Abstract
Abstract 680
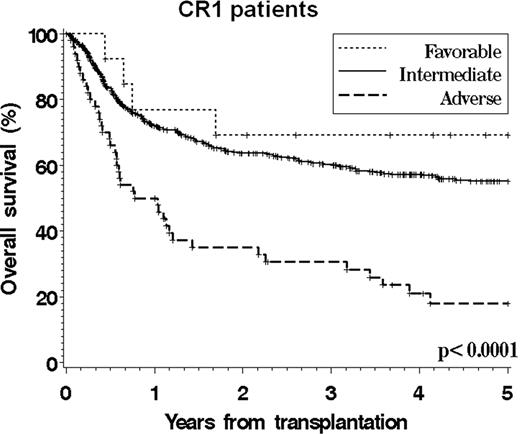
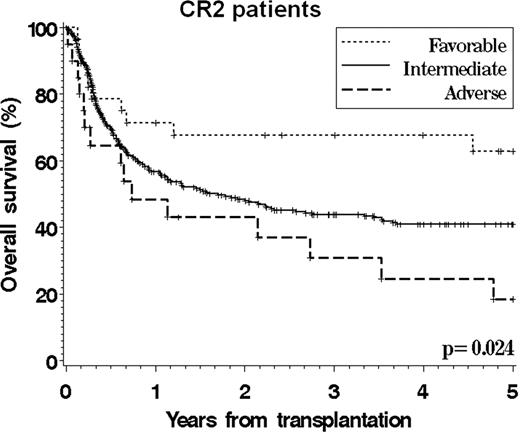
Cytogenetics play an essential role in determining the prognosis of patients with AML. However, there is still no validated cytogenetics grouping scheme that specifically applies to patients undergoing allogeneic stem cell transplantation, which hampers accurate prognostication and risk stratification. We studied 821 adult patients (median age 41, range 18–74) who underwent SCT between 1999 and 2004 for AML (excluding APL) in CR1 or CR2 and who were reported to the CIBMTR from centers with >20 patients meeting study criteria. 75% of patients received a myeloablative conditioning. 496 patients had a normal karyotype. The primary cytogenetics reports were manually reviewed for 92% of the patients with an abnormal karyotype. We compared the ability of the 6 existing grouping schemes (MRC, CALGB, EORTC/GIMEMA, SWOG/ECOG, DFCI, and Monosomal Karyotype (MK) classifications) to stratify patients, using both the Akaike Information Criterion in multivariable models and the C-statistic. Among all existing schemes, the DFCI system provided a marginally superior stratification for overall and leukemia-free survival. We also built a new classification using individual cytogenetic abnormalities in a Cox model that included other significant covariates (performance status, therapy-related disease, conditioning intensity, graft source, donor match, duration of CR1, and gender match). This CIBMTR scheme (see Table), which is similar to the DFCI scheme except for the inclusion of patients with t(8;21) in the intermediate group, could stratify patients into 3 groups with similar treatment-related mortality but significantly different overall survival, leukemia-free survival, and incidence of relapse. This scheme appeared to apply to both patients in CR1 and in CR2 (see Figures,F2). This transplant-specific scheme could be adopted for prognostication purposes and to stratify patients with karyotypic abnormalities entering transplantation clinical trials. Future studies may clarify the relative outcome of patients with t(8;21) and refine this scheme with the inclusion of molecular abnormalities.
| Group . | Abnormality . | % of patients with abnormal karyotype . | 5y OS CR1 . | 5y OS CR2 . |
|---|---|---|---|---|
| Favorable | Inv(16) | 12% | 73% | 62% |
| Intermediate | Normal karyotype | n/a | 58% | 38% |
| t(8;21) 11q23 abnormality Trisomy/tetrasomy 8 Abnormal 5 or 7 Other abnormalitiesa | 66% | 50% | 41% | |
| Adverse | Complex (≥4 abns) | 22% | 18% | 18% |
| Group . | Abnormality . | % of patients with abnormal karyotype . | 5y OS CR1 . | 5y OS CR2 . |
|---|---|---|---|---|
| Favorable | Inv(16) | 12% | 73% | 62% |
| Intermediate | Normal karyotype | n/a | 58% | 38% |
| t(8;21) 11q23 abnormality Trisomy/tetrasomy 8 Abnormal 5 or 7 Other abnormalitiesa | 66% | 50% | 41% | |
| Adverse | Complex (≥4 abns) | 22% | 18% | 18% |
Except for abn12p, abn3q, del(9q), t(1;19), and t(6;9), which are not classified in this scheme.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal