Abstract
Abstract 2980
Deletion of chromosome 13, detected by conventional chromosome analysis (CC), has been suggested to be an independent indicator of poor prognosis in myeloma (MM). With the exception of a single study using conventional therapy this has not been confirmed in multicenter randomized controlled trials which have mostly used FISH analysis. We set out to assess the prognostic value of del(13) and compare its detection by FISH and CC.
We have examined the effect of del(13) in a large multicenter randomized controlled phase III trial, MRC Myeloma IX (ISRCTN68454111), which tested the effect of induction therapy with a thalidomide combination in both an intensive (CTD vs CVAD prior to single HDM plus ASCT) and a non-intensive (CTDa vs MP) setting. From June 2003 to November 2007 samples from newly presenting MM patients were sent from hospitals throughout the UK to a central laboratory where CD138 purification for FISH was performed, with conventional cytogenetic culture(s), using unpurified cells, being set up wherever possible.
1960 evaluable patients were entered into the trial (1111 intensive, 849 non-intensive) of which 1036 were evaluable for del(13) by FISH (613 intensive, and 423 non-intensive); del(13) was seen in 45% (470/1036) with a similar incidence in both pathways. With a median follow up of 3.7 years del(13) detected by FISH was associated with impaired OS (median, 42 vs 52 mo, p=0.004). The effect was stronger in the non-intensive pathway (median OS 25 vs 37 mo, p=0.001) than in the intensive one (median 58 mo vs not reached, p=0.095). However, this adverse prognostic impact was negated when the strong association of del(13) with the bad prognosis IGH translocations t(4;14), t(14;16) and t(14;20) (bad IGH) was taken into account.
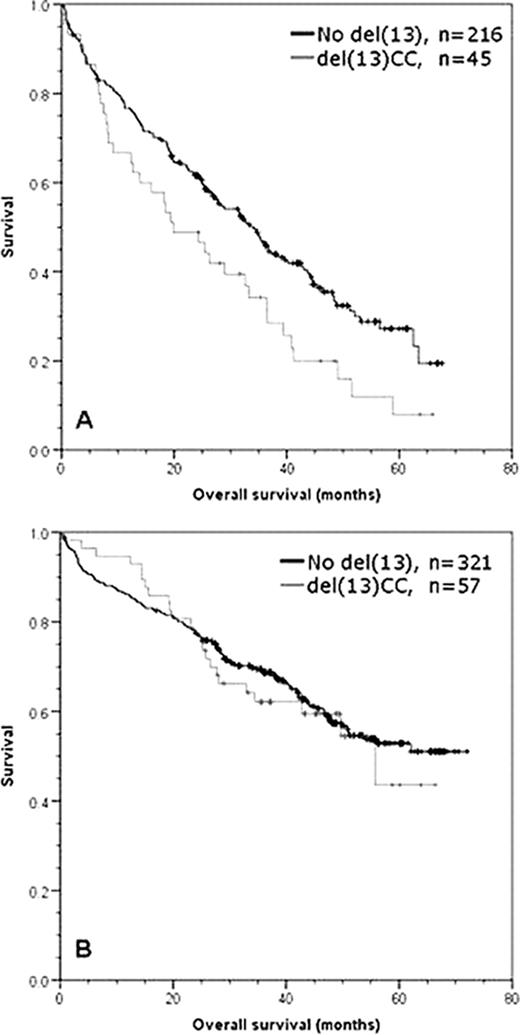
In comparison, 639 patients were evaluable for CC results (378 intensive, 261 non-intensive) with an overall abnormality rate of 35% (224/639). We note that the impact of detecting any abnormal karyotype was not significantly different from a normal/failed karyotype irrespective of intensive or non-intensive treatment being used (whole trial p=0.213, intensive p=0.249, non-intensive p=0.252) suggesting that the capacity to generate abnormal metaphases alone is not an adverse prognostic factor. Del(13) was seen in 45% of abnormal karyotype cases (102/224) and 16% of all cases tested for CC. Overall the detection of del(13) by CC was associated with an adverse prognostic effect on OS (median 34 vs 47 mo, p=0.039). However, the entire effect was in the non-intensive pathway (median 20 vs 34 mo, p=0.018) (Fig1A), with no detectable effect in the intensive pathway (median 69 vs not reached, p=0.679) (Fig1B). We addressed the impact of the treatment used, and in a comparison of the thalidomide and non-thalidomide regimes for all del(13) CC patients there was no significant impact on OS (p=0.538). However, in the non-intensive pathway there was a significant improvement in favour of thalidomide (median 33 vs 18 mo p=0.03). Multivariate analysis of the prognostic impact of genetic factors within abnormal metaphases showed that bad IGH (p=0.001), gain of 1q (p=0.001) and deletion of 17p (p=0.001) were the only independent prognostic factors. In further analyses including all FISH-detected abnormalities, the markers bad IGH (p<0.001) and 1q gain (p<0.001) retained statistical significance when ISS, performance status and age were added into the model.
We demonstrate that in a large multicenter trial carried out across the UK del 13 detected by FISH does not have adverse prognostic impact if its association with the poor prognosis translocations is taken into account. Similarly del(13) detected by CC is not an independent prognostic factor when bad IGH and gain of 1q are taken into acount. The trial shows that thalidomide improves the outcome for del(13) by CC in patients treated with conventional dose therapy. However, this beneficial effect of thalidomide is not enough to account for the lack of independent significance of this marker across the study. On the basis of these results we cannot recommend the use of conventional cytogenetic testing methods as a routine for patients entered into multicenter clinical trials.
Graphs of overall survival for del(13) detected by CC in (A) non-intensive pathway patients, (B) intensive pathway patients
Graphs of overall survival for del(13) detected by CC in (A) non-intensive pathway patients, (B) intensive pathway patients
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal