15 in a similar way to the inflammatory chemokine scavenger D6.
It has recently been demonstrated that chemokine abundance and bioavailability is regulated not only by specialized chemokine scavengers such as D6 and DARC,35,36 but also in some circumstances by typical chemokine receptors such as CCR5,37 CCR2, CXCR2, CXCR3, and CX3CR1.38 Importantly, regulation of CCR7-ligand abundance seems to be much more dependent on the specialized scavenger CCX-CKR than on CCR7 as our analysis of CCR7−/− mice revealed that these animals have a more modest or no increase in CCL21 abundance in the serum and exhibit no difference in secondary lymphoid organs. Together, these data suggest that CCX-CKR plays a more important role than CCR7 in regulating CCL19 and CCL21 bioavailability in vivo, consistent with our predictions based on in vitro experimentation.15
That higher levels of CCR7-ligand chemokines in LNs did not lead to hypercellularity in these structures was a curious and unexpected observation. Indeed, we measured significantly fewer cells in LNs of resting CCX-CKR−/− than wild-type mice (data not shown) and fewer cells in these structures after immunization. This decrease in LN cellularity is likely a result of reduced trafficking of CCR7+ cells either by desensitization of CCR7 after overexposure to CCL21 in the peripheral blood or failure to establish a CCR7-ligand gradient required for the cells to traffic as effectively into the LN. Alternatively, it could be that, despite the increase in CCR7-ligand levels in peripheral blood and LN in CCX-CKR−/− mice, these chemokines are not present in the appropriate abundance at appropriate microenvironmental locations, such as on high endothelial venules, in the absence of CCX-CKR. Experiments testing these hypotheses are underway and will be the focus of a follow-up study, which may require development of more sensitive means of in situ chemokine detection than is currently available.
Presently, it is not clear how the absence of CCX-CKR leads to an increase in the level of CCL21 in LN and blood. CCX-CKR scavenging of CCL21 produced by lymphatic endothelial cells resident in peripheral tissues may play an important role in regulating the level of CCL21 in LN. CCX-CKR is abundantly expressed by keratinocytes, according to the work of Heinzel et al,16 so its absence at this site may result in more CCL21 draining from the skin into the LN and, subsequently, via the efferent lymphatics into the blood. In support, analysis of punch biopsies from CCX-CKR−/− skin revealed elevated levels of bioavailable CCL21 (data not shown). However, regardless of mechanism, it is clear that serum CCL21 is not able to then diffuse into the spleen, as no differences in splenic CCL21 levels are evident in CCX-CKR−/− mice despite the large increase in CCL21 in peripheral blood. Indeed, it appears from our observations that the total amount of CCL21 in peripheral blood is significantly greater than that in the spleen in both wild-type and CCX-CKR−/− mice, suggesting either that CCL21 transport from peripheral blood to the spleen does not occur efficiently or that splenic CCL21 levels are regulated independently of CCX-CKR.
Another major issue that requires significant future investigation lies in the nature of the cell types expressing CCX-CKR in vivo and how this expression is regulated. Such investigations are currently hampered by the lack of suitable assays for detection of this receptor, a technical hurdle we are currently addressing. RT-PCR and Northern blot have revealed that CCX-CKR is widely expressed in most tissues, with high-level expression in heart, lung, and intestine.14 In regard to the cell types, it has been shown that immature human DCs and human T cells express CCX-CKR mRNA.13 Conversely, studies using the GFP-reporter mouse suggested that CCX-CKR is expressed by thymic epithelial cells, skin epidermal cells, and stromal cells within the marginal sinus of skin-draining LN.16 Despite this data, the CCX-CKR-GFP reporter mouse did not indicate expression in hematopoietic cells. However, we have recently generated RT-PCR data indicating that certain leukocyte subpopulations express high levels of CCX-CKR (data not shown), indicating that these issues require in-depth clarification in the future.
CCX-CKR−/− animals developed EAE more rapidly than wild-type mice after immunization, and clinical disease was more severe. This was associated with increases in CCL21 in the CNS. In addition, neutralization of CCL21 reversed the enhanced disease onset in CCX-CKR−/− mice, indicating that the phenotype in CCX-CKR−/− mice is dependent on excess CCL21. Furthermore, disease in wild-type animals was also delayed by neutralizing CCL21. The observation that CCL21 plays an important role in EAE is consistent with a recent report demonstrating that the development of EAE is dependent on CCR7 and CCL19-CCL21 and that this axis is important for the development of efficient Th17-dependent autoimmune response.39 In addition, recent findings by Buonamici et al40 have indicated that leukemic T cells navigate to the CNS via CCR7. It is plausible that encephalitogenic T cells also use CCR7 for entry into the CNS and that the increase in CCL21 in this site caused by deletion of CCX-CKR contributes to the process by which CCX-CKR−/− mice develop more rapid and severe disease.
The earlier onset of EAE in CCX-CKR−/− mice was associated with a more rapid appearance of autoimmune antibody in the serum. However, the magnitude of T-cell responses in the draining LNs for these mice was reduced, a phenotype similar to that seen in CCR7−/− mice that have been reported to develop EAE pathology normally but in which T-cell priming occurs in the spleen rather than in the draining LN due to defects in T cell and DC migration26 —although a more profound phenotype was evident when CCR7−/− mice were used in the same model in a different study.39 Similar to the first of these reports, we measured increased numbers of T cells with an activated phenotype in the spleens of CCX-CKR−/− mice. While we did not measure any significant differences in the frequencies of CD11c+/major histocompatibility complex class II+ DCs in either LN or spleen of CCX-CKR−/− compared with wild-type mice on day 12 after immunization (data not shown), it remains possible that the alteration in the site of immune priming in CCX-CKR−/− mice is due to subtle defects in recruitment of subsets of DCs to the draining LN but not the spleen. Detailed investigation examining the recruitment of various DC subsets over an extended time-course is warranted and may yet reveal a role for CCX-CKR in regulating traffic of specific DC subsets to both the spleen and LNs. These studies are currently underway in our laboratory.
Examination of T-cell subsets after immunization provided further insights into the mechanism driving the enhanced onset of EAE. Signaling through CCR7 has been reported to play an important role in the function of CD4+/FoxP3+ regulatory T cells,41 which have been shown to limit EAE pathology.28,42 However, no difference in frequency of these cells was detected in the secondary lymphoid organs between wild-type and CCX-CKR−/− mice after immunization. Examination of the nature of pathogenic Th responses during EAE revealed that the balance between Th1 and Th17 responses was tipped further toward Th17 in CCX-CKR−/− mice than in wild-type counterparts. Significantly increased numbers of Th17 cells per Th1 cell were measured in the spleen and spinal cord of CCX-CKR−/− mice. Recent work using plt mice shows that CCL19 and CCL21 are important for the production of IL-23 by DCs and the subsequent expansion of Th17 cells in vivo.39 Therefore, it is possible that, in CCX-CKR−/− animals, Th17 responses are enhanced as a result of increased CCL19/21-driven IL-23 production. Indeed, our data show increased IL-23 in the spleen of CCX-CKR−/− mice, possibly as a result of exposure of DCs or DC precursors to higher levels of CCL19/21 in peripheral blood or tissues. Future studies should address these possibilities and define the precise molecular basis of these novel findings to show CCX-CKR-dependent alterations in T-cell activation.
In summary, this study provides unequivocal evidence demonstrating that CCX-CKR functions in vivo to regulate the abundance of homeostatic chemokines in blood and tissues. Deletion of CCX-CKR leads to dysregulation of CCL21 and CCL19, enhancing Th17 responses and accelerating a Th17-dependent model of immunopathogenesis. These data highlight the importance of CCX-CKR in the regulation of homeostatic chemokine biology and suggest that it plays an important role in the generation and control of Th17 responses, which has implications for numerous human pathologies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Marina Kochetkova for critical reading of the manuscript.
This work was supported by the National Health and Medical Research Council of Australia, the Multiple Sclerosis Society–USA, and Cancer Research UK.
Authorship
Contribution: I.C. wrote the manuscript, designed and carried out experiments, prepared the figures, and made the CCX-CKR−/− mice; R.J.B.N. wrote the manuscript, designed experiments, provided project direction, and made the CCX-CKR−/− mice; W.L. designed and carried out experiments and edited the manuscript; M.B. designed and carried out experiments and edited the manuscript; Y.H.-L. carried out experiments and edited the manuscript; S.H.-J. carried out experiments and edited the manuscript; S.F. made the CCX-CKR−/− mice; H.K. edited the manuscript and contributed reagents; and S.R.M. wrote the manuscript, designed experiments, and directed the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Professor Shaun R. McColl, University of Adelaide, Rm 5.14, Molecular Life Sciences Bldg, North Terrace, Adelaide, 5005 SA, Australia; e-mail: shaun.mccoll@adelaide.edu.au.
Address requests for CCX-CKR−/− mice to Dr R.J.B. Nibbs, Division of Immunology, Infection and Inflammation, University of Glasgow, United Kingdom; e-mail: r.nibbs@clinmed.gla.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal