Abstract
Abstract 3388
We evaluated BCR-ABL1 mutational status in 70 pts with chronic myeloid leukemia in chronic phase after imatinib failure and during dasatinib therapyby DNA expansion of specific clones followed by DNA sequencing of ≥10 clones. Prior to dasatinib, 125 ABL1 kinase domain mutations at 113 amino acid positions were detected in 61/70 (87%) pts, including 38 (54%) with mutations in ≥20% of sequenced clones. Mutations conferring resistance to >1μ M imatinib (M244V, G250E, Q252H, Y253H, E255K/V, F359V, H396R, and T315I) were detected in 30 (43%) pts. Two or more mutations within the same clone (polymutants) were detected in 29/70 (41%) pts. These patients received dasatinib for a median of 19 mos (range, 2–52), during which dasatinib-resistant mutations (L248V/R, Q252H, E255K, V299L, T315I/A, and F317L/C/I/S/V) were detected in 10/32 (31%) assessable cases (5 with T315I). However, polymutants were observed in 16/32 (50%) pts, of whom 13 died in BP and 3 are alive in CP. Given the high frequency and variety of highly resistant polymutants observed in pts failing sequential imatinib-dasatinib therapy, we next performed 3D structural analyses to model the activity of dasatinib, AP23534, and PHA739358 against the most frequently encountered polymutants (Table1).
Free energy of binding (AGbind, kcal/mol), IC50 (nM), and IC50 fold change with respect to unmutated BCR-ABL1 (FCIC50) for dasatinib, AP23534, and PHA739358 in complex with BCR-ABL1 and some single/double mutant isoforms.
| BCR-ABL1 isoform . | DASATINIB . | AP24534 . | PHA739358 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| DGbind . | IC50 . | FC IC50 . | DGbind . | IC50 . | FC IC50 . | DGbind . | IC50 . | FC IC50 . | |
| Unmutated | −12.39 | 0.80 | – | 0.52 | −12.67 | – | 18 | −10.6 | – |
| T315I | −6.44 | 19000 | 23750 | 7.4 | −11.1 | 14 | 7 | −11.13 | 0.39 |
| V299L | −8.21 | 960 | 1200 | 1.3 | −12.13 | 2.5 | 42 | −10.07 | 2.3 |
| F317L | −9.23 | 170 | 212.5 | 12 | −10.81 | 23 | 53 | −9.93 | 2.9 |
| E255K | −10.77 | 12.8 | 16 | 8.8 | −10.99 | 17 | 45 | −10.03 | 2.5 |
| Q252H | −10.85 | 11.2 | 14 | 0.79 | −12.42 | 1.5 | 25 | −10.38 | 1.4 |
| Y253H | −11.08 | 7.6 | 9.5 | 0.55 | −12.64 | 1.1 | 20 | −10.51 | 1.1 |
| H396R | −11.98 | 1.7 | 2.125 | 1.3 | −12.13 | 2.5 | 23 | −10.42 | 1.3 |
| F359V | −12 | 1.6 | 2 | 0.81 | −12.41 | 1.6 | 26 | −10.35 | 1.4 |
| M244V | −12.22 | 1.4 | 1.75 | 1.1 | −12.23 | 2.1 | 16 | −10.64 | 0.89 |
| V304D | −12.27 | 1 | 1.25 | 0.72 | −12.48 | 1.4 | 22 | −10.45 | 1.2 |
| T315I/F317L | −5.47 | 970000 | 1212500 | 21 | −10.48 | 40 | 68 | −9.78 | 3.1 |
| T315I/V299L | −4.83 | 290000 | 362500 | 10 | −10.92 | 19 | 50 | −9.96 | 2.7 |
| T315I/V304D | −6.02 | 38000 | 47500 | 8.8 | −11.01 | 16 | 12 | −10.81 | 0.66 |
| V299L/F317L | −7.42 | 3650 | 4563 | 18 | −10.56 | 34 | 109 | −9.5 | 5 |
| V304D/V299L | −8.05 | 1250 | 1563 | 2.1 | −11.84 | 4 | 46 | −10.01 | 2.5 |
| F317L/V304D | −9.01 | 250 | 313 | 15 | −10.68 | 29 | 51 | −9.95 | 2.8 |
| BCR-ABL1 isoform . | DASATINIB . | AP24534 . | PHA739358 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| DGbind . | IC50 . | FC IC50 . | DGbind . | IC50 . | FC IC50 . | DGbind . | IC50 . | FC IC50 . | |
| Unmutated | −12.39 | 0.80 | – | 0.52 | −12.67 | – | 18 | −10.6 | – |
| T315I | −6.44 | 19000 | 23750 | 7.4 | −11.1 | 14 | 7 | −11.13 | 0.39 |
| V299L | −8.21 | 960 | 1200 | 1.3 | −12.13 | 2.5 | 42 | −10.07 | 2.3 |
| F317L | −9.23 | 170 | 212.5 | 12 | −10.81 | 23 | 53 | −9.93 | 2.9 |
| E255K | −10.77 | 12.8 | 16 | 8.8 | −10.99 | 17 | 45 | −10.03 | 2.5 |
| Q252H | −10.85 | 11.2 | 14 | 0.79 | −12.42 | 1.5 | 25 | −10.38 | 1.4 |
| Y253H | −11.08 | 7.6 | 9.5 | 0.55 | −12.64 | 1.1 | 20 | −10.51 | 1.1 |
| H396R | −11.98 | 1.7 | 2.125 | 1.3 | −12.13 | 2.5 | 23 | −10.42 | 1.3 |
| F359V | −12 | 1.6 | 2 | 0.81 | −12.41 | 1.6 | 26 | −10.35 | 1.4 |
| M244V | −12.22 | 1.4 | 1.75 | 1.1 | −12.23 | 2.1 | 16 | −10.64 | 0.89 |
| V304D | −12.27 | 1 | 1.25 | 0.72 | −12.48 | 1.4 | 22 | −10.45 | 1.2 |
| T315I/F317L | −5.47 | 970000 | 1212500 | 21 | −10.48 | 40 | 68 | −9.78 | 3.1 |
| T315I/V299L | −4.83 | 290000 | 362500 | 10 | −10.92 | 19 | 50 | −9.96 | 2.7 |
| T315I/V304D | −6.02 | 38000 | 47500 | 8.8 | −11.01 | 16 | 12 | −10.81 | 0.66 |
| V299L/F317L | −7.42 | 3650 | 4563 | 18 | −10.56 | 34 | 109 | −9.5 | 5 |
| V304D/V299L | −8.05 | 1250 | 1563 | 2.1 | −11.84 | 4 | 46 | −10.01 | 2.5 |
| F317L/V304D | −9.01 | 250 | 313 | 15 | −10.68 | 29 | 51 | −9.95 | 2.8 |
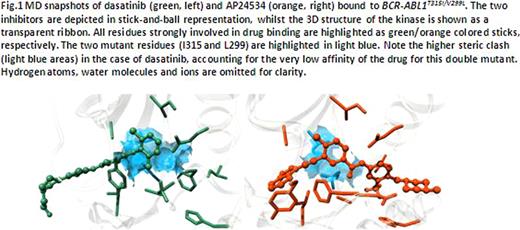
All polymutants are predicted to be highly resistant to dasatinib. AP24534 and PHA739358 are predicted to maintain a notable affinity toward the clinically relevant BCR-ABL1 polymytants T315I/V299L and V299L/F317L. Figure 1 depicts dasatinib and AP24534 in complex with the double mutant BCR-ABL1T315I/V299L, as extracted from the corresponding equilibrated molecular dynamics (MD) simulations performed in this study. AP24534 accommodates the great steric hindrance imposed by the polymutant to a much greater extent than dasatinib. BCR-ABL1T315I/V299L results in a dramatic conformational distortion of the entire binding site, leading to a less tight dasatinib binding and lower affinity of the inhibitor for the kinase.
MD snapshots of dasatinib (green, left) and AP24534 (orange, right) bound to BCR-ABL1T315I/V299L. The two inhibitors are depicted in stick-and-ball representation, whilst the 3D structure of the kinase is shown as a transparent ribbon. All residues strongly involved in drug binding are highlightedas green/orange colored sticks respectively.The two mutant residues (1315 and L299)are highlighted inlight blue. Note the higher steric clash (light blue areas) in the case of dasatinib, accounting for the very low affinity of the drug for this double mutant. Hydrogenatoms, water molecutes and ions are omitted for clarity.
MD snapshots of dasatinib (green, left) and AP24534 (orange, right) bound to BCR-ABL1T315I/V299L. The two inhibitors are depicted in stick-and-ball representation, whilst the 3D structure of the kinase is shown as a transparent ribbon. All residues strongly involved in drug binding are highlightedas green/orange colored sticks respectively.The two mutant residues (1315 and L299)are highlighted inlight blue. Note the higher steric clash (light blue areas) in the case of dasatinib, accounting for the very low affinity of the drug for this double mutant. Hydrogenatoms, water molecutes and ions are omitted for clarity.
In conclusion, mutational analysis of patients failing imatinib reveals a high rate of dasatinib resistant polymutants that explain, at least in part, why only ~40% of pts carrying unmutated BCR-ABL1 by direct sequencing achieve CCyR with second generation TKIs. AP24534 and PHA739358 are highly active against these complex polymutants in silico. Preliminary clinical data with these compounds against T315I appear to validate our modeling. Full modeling data for the most prevalent polymutants in complex with a panel of novel TKIs will be presented.
Kantarjian:Bristol Myers Squibb: Research Funding; ARIAD: Research Funding; Nerviano: Research Funding. Cortes:Bristol Myers Squibb: Research Funding; ARIAD: Research Funding; Nerviano: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal