Abstract
Abstract 922
Patients with fludarabine refractory chronic lymphocytic leukemia (CLL) have a very poor prognosis with conventional chemotherapy with a median survival of 10 months and <10% surviving for 5 years. In 2006 the results of the response assessment of a multicenter phase II (UKCLL02) trial of subcutaneous (SC) alemtuzumab in fludarabine refractory CLL (defined as failing to respond to, or relapsing within 6 months of, fludarabine containing therapy) were presented, showing an overall response rate of 49%. We now report the survival data after a median follow up period of 64.6 months. SC alemtuzumab was given at a dose of 30mg 3x a week (after dose escalation) for up to 24 weeks depending on 6-weekly marrow assessments. Patients failing to respond to alemtuzumab could receive oral fludarabine (40mg/m2/day for 3 days every 4 weeks) combined with SC alemtuzumab.
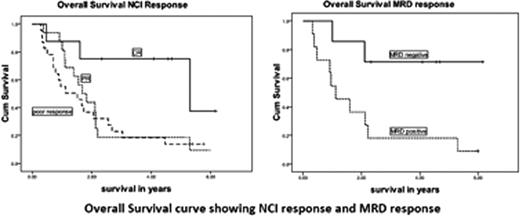
53 patients were enrolled but 2 patients died before receiving alemtuzumab and 1 withdrew consent. 50 patients have completed therapy (median duration of alemtuzumab was 18.8 months with a median dose of 1370mg [106-2323mg]). Response assessment was possible in 49 patients of which 8 patients achieved a complete remission (CR) and 16 patients achieved a partial remission (PR) giving an overall response rate of 49%. 17 patients had fludarabine added to the alemtuzumab (6 PR, 11 non-responders [NR]) and 3 had an improvement in response (1 from PR to CR and 2 from NR to PR). Median overall survival (OS) was 20.2 mo (CI 95%, 13.2–27.1) and progression free survival (PFS) was 13.0 mo (CI95%, 10.3–15.7). The 5-year OS and PFS rates were 24% and 9% respectively. Patients who achieved a CR, PR and less than a PR had a median OS of 63.6 mo, 20.2 mo and 18.2 mo, respectively and a median PFS of 47.1 mo, 13.8 mo and 7.5 mo respectively. Of 18 patients evaluable for MRD status, 7/18 achieved an MRD negative response and 11 were MRD positive with OS of not reached vs 13 mo (p=.02) and PFS of 47.1 vs 22.3 mo (p=.02), respectively. Of the 7 patients achieving MRD negativity, 5/7 (71%) were alive at 5 years. The IGHV gene was unmutated (>98% germ line homology) in 23/31 evaluable patients. Comparing unmutated with mutated for OS gave 18.2 and 13.1 mo (p=0.3) and PFS of 16.6 and 12.3 mo (p=0.08). FISH revealed poor risk deletions (11q and/or 17p) in 21/45 patients (17p- in 10, 11q- in 8, both in 3). 25/38 patients had dysfunction p53 in a functional assay previously reported by Pettitt et al (Blood, 2001; 98: 814–822). The table shows the lack of impact on PFS and OS.
| Abnormality . | status . | Overall Survival . | Progression Free Survival . | ||
|---|---|---|---|---|---|
| p53 | Deleted (n=13) | 13.4 | P=0.5 | 12.3 | P=0.8 |
| Normal (n=32) | 19.2 | 13.0 | P=0.4 | ||
| p53 | Dysfunctional (n=25) | 16.7 | P=0.2 | 12.3 | |
| Wild type (n=13) | 20.8 | 12.0 | P=0.8 | ||
| ATM | Deleted (n=11) | 18.6 | P=0.8 | 11.3 | |
| Normal (n=34) | 20.8 | 12.6 | P=0.3 | ||
| All poor risk | Poor risk (n=32) | 19.9 | P=0.8 | 12 | |
| Without (n=16) | 20.8 | 13 | |||
| Abnormality . | status . | Overall Survival . | Progression Free Survival . | ||
|---|---|---|---|---|---|
| p53 | Deleted (n=13) | 13.4 | P=0.5 | 12.3 | P=0.8 |
| Normal (n=32) | 19.2 | 13.0 | P=0.4 | ||
| p53 | Dysfunctional (n=25) | 16.7 | P=0.2 | 12.3 | |
| Wild type (n=13) | 20.8 | 12.0 | P=0.8 | ||
| ATM | Deleted (n=11) | 18.6 | P=0.8 | 11.3 | |
| Normal (n=34) | 20.8 | 12.6 | P=0.3 | ||
| All poor risk | Poor risk (n=32) | 19.9 | P=0.8 | 12 | |
| Without (n=16) | 20.8 | 13 | |||
In addition to prospective FISH evaluation we have performed retrospective SNP microarray using Illumina human CytoSNP-12 BeadChip on stored bone marrow and peripheral blood slides. Each BeadChip includes a complete panel of genome-wide tag SNPs and markers (>330,000 in total) targeting all regions of known cytogenetic importance. 36 patients were tested, SNP analysis identified all of the deletions present in more than 20% of CLL cells detected by FISH except in 1 patient due to technical issues. SNP analysis identified 2 additional patients with abnormalities in 17p (one deletion and one copy neutral loss of heterozygosity) and 2 more with deletions in 11q. A number of other chromosomal aberrations (average number per patient =2.6, with a maximum of 12 in one patient) were also detected although no recurrent abnormalities were identified.
We report that subcutaneous alemtuzumab is effective in poor-risk fludarabine-refractory CLL. In this group, patients with poor risk deletions havenÕt got a survival disadvantage over the non deleted group presumably because all of the patients were resistant to conventional chemotherapy regardless of whether a p53 pathway abnormality could be detected. SNP microarray is a useful research tool in studying genomic aberration from archived bone marrow and blood samples including in stored blood and marrow slides. The long term survival data shows that subcutaneous alemtuzumab improves the OS and PFS in fludarabine refractory CLL particularly in patients achieving MRD negative remissions.
Rawstron:Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees; BD Bioscience: Honoraria, Membership on an entity's Board of Directors or advisory committees, Patents & Royalties; Genzyme: Honoraria. Dearden:Roche Pharmaceuticals: Honoraria, Membership on an entity's Board of Directors or advisory committees; Genzyme: Honoraria, Membership on an entity's Board of Directors or advisory committees. Pettitt:Glaxo Smith Kline: Research Funding. Kennedy:Roche Pharmaceuticals: Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal