Epstein-Barr virus (EBV) is present in B cells in the blood of healthy people; few studies have looked for EBV in other cell types in blood from patients with lymphoproliferative disorders. We use a new technique combining immunofluorescent cell-surface staining and fluorescent in situ hybridization to quantify both EBV copy number per cell and cell types in blood from patients with high EBV DNA loads. In addition to CD20+ B cells, EBV was present in plasmablast/plasma cells in the blood of 50% of patients, in monocytes or T cells in a small proportion of patients, and in “non-B, non-T, non-monocytes” in 69% of patients. The mean EBV copy number in B cells was significantly higher than in plasmablast/plasma cells. There was no correlation between EBV load and virus copy number per cell. Although we detected CD21, the EBV B-cell receptor, on EBV-infected B cells, we could not detect it on virus-infected T cells. These findings expand the range of cell types infected in the blood. Determining the number of EBV genomes per cell and the type of cells infected in patients with high EBV loads may provide additional prognostic information for the development of EBV lymphoproliferative diseases.
Introduction
Epstein-Barr virus (EBV) infects more than 90% of the human population.1 In immunocompetent hosts, the virus is latent in B cells of the peripheral blood and is not associated with disease.2,–4 However, in immunocompromised patients, immune surveillance to the virus is often impaired, a larger number of B cells are infected with EBV, and the virus can contribute to lymphoproliferative disease. Approximately 1%-20% of transplant recipients can develop posttransplantation lymphoproliferative disease (PTLD) during the first year after transplantation, and approximately 90% of these cases are EBV positive.5 Persons with AIDS have a 60-fold increased risk of developing lymphoma, compared with the general population, and virtually all Hodgkin and non-Hodgkin lymphomas that occur in the late stages of HIV infection are EBV positive.6
Although EBV establishes a latent infection in peripheral blood B cells of healthy people, less is known about the phenotype of virus-infected cells in the blood of immunocompromised persons with high EBV DNA loads. Most studies have focused on the phenotype of virus-infected B cells in transplant recipients.2,7,,,–11 However, EBV can infect cells other than B cells, including T cells, natural killer (NK) cells, monocytes, and pre-Langerhans cells.12,,,–16
Several techniques have been developed to detect EBV in cells. In situ hybridization using a probe that detects the EBV-encoded RNAs (EBERs) is considered the best test for localizing latent EBV in tissue samples.17 Combined staining for EBERs and antibodies to cell-surface markers for tissues on microscope slides, or for peripheral blood by flow cytometry,18 has been used to determine the phenotype of the EBV-infected cells. Although detection of EBERs indicates that cells are infected with EBV, this test cannot provide an estimate of the number of EBV genomes present per cell.
We describe a new technique (Immuno-FISH) that combines immunofluorescent staining for surface proteins (using antibodies directly conjugated to fluorochromes) and fluorescent in situ hybridization for EBV DNA. This technique allows the simultaneous determination of the cell type infected by EBV and quantification of EBV copy number in the infected cell. We show that EBV is present not only in B cells, but also in a large percentage of other cell types in the peripheral blood of patients with high EBV DNA loads. In addition, we correlate the number of EBV genomes per cell with the phenotype of the infected cells.
Methods
Study participants
Patients had blood drawn after informed consent was obtained in accordance with the Declaration of Helsinki under protocols approved by the Institutional Review Boards of the National Institute of Allergy and Infectious Diseases, the National Cancer Institute, the National Heart, Lung, and Blood Institute (patients 1-23), Nagoya University Hospital (patients 24-27), or the University of Maryland and the National Institute of Allergy and Infectious Diseases (patients 28-29). For patients from the United States, we selected those whose EBV DNA loads were more than 5000 copies per million cells (normal is < 200 copies/million cells) and, for patients from Japan, more than 50 000 copies per μg of DNA.
Measurement of EBV DNA in blood
For patients 1-23 and 28-29, the EBV DNA load data were reported as the number of EBV genomes per 106 cells. Peripheral blood mononuclear cells (PBMCs) were lysed and EBV quantitative real-time polymerase chain reaction (qPCR) was performed as previously described19 (see supplemental data, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Immuno-FISH procedure
Cryopreserved PBMCs were thawed at 37°C, washed once in media and once in phosphate-buffered saline (PBS), and then resuspended in a solution of 0.2N acetic acid, 0.02N HCl, in Tris HCl 0.1M (adjusted to pH 3.7) for 15 minutes at room temperature. The cells were washed twice in PBS and incubated with fluorochrome-conjugated monoclonal antibodies (to stain cell-surface proteins) in suspension for 30 minutes at 4°C. Cells were washed 2× with PBS and applied to Superfrost Plus microscope slides (Thermo Fisher Scientific) by cytospin centrifugation at 500g for 5 minutes.
The cells were then fixed with 5mM BS3 (Thermo Fisher Scientific) for 30 minutes at room temperature and permeabilized with Triton X-100 (Sigma-Aldrich) at 0.5% in PBS for 10 minutes at room temperature. The slides were washed with PBS and incubated at 37°C for 1 hour in a solution of 2× saline-sodium citrate (SSC) containing 10 U/mL RNAse T1 (Roche Applied Science) and 15 U/mL RNAse A (Sigma-Aldrich). The slides were then washed once in 0.1M Tris-HCl (pH 7.5), incubated for 5 minutes in a solution of 0.1M Tris-HCl (pH 7.5)/0.1M glycine, and then washed in 2× SSC.
FISH was performed with a biotinylated EBV BioProbe (Enzo Life Sciences) specific for the BamHI W region of EBV. There are 5-12 BamH1 W repeats per EBV genome, depending on the virus strain.20 The probe was diluted to a final concentration of 0.25 ng/μL in hybridization buffer (Enzo Life Sciences) and denatured at 94°C for 10 minutes. The slides were then incubated with the denatured probe at 85°C for 6 minutes under a coverslip sealed with rubber cement and then hybridized overnight at 37°C. Slides were washed 2× for 15 minutes in a solution of 0.1× SSC, 0.1% Tween, and 1% BSA at 42°C, washed once in 0.1× SSC, and washed 3 times in Tris-HCl (0.1M; pH 7.5). Slides were incubated with streptavidin-conjugated Alexa 488 or streptavidin-conjugated Alexa 594 for 15 minutes at a final concentration of 1 μg/mL, washed in PBS/Tween 0.1% for 5 minutes, washed 3× in Tris-HCl (0.1M; pH 7.5), and mounted with Fluoromount-G medium (Southern-Biotech). The slides were visualized with a Leica SP5 confocal microscope (Carl Zeiss), with a 63×/1.4 numerical aperture magnification objective. For each field, photographs of serial z-stack sections (0.5 μm thick) were obtained throughout the cells and then overlapped to quantify the total number of fluorescent spots (each spot corresponds to one EBV genome copy) in the cells.
When FISH alone was performed, the cells were incubated for 15 minutes with a solution of 0.2N acetic acid, 0.02N HCl, in Tris HCl (0.1M; adjusted to pH 3.7), then applied to the slides, fixed with BS3, permeabilized with Triton X-100, and FISH was performed as described above.
Cell-surface markers, fluorochromes, and EBV antibodies
After incubation with acetic acid and HCl, cells were resuspended in PBS at a concentration of 1 × 106 cells per 100 μL and stained with mouse anti–human monoclonal antibodies to label cell-surface markers.
For the sorting procedure, the CD19 phycoerythrin (PE)–Cy7 antibody was used to identify B cells. The CD3 allophycocyanin (APC)–H7 and the CD56 APC antibodies were used to identify the T and NK subpopulations, respectively. All of these antibodies were purchased from BD Biosciences.
For each patient sample, cells were stained with different combinations of monoclonal antibodies. The first combination contained antibodies that recognize CD3 T cells (Alexa 594), B cells (Alexa 647), and monocytes (V450); the second combination included antibodies that recognize CD3 T cells (Alexa 594), NK cells (Alexa 647), and monocytes (V450), and the third combination contained antibodies that recognize the B-cell surface marker, CD20 (Alexa 488), together with the κ and λ light chains of immunoglobulin (Ig; Alexa 647). Staining with antibodies that recognize the κ and λ chains was performed after permeabilization of the cells to allow staining of intracytoplasmic immunoglobulin. The biotinylated FISH EBV probe was detected using streptavidin-conjugated Alexa 488 for the first and second combinations, and with streptavidin-conjugated Alexa 594 for the third combination.
For samples in which EBV was detected in CD3 T cells, another aliquot of cells was stained with Alexa 647–conjugated anti-CD4 antibody and Alexa 488–conjugated anti-CD8 antibody, and the FISH probe was detected using streptavidin-conjugated Alexa 594.
To detect lytic replication of EBV, PBMCs were stained with a combination of antibodies for B and T cells (CD3 Alexa 594 and CD19/CD20 Alexa 647), applied to Superfrost Plus microscope slides, and fixed and permeabilized as previously described. Cells were then incubated for 30 minutes with BZLF1 monoclonal antibody (Meridian Life Science) conjugated with Alexa 488. Slides were then washed 3× in PBS and mounted with Fluoromount-G medium. B95-8 and BJAB cells were stained with BZLF1 antibody as positive and negative controls for virus lytic replication, respectively.
Results
FISH and Immuno-FISH assays are sensitive and specific
To determine the sensitivity and specificity of our FISH assay, we diluted EBV-positive X50-7 cells with EBV-negative BJAB cells and performed FISH. A mixture of 10% of X50-7 cells and 90% BJAB cells yielded 9.6% (11/114) EBV-positive cells by FISH, whereas a mixture of 1% X50-7 cells and 99% BJAB cells showed 1.1% (8/745) EBV-positive cells by FISH (representative example in supplemental Figure S1A-B). As controls, one slide of 100% EBV-positive X50-7 cells and one of 100% EBV-negative BJAB cells were also stained by FISH, and EBV episomes were detected in all of the EBV-positive cells and none of the EBV-negative cells (supplemental Figure 1C-D). As an additional negative control, Immuno-FISH staining of cells from patients with very low or undetectable EBV viral loads by PCR did not detect any FISH-positive cells (data not shown). Combining Immuno-FISH with DAPI (4′,6′-diamidino-2-phenylindole) staining, we found EBV DNA only in the nucleus of the cells (data not shown). These results indicate that the FISH procedure is specific, and that it can detect at least 1% of EBV-positive cells in a mixture of EBV-negative cells.
We also quantified the number of EBV genomes per cell using the FISH procedure. EBV-positive cells were analyzed by confocal microscopy; serial z-stack sections were imaged every 0.5 μm, overlapped, and the number of fluorescent spots, each corresponding to a single EBV episome, was counted. Analysis of 65 individual X-50-7 cells from 5 separate experiments showed a median of 7 EBV genomes per cell, with a mean of 9.4 genomes per cell (range, 2-18 genomes per cell). These results are similar to those previously reported, indicating that X50-7 cells contain approximately 5 EBV genomes per cell.21 To confirm the sensitivity of the FISH assay, we tested the Namalwa cell line, which contains 2 copies of EBV integrated in each cellular genome.22 Analysis of 411 cells in 5 separate experiments showed a median of 1 EBV genome per cell and a mean of 0.76 genomes per cell (range, 0-4 copies per cell) (supplemental Figure 1E). This value is slightly lower than the expected number and could be a result of the reduced accessibility of the probe to hybridize to the target when integrated in genomic DNA. Moreover, the 2 copies of EBV are integrated in tandem in the cellular genome of Namalwa cells,23 and we may not have been able to resolve the 2 genomes using our Immuno-FISH procedure.
To determine whether the immunofluorescent staining component of Immuno-FISH was sensitive and specific when followed by FISH, we isolated fresh PBMCs and sorted B cells (CD19+), NK cells (CD56+/CD3−) and T cells (CD3+) by flow cytometry. We stained aliquots of each of the 4 subpopulations with Alexa-conjugated antibodies specific for each subpopulation (eg, T cells with anti-CD3 antibody) and with antibodies that should not recognize the subpopulation (eg, T cells with anti-CD19/anti-CD20). The purity of the staining was more than 97% in all of the combinations tested (supplemental Table 1).
EBV is detected in circulating B cells
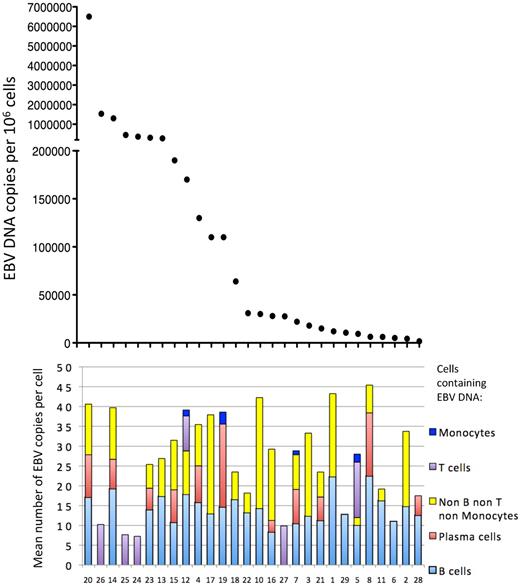
Immuno-FISH was used to characterize the phenotype of the cells infected by EBV from 29 patients with elevated EBV DNA levels in the blood (Table 1). We defined B cells as cells recognized by a combination of anti-CD19 and anti-CD20 antibodies. EBV was detected in B lymphocytes from all 29 patients tested (Figure 1; Table 1), except for the 4 patients with T-cell chronic active EBV (CAEBV) from Japan. In the other 25 patients, B cells represented the majority of cells containing EBV, except for one patient (patient 5) with HIV infection. The median number of EBV genomes in B cells for all the other patients was 13.9 per cell, with a mean of 14.2 per cell (range, 1-40) (Table 2; Figure 2).
Phenotype of EBV-infected PBMCs from subjects
| Patient . | Diagnosis . | EBV DNA load/106 cells . | Percent of EBV-positive cells (no. positive/total no. cells) . | ||||
|---|---|---|---|---|---|---|---|
| B cells . | Plasmablast/plasma cells . | Non-B, non-T, nonmonocyte cells . | T cells . | Monocytes . | |||
| 1 | HIV | 12 000 | 75 (3/4) | ND | 25 (1/4) | 0 | 0 |
| 2 | HIV | 4300 | 89 (8/9) | 0 | 11 (1/9) | 0 | 0 |
| 3 | HIV | 18 000 | 91 (10/11) | 0 | 9 (1/11) | 0 | 0 |
| 4 | HIV | 130,000 | 71 (12/17) | 67 (6/9) | 29 (5/17) | 0 | 0 |
| 5 | HIV | 9400 | 25 (1/4) | 0 | 25 (1/4) | 25 (1/4) | 25 (1/4) |
| 6 | HIV | 5100 | 100 (11/11) | 0 | 0 | 0 | 0 |
| 7 | HIV | 22 000 | 67 (10/15) | 50 (4/8) | 20 (3/15) | 0 | 13 (2/15) |
| 8 | HIV | 6300 | 82 (9/11) | 17 (1/6) | 18 (2/11) | 0 | 0 |
| 9 | Tx | NA | 100 (4/4) | 0 | 0 | 0 | 0 |
| 10 | Tx | 30 000 | 92 (12/13) | 0 | 8 (1/13) | 0 | 0 |
| 11 | Tx | 6200 | 83 (5/6) | ND | 17 (1/6) | 0 | 0 |
| 12 | LyG/HL | 170 000 | 64 (49/77) | 0 | 6 (5/77) | 27 (21/77) | 3 (2/77) |
| 13 | Severe primary EBV infection | 280 000 | 90 (30/33) | 0 | 10 (3/33) | 0 | 0 |
| 14 | PTLD | 1 300 000 | 55 (11/20) | 25 (2/8) | 45 (9/20) | 0 | 0 |
| 15 | ATL EBV-LPD | 190 000 | 79 (19/24) | 21 (3/14) | 21 (5/24) | 0 | 0 |
| 16 | LyG | 28 000 | 79 (15/19) | 14 (1/7) | 21 (4/19) | 0 | 0 |
| 17 | ICL | 110 000 | 97 (28/29) | 0 | 3 (1/29) | 0 | 0 |
| 18 | AA | 64 000 | 80 (8/10) | ND | 20 (2/10) | 0 | 0 |
| 19 | AA | 110 000 | 89 (17/19) | 20 (1/5) | 0 | 0 | 5 (1/19) |
| 20 | AA | 6 500 000 | 52 (24/46) | 37 (6/16) | 48 (22/46) | 0 | 0 |
| 21 | B-cell CAEBV | 15 000 | 73 (11/15) | 20 (1/5) | 27 (4/15) | 0 | 0 |
| 22 | B-cell CAEBV | 30 900 | 97 (29/30) | 0 | 3 (1/30) | 0 | 0 |
| 23 | B-cell CAEBV | 310 000 | 80 (12/15) | 25 (2/8) | 20 (3/15) | 0 | 0 |
| 24 | T-cell CAEBV | 54 666/μg | 0 | ND | 0 | 100 (88/88) | 0 |
| 25 | T-cell CAEBV | 67 612/μg | 0 | ND | 0 | 100 (21/21) | 0 |
| 26 | T-cell CAEBV | 230 000/μg | 0 | ND | 0 | 100 (78/78) | 0 |
| 27 | T-cell CAEBV | 4154/μg | 0 | ND | 0 | 100 (18/18) | 0 |
| 28 | IM | 1700 | 83 (10/12) | 17 (1/6) | 0 | 0 | 0 |
| 29 | IM | 10 651 | 100 (11/11) | 0 | 0 | 0 | 0 |
| Patient . | Diagnosis . | EBV DNA load/106 cells . | Percent of EBV-positive cells (no. positive/total no. cells) . | ||||
|---|---|---|---|---|---|---|---|
| B cells . | Plasmablast/plasma cells . | Non-B, non-T, nonmonocyte cells . | T cells . | Monocytes . | |||
| 1 | HIV | 12 000 | 75 (3/4) | ND | 25 (1/4) | 0 | 0 |
| 2 | HIV | 4300 | 89 (8/9) | 0 | 11 (1/9) | 0 | 0 |
| 3 | HIV | 18 000 | 91 (10/11) | 0 | 9 (1/11) | 0 | 0 |
| 4 | HIV | 130,000 | 71 (12/17) | 67 (6/9) | 29 (5/17) | 0 | 0 |
| 5 | HIV | 9400 | 25 (1/4) | 0 | 25 (1/4) | 25 (1/4) | 25 (1/4) |
| 6 | HIV | 5100 | 100 (11/11) | 0 | 0 | 0 | 0 |
| 7 | HIV | 22 000 | 67 (10/15) | 50 (4/8) | 20 (3/15) | 0 | 13 (2/15) |
| 8 | HIV | 6300 | 82 (9/11) | 17 (1/6) | 18 (2/11) | 0 | 0 |
| 9 | Tx | NA | 100 (4/4) | 0 | 0 | 0 | 0 |
| 10 | Tx | 30 000 | 92 (12/13) | 0 | 8 (1/13) | 0 | 0 |
| 11 | Tx | 6200 | 83 (5/6) | ND | 17 (1/6) | 0 | 0 |
| 12 | LyG/HL | 170 000 | 64 (49/77) | 0 | 6 (5/77) | 27 (21/77) | 3 (2/77) |
| 13 | Severe primary EBV infection | 280 000 | 90 (30/33) | 0 | 10 (3/33) | 0 | 0 |
| 14 | PTLD | 1 300 000 | 55 (11/20) | 25 (2/8) | 45 (9/20) | 0 | 0 |
| 15 | ATL EBV-LPD | 190 000 | 79 (19/24) | 21 (3/14) | 21 (5/24) | 0 | 0 |
| 16 | LyG | 28 000 | 79 (15/19) | 14 (1/7) | 21 (4/19) | 0 | 0 |
| 17 | ICL | 110 000 | 97 (28/29) | 0 | 3 (1/29) | 0 | 0 |
| 18 | AA | 64 000 | 80 (8/10) | ND | 20 (2/10) | 0 | 0 |
| 19 | AA | 110 000 | 89 (17/19) | 20 (1/5) | 0 | 0 | 5 (1/19) |
| 20 | AA | 6 500 000 | 52 (24/46) | 37 (6/16) | 48 (22/46) | 0 | 0 |
| 21 | B-cell CAEBV | 15 000 | 73 (11/15) | 20 (1/5) | 27 (4/15) | 0 | 0 |
| 22 | B-cell CAEBV | 30 900 | 97 (29/30) | 0 | 3 (1/30) | 0 | 0 |
| 23 | B-cell CAEBV | 310 000 | 80 (12/15) | 25 (2/8) | 20 (3/15) | 0 | 0 |
| 24 | T-cell CAEBV | 54 666/μg | 0 | ND | 0 | 100 (88/88) | 0 |
| 25 | T-cell CAEBV | 67 612/μg | 0 | ND | 0 | 100 (21/21) | 0 |
| 26 | T-cell CAEBV | 230 000/μg | 0 | ND | 0 | 100 (78/78) | 0 |
| 27 | T-cell CAEBV | 4154/μg | 0 | ND | 0 | 100 (18/18) | 0 |
| 28 | IM | 1700 | 83 (10/12) | 17 (1/6) | 0 | 0 | 0 |
| 29 | IM | 10 651 | 100 (11/11) | 0 | 0 | 0 | 0 |
For some patients, the percentages for each subpopulation total to > 100%, because the same cell marker (eg, CD20 for B cells) was used in more than one combination (eg, CD20 with CD3 and CD14 in one combination, CD20 with kappa/lambda in another combination). Patients with HIV infection had CD4 T-cell counts < 400/μL and no EBV-positive lymphomas. Patient 21 received rituximab therapy 1 month before blood was drawn; patients 10, 11, 18, 19, and 20 received anti-thymocyte globulin 5 days to 1 month before blood was drawn (patient 18 received rabbit anti-thymocyte globulin, and patients 10, 11, 18, 19, and 20 received horse anti-thymocyte globulin).
ND indicates not done; Tx, allogeneic hematopoietic stem cell transplant recipient; LyG/HL, lymphomatoid granulomatosis/Hodgkin lymphoma; ATL EBV-LPD, adult T-cell leukemia and EBV-lymphoproliferative disease; ICL, idiopathic CD4 lymphopenia; AA, aplastic anemia receiving antithymocyte globulin; and IM, EBV infectious mononucleosis.
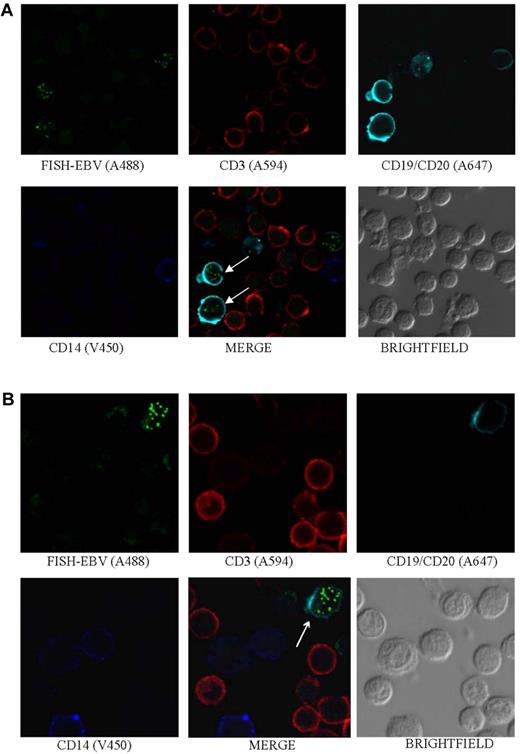
EBV is detected in B cells. Photomicrographs of PBMCs stained by Immuno-FISH from patients 17 (A) and 8 (B) show EBV in the nucleus of B cells (arrows). EBV DNA was identified with the biotinylated FISH probe and streptavidin-conjugated Alexa 488, B cells using a combination of Alexa 647–conjugated anti-CD19 and anti-CD20 antibodies (Biolegend), T cells with Alexa 594–conjugated anti-CD3 antibody (Invitrogen), and monocytes with V450-conjugated anti-CD14 antibody (BD Biosciences). Arrows indicate EBV-positive cells.
EBV is detected in B cells. Photomicrographs of PBMCs stained by Immuno-FISH from patients 17 (A) and 8 (B) show EBV in the nucleus of B cells (arrows). EBV DNA was identified with the biotinylated FISH probe and streptavidin-conjugated Alexa 488, B cells using a combination of Alexa 647–conjugated anti-CD19 and anti-CD20 antibodies (Biolegend), T cells with Alexa 594–conjugated anti-CD3 antibody (Invitrogen), and monocytes with V450-conjugated anti-CD14 antibody (BD Biosciences). Arrows indicate EBV-positive cells.
Mean number of EBV genomes per cell for each cell subpopulation
| Patient . | Diagnosis . | EBV DNA load/106 cells . | Average number of EBV genomes per cell (range) . | ||||
|---|---|---|---|---|---|---|---|
| B cells . | Plasmablast/plasma cells . | Non-B, non-T, non-monocyte cells . | T cells . | Monocytes . | |||
| 1 | HIV | 12 000 | 22.25 (5-40) | ND | 21 (21) | 0 | 0 |
| 2 | HIV | 4300 | 14.75 (6-23) | 0 | 19 (19) | 0 | 0 |
| 3 | HIV | 18 000 | 12.3 (4-24) | 0 | 21 (21) | 0 | 0 |
| 4 | HIV | 130 000 | 15.75 (1-38) | 9.3 (6-16) | 10.4 (6-15) | 0 | 0 |
| 5 | HIV | 9400 | 10 (10) | 0 | 2 (2) | 14 (14) | 2 (2) |
| 6 | HIV | 5100 | 10.9 (4-20) | 0 | 0 | 0 | 0 |
| 7 | HIV | 22 000 | 10.4 (6-17) | 8.7 (4-16) | 8.75 (6-14) | 0 | 1 (1) |
| 8 | HIV | 6300 | 22.4 (5-35) | 16 (16) | 7 (7) | 0 | 0 |
| 9 | Tx | NA | 7.3 (4-11) | 0 | 0 | 0 | 0 |
| 10 | Tx | 30 000 | 14.25 (6-30) | 0 | 28 (28) | 0 | 0 |
| 11 | Tx | 6200 | 16.2 (2-27) | ND | 3 (3) | 0 | 0 |
| 12 | LyG/HL | 170 000 | 17.8 (4-38) | 0 | 11.2 (7-21) | 8.8 (2-40) | 1.5 (1-2) |
| 13 | Severe primary EBV infection | 280 000 | 17.3 (5-35) | 0 | 9.6 (9-10) | 0 | 0 |
| 14 | PTLD | 1 300 000 | 19.2 (3-35) | 7.5 (3-12) | 13 (7-27) | 0 | 0 |
| 15 | ATL EBV-LPD | 190 000 | 10.7 (2-23) | 8.3 (7-10) | 12.5 (10-15) | 0 | 0 |
| 16 | LyG | 28 000 | 8.26 (2-16) | 3 (3) | 18 (8-34) | 0 | 0 |
| 17 | ICL | 110 000 | 12.9 (3-28) | 0 | 25 (25) | 0 | 0 |
| 18 | AA | 64 000 | 16.5 (7-26) | ND | 7 (7) | 0 | 0 |
| 19 | AA | 110 000 | 14.58 (2-34) | 21 (21) | 0 | 0 | 3 (3) |
| 20 | AA | 6 500 000 | 17.04 (3-35) | 10.8 (4-17) | 12.75 (5-21) | 0 | 0 |
| 21 | B-cell CAEBV | 15 000 | 11.18 (4-22) | 6 (6) | 6.3 (2-9) | 0 | 0 |
| 22 | B-cell CAEBV | 30 900 | 13.17 (4-33) | 0 | 5 (5) | 0 | 0 |
| 23 | B-cell CAEBV | 310 000 | 13.9 (6-23) | 5.5 (5-6) | 6 (6) | 0 | 0 |
| 24 | T-cell CAEBV | 54 666/μg | 0 | ND | 0 | 7.25 (1-22) | 0 |
| 25 | T-cell CAEBV | 67 612/μg | 0 | ND | 0 | 7.66 (1-17) | 0 |
| 26 | T-cell CAEBV | 230 000/μg | 0 | ND | 0 | 10.24 (2-26) | 0 |
| 27 | T-cell CAEBV | 4154/μg | 0 | ND | 0 | 9.88 (3-21) | 0 |
| 28 | IM | 1700 | 12.5 (9-22) | 5 (4-6) | 0 | 0 | 0 |
| 29 | IM | 10 651 | 12.63 (5-27) | 0 | 0 | 0 | 0 |
| Patient . | Diagnosis . | EBV DNA load/106 cells . | Average number of EBV genomes per cell (range) . | ||||
|---|---|---|---|---|---|---|---|
| B cells . | Plasmablast/plasma cells . | Non-B, non-T, non-monocyte cells . | T cells . | Monocytes . | |||
| 1 | HIV | 12 000 | 22.25 (5-40) | ND | 21 (21) | 0 | 0 |
| 2 | HIV | 4300 | 14.75 (6-23) | 0 | 19 (19) | 0 | 0 |
| 3 | HIV | 18 000 | 12.3 (4-24) | 0 | 21 (21) | 0 | 0 |
| 4 | HIV | 130 000 | 15.75 (1-38) | 9.3 (6-16) | 10.4 (6-15) | 0 | 0 |
| 5 | HIV | 9400 | 10 (10) | 0 | 2 (2) | 14 (14) | 2 (2) |
| 6 | HIV | 5100 | 10.9 (4-20) | 0 | 0 | 0 | 0 |
| 7 | HIV | 22 000 | 10.4 (6-17) | 8.7 (4-16) | 8.75 (6-14) | 0 | 1 (1) |
| 8 | HIV | 6300 | 22.4 (5-35) | 16 (16) | 7 (7) | 0 | 0 |
| 9 | Tx | NA | 7.3 (4-11) | 0 | 0 | 0 | 0 |
| 10 | Tx | 30 000 | 14.25 (6-30) | 0 | 28 (28) | 0 | 0 |
| 11 | Tx | 6200 | 16.2 (2-27) | ND | 3 (3) | 0 | 0 |
| 12 | LyG/HL | 170 000 | 17.8 (4-38) | 0 | 11.2 (7-21) | 8.8 (2-40) | 1.5 (1-2) |
| 13 | Severe primary EBV infection | 280 000 | 17.3 (5-35) | 0 | 9.6 (9-10) | 0 | 0 |
| 14 | PTLD | 1 300 000 | 19.2 (3-35) | 7.5 (3-12) | 13 (7-27) | 0 | 0 |
| 15 | ATL EBV-LPD | 190 000 | 10.7 (2-23) | 8.3 (7-10) | 12.5 (10-15) | 0 | 0 |
| 16 | LyG | 28 000 | 8.26 (2-16) | 3 (3) | 18 (8-34) | 0 | 0 |
| 17 | ICL | 110 000 | 12.9 (3-28) | 0 | 25 (25) | 0 | 0 |
| 18 | AA | 64 000 | 16.5 (7-26) | ND | 7 (7) | 0 | 0 |
| 19 | AA | 110 000 | 14.58 (2-34) | 21 (21) | 0 | 0 | 3 (3) |
| 20 | AA | 6 500 000 | 17.04 (3-35) | 10.8 (4-17) | 12.75 (5-21) | 0 | 0 |
| 21 | B-cell CAEBV | 15 000 | 11.18 (4-22) | 6 (6) | 6.3 (2-9) | 0 | 0 |
| 22 | B-cell CAEBV | 30 900 | 13.17 (4-33) | 0 | 5 (5) | 0 | 0 |
| 23 | B-cell CAEBV | 310 000 | 13.9 (6-23) | 5.5 (5-6) | 6 (6) | 0 | 0 |
| 24 | T-cell CAEBV | 54 666/μg | 0 | ND | 0 | 7.25 (1-22) | 0 |
| 25 | T-cell CAEBV | 67 612/μg | 0 | ND | 0 | 7.66 (1-17) | 0 |
| 26 | T-cell CAEBV | 230 000/μg | 0 | ND | 0 | 10.24 (2-26) | 0 |
| 27 | T-cell CAEBV | 4154/μg | 0 | ND | 0 | 9.88 (3-21) | 0 |
| 28 | IM | 1700 | 12.5 (9-22) | 5 (4-6) | 0 | 0 | 0 |
| 29 | IM | 10 651 | 12.63 (5-27) | 0 | 0 | 0 | 0 |
Mean EBV copy number per cell for each cell subpopulation for each patient. P values were obtained with the t test. The difference in mean EBV copy number between B cells and non-B, non-T, non-monocyte cells (P = .2878) and between B and T cells (P = .77) was not significant.
Mean EBV copy number per cell for each cell subpopulation for each patient. P values were obtained with the t test. The difference in mean EBV copy number between B cells and non-B, non-T, non-monocyte cells (P = .2878) and between B and T cells (P = .77) was not significant.
EBV is detected in circulating plasmablasts/plasma cells
Although EBV has been detected in latently infected plasmablastic lymphomas24 and plasmacytomas in immunocompromised patients with HIV,25 EBV has not been reported in circulating plasmablasts or plasma cells. Plasmablasts and plasma cells have a prominent cytoplasm containing abundant κ or λ light-chain Ig and lack CD20 on their surface. To determine whether EBV is present in circulating plasmablasts/plasma cells, Immuno-FISH was performed using PBMCs stained with a combination of antibodies to κ and λ chains of Ig and CD20. Surprisingly, we detected EBV in plasmablast/plasma cells in 50% (11 of 22) of the patients tested (Figure 3A; Table 1). Nearly all of the patient groups had some patients with EBV-positive plasmablast/plasma cells. The median number of EBV genomes in plasmablast/plasma cells was 8.3 copies per cell, with a mean of 9.2 copies per cell (range, 3-21), which was lower than the median (13.9) or mean (14.2) number of EBV genomes in B cells (Table 2; Figure 2).
EBV is detected in plasmablasts/plasma cells and T cells. (A) Photomicrographs of PBMCs stained by Immuno-FISH from patient 4 shows EBV in plasmablasts/plasma cells identified using Alexa 647–conjugated anti-κ and anti-λ chain immunoglobulin antibodies. The EBV-positive cells are negative for CD20 using Alexa 488–conjugated anti-CD20 antibody. EBV DNA was identified with the biotinylated FISH probe and streptavidin-conjugated Alexa 594 and monocytes with V450-conjugated anti-CD14 antibody. Although most of the EBV DNA was detected in the nucleus, some viral DNA was detected in the cytoplasm, suggesting that EBV lytic infection was present in the cell. (B) Photomicrographs of PBMCs stained by Immuno-FISH from patient 12 shows EBV in CD3+ T cells. EBV DNA was identified with biotinylated FISH probe and streptavidin-conjugated Alexa 488, B cells using a combination of Alexa 647–conjugated anti-CD19 and anti-CD20 antibodies, T cells with Alexa 594–conjugated anti-CD3 antibody, and monocytes with V450-conjugated anti-CD14 antibody. Arrows indicate EBV-positive cells.
EBV is detected in plasmablasts/plasma cells and T cells. (A) Photomicrographs of PBMCs stained by Immuno-FISH from patient 4 shows EBV in plasmablasts/plasma cells identified using Alexa 647–conjugated anti-κ and anti-λ chain immunoglobulin antibodies. The EBV-positive cells are negative for CD20 using Alexa 488–conjugated anti-CD20 antibody. EBV DNA was identified with the biotinylated FISH probe and streptavidin-conjugated Alexa 594 and monocytes with V450-conjugated anti-CD14 antibody. Although most of the EBV DNA was detected in the nucleus, some viral DNA was detected in the cytoplasm, suggesting that EBV lytic infection was present in the cell. (B) Photomicrographs of PBMCs stained by Immuno-FISH from patient 12 shows EBV in CD3+ T cells. EBV DNA was identified with biotinylated FISH probe and streptavidin-conjugated Alexa 488, B cells using a combination of Alexa 647–conjugated anti-CD19 and anti-CD20 antibodies, T cells with Alexa 594–conjugated anti-CD3 antibody, and monocytes with V450-conjugated anti-CD14 antibody. Arrows indicate EBV-positive cells.
EBV is detected in circulating T cells in some patients, and in CD3+, CD4−, and CD8− T cells in Japanese patients with T-cell CAEBV
EBV has been reported in circulating T cells from patients with HIV infection in 1 study based on EBV PCR of sorted T cells26 and in patients with T-cell CAEBV.12 Excluding the patients with T-cell CAEBV, we detected EBV in T (CD3+) cells from 2 patients: 1 with HIV (patient 5) and 1 with Hodgkin lymphoma (patient 12) (Figure 3B; Table 1). Interestingly, in these patients, the EBV-positive T cells represented more than 25% of their EBV-positive cells. These cells contained a median of 7.85 genomes per cell, with a mean of 9 genomes per cell (range, 2-40). Sufficient cells were available from the patient with Hodgkin lymphoma to perform additional staining of CD4 and CD8 T cells; EBV was only detected in CD4+ T cells, each of which had more than 30 EBV genomes per cell (data not shown). EBV was not detected in T cells from the 7 other patients with HIV.
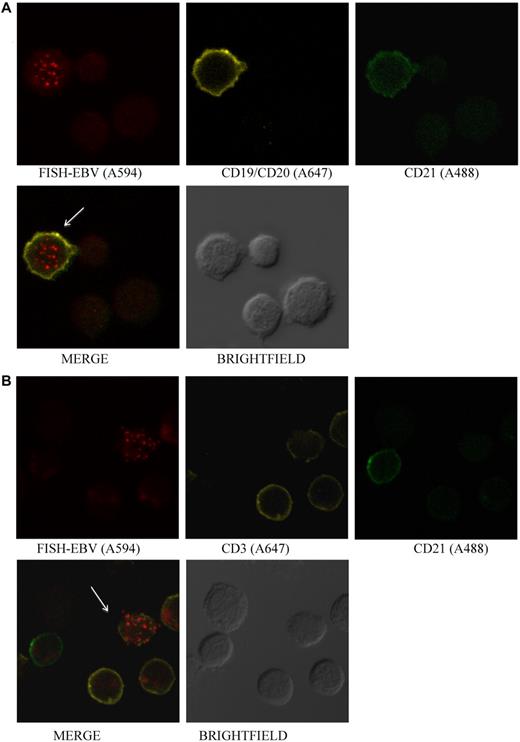
The EBV receptor in B cells is CD21,27 whereas in T cells, the virus receptor has not been identified. Although EBV was detected in CD21+ B cells from patient 12 (Figure 4A), none of the EBV-positive, CD3+ cells were also positive for CD21 (Figure 4B).
EBV is detected in CD21+ B cells and in CD21− T cells.(A) Photomicrographs of PBMCs stained by Immuno-FISH from patient 12. EBV is detected in B cells that are CD21+. (B) EBV is detected in CD3+ T cells that do not express CD21. EBV DNA was identified with the biotinylated FISH probe and streptavidin-conjugated Alexa 594, B cells using a combination of Alexa 647–conjugated anti-CD19 and anti-CD20 antibodies, T cells with Alexa 647–conjugated anti-CD3 antibody, and CD21 with Alexa 488–conjugated anti-CD21 antibody. Arrows indicate EBV-positive cells.
EBV is detected in CD21+ B cells and in CD21− T cells.(A) Photomicrographs of PBMCs stained by Immuno-FISH from patient 12. EBV is detected in B cells that are CD21+. (B) EBV is detected in CD3+ T cells that do not express CD21. EBV DNA was identified with the biotinylated FISH probe and streptavidin-conjugated Alexa 594, B cells using a combination of Alexa 647–conjugated anti-CD19 and anti-CD20 antibodies, T cells with Alexa 647–conjugated anti-CD3 antibody, and CD21 with Alexa 488–conjugated anti-CD21 antibody. Arrows indicate EBV-positive cells.
We also studied 4 patients with T-cell CAEBV from Japan (patients 24-27). Three of 4 patients showed EBV only in CD3+, CD4−, and CD8− cells (supplemental Figure 2A-B; Table 1). Although the staining for CD4 and CD8 is performed separately from staining for CD3, we can assume that the CD4−, CD8−, and EBV-positive cells (supplemental Figure 2B) are CD3+, because no EBV-positive B cells were detected in these patients. One patient had EBV in CD3+, CD8+, and CD4− cells (supplemental Figure 2C). The median number of EBV genomes in T cells for the T-cell CAEBV patients was 8.5 copies per cell, with a mean of 9.1 copies per cell (range, 1-26). Combining the results from all 6 patients in whom we detected EBV in T cells, the mean copy number of EBV genomes in T cells was lower than the mean copy number for B cells; however, the difference was not significant (paired t test, P = .77; Figure 2).
EBV is detected in circulating monocytes
EBV has been reported to infect circulating monocytes of patients with HIV.15 We detected EBV in monocytes (CD14+ cells) from 4 patients: 2 HIV-positive patients, 1 with Hodgkin lymphoma, and 1 receiving antithymocyte globulin for aplastic anemia (Figure 5A; Table 1). The median number of EBV genomes in monocytes for the 4 patients was 1.75 copies per cell, with a mean of 1.85 copies per cell (range, 1-3). This was significantly lower than the mean copy number for B cells (paired t test; P = .0083) (Figure 2).
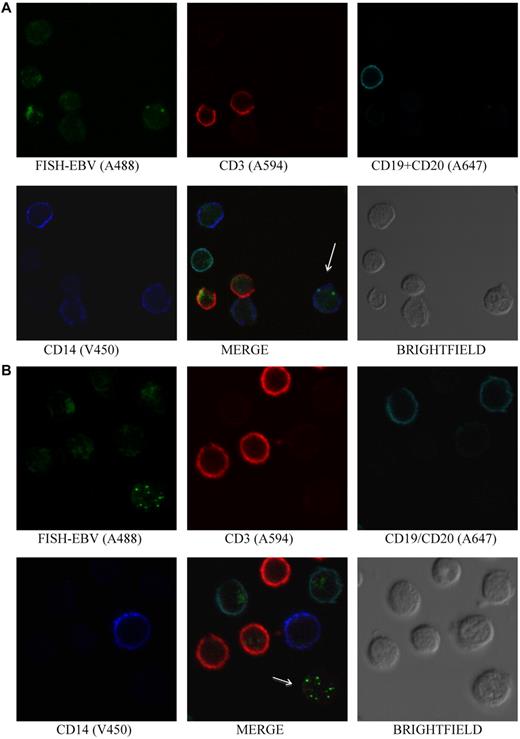
EBV is detected in monocytes and in non-B, non-T, and non-monocyte cells. (A) PBMCs stained by Immuno-FISH from patient 5 shows EBV in monocytes. EBV DNA was identified with the biotinylated FISH probe and streptavidin-conjugated Alexa 488, T cells using Alexa 594–conjugated anti-CD3, B cells using a combination of Alexa 647–conjugated anti-CD19 and anti-CD20 antibodies, and monocytes with V450-conjugated anti-CD14 antibody. Arrow indicates EBV-positive cell. (B) PBMCs stained by Immuno-FISH from patient 6 shows EBV in non-B, non-T, and non-monocyte cells. Arrow indicates EBV-positive cell.
EBV is detected in monocytes and in non-B, non-T, and non-monocyte cells. (A) PBMCs stained by Immuno-FISH from patient 5 shows EBV in monocytes. EBV DNA was identified with the biotinylated FISH probe and streptavidin-conjugated Alexa 488, T cells using Alexa 594–conjugated anti-CD3, B cells using a combination of Alexa 647–conjugated anti-CD19 and anti-CD20 antibodies, and monocytes with V450-conjugated anti-CD14 antibody. Arrow indicates EBV-positive cell. (B) PBMCs stained by Immuno-FISH from patient 6 shows EBV in non-B, non-T, and non-monocyte cells. Arrow indicates EBV-positive cell.
EBV is not detected in circulating NK cells in the patients in this study
EBV has been detected in circulating NK cells of patients with NK-cell CAEBV.12 When PBMCs from the patients were stained with a combination of antibodies to CD16 and CD56, NK cells (CD16+/CD56+ and CD3−) were detected in each of the patients; however, none contained EBV.
EBV is detected in circulating “non-B, non-T, and non-monocyte cells”
Surprisingly, in 69% of the patients, we detected EBV in PBMCs that were not recognized by B-cell (CD19, CD20), T-cell (CD3), or monocyte (CD14) monoclonal antibodies (Figure 5B; Table 1). Several patients, including some with HIV (patients 1, 4, 5, and 7), PTLD (patient 14), aplastic anemia receiving antithymocyte globulin (patients 18 and 20), and B-cell CAEBV (patients 21 and 23) had EBV in non-B, non-T, and non-monocyte cells that represented more than 20% of EBV-positive cells. EBV was detected in this cell population in each of the groups of patients we studied, except for patients with T-cell CAEBV from Japan and patients with infectious mononucleosis (IM). The median number of EBV genomes in non-B, non-T, non-monocyte cells for all the patients was 10.8 copies per cell, with a mean of 12.3 copies per cell (range, 2-34). Comparison of the mean copy number in EBV-positive B cells versus EBV-positive non-B, non-T, non-monocyte cells showed no significant difference (paired t test; P = .197; Figure 2).
EBV lytic protein BZLF1 is detected in circulating PBMCs
To determine whether we could detect evidence for lytic replication of EBV in virus-infected PBMCs, we performed immunofluorescence staining for EBV BZLF1 (an immediate-early protein that mediates the switch between the latent and the lytic forms of EBV infection), together with T- and B-cell markers (CD3 and CD19/CD20). Sufficient cells were available to assay 15 of the 29 patients (patients 8, 11, 12, 14, 15, 19-23, and 25-29). We detected BZLF1 in 87% (13 of 15) of these patients tested, indicating lytic EBV replication. The cell-surface phenotype of the BZLF1-positive cells was variable: in 4 patients, some of the BZLF1-positive cells were CD19+/CD20+, whereas other cells were negative for both CD19/CD20 and CD3 (patients 8, 14, 15, and 21); in 3 patients, some BZLF1-positive cells were CD3+, whereas others were negative for both CD19/CD20 and CD3 (patients 12, 26, and 27) (Figure 6). In 6 patients (patients 11, 19, 22, 23, 28, and 29), all of the BZLF1-positive cells were negative for CD3 and CD19/CD20.
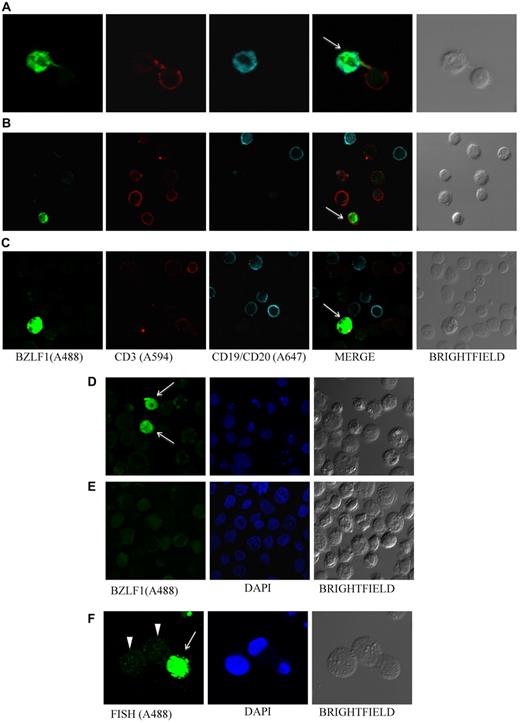
EBV lytic protein BZLF1 is detected in PBMCs. Photomicrographs of PBMCs stained for EBV BZLF1 shows virus lytic infection in B cells (patient 21, A), T cells (patient 12, B), and non-B, non-T cells (patient 26, C). BZLF1 was directly conjugated to Alexa 488. T cells were stained by using Alexa 594–conjugated anti-CD3, and B cells by using a combination of Alexa 647–conjugated anti-CD19 and anti-CD20 antibodies. BZLF1 staining of EBV-positive B95-8 (D) and EBV-negative BJAB (E) cells served as positive and negative controls, respectively, for EBV lytic infection. Arrows indicate BZLF1-positive cells. (F) B95-8 cells stimulated with butyric acid and TPA (phorbol 12-myristate 13-acetate) served as an additional control for lytic infection; cells undergoing lytic infection show a homogeneous pattern with high levels of EBV DNA (arrow), and latently infected cells show a punctate pattern (arrowheads).
EBV lytic protein BZLF1 is detected in PBMCs. Photomicrographs of PBMCs stained for EBV BZLF1 shows virus lytic infection in B cells (patient 21, A), T cells (patient 12, B), and non-B, non-T cells (patient 26, C). BZLF1 was directly conjugated to Alexa 488. T cells were stained by using Alexa 594–conjugated anti-CD3, and B cells by using a combination of Alexa 647–conjugated anti-CD19 and anti-CD20 antibodies. BZLF1 staining of EBV-positive B95-8 (D) and EBV-negative BJAB (E) cells served as positive and negative controls, respectively, for EBV lytic infection. Arrows indicate BZLF1-positive cells. (F) B95-8 cells stimulated with butyric acid and TPA (phorbol 12-myristate 13-acetate) served as an additional control for lytic infection; cells undergoing lytic infection show a homogeneous pattern with high levels of EBV DNA (arrow), and latently infected cells show a punctate pattern (arrowheads).
Correlation between EBV copy number per cell and underlying disease
We analyzed the distribution of EBV copy numbers per cell for each of the 29 patients (Tables 2–3). Half of the patients infected with HIV had cells carrying more than 30 EBV genomes per cell; however, high copy numbers per cell did not correlate with the EBV DNA load in the blood (supplemental Figure S3). Some patients with high EBV DNA loads (patients 7 and 10 with 22 000 and 30 000 EBV genome copies per million cells, respectively) had no cells with more than 30 EBV genome copies, whereas others with lower EBV DNA loads (patients 6 and 8 with 5100 and 6300 EBV genome copies per million cells, respectively) had cells with more than 30 EBV genome copies (Table 3). Moreover, the percentage of cells with more than 30 EBV genome copies in some patients with a lower EBV DNA load (patient 1 with 20% of cells with more than 30 EBV genomes and 12 000 copies of EBV per million cells) was higher than in patients with a high EBV DNA load (patient 4 with 8% of cells with more than 30 EBV genomes and 130 000 copies of EBV per million cells). Only one patient in our study, HIV patient 1, had a majority of EBV-positive cells with more than 20 EBV genomes per cell.
Distribution of the number of EBV genomes per cell
| Patient . | Diagnosis . | EBV DNA load/106 cells . | % of range of EBV genomes per cell . | Total EBV+ cells counted . | |||
|---|---|---|---|---|---|---|---|
| 1-10 . | 11-20 . | 21-30 . | > 30 . | ||||
| 1 | HIV | 12 000 | 10 | 30 | 40 | 20 | 10 |
| 2 | HIV | 4300 | 34 | 44 | 22 | 0 | 9 |
| 3 | HIV | 18 000 | 31 | 31 | 38 | 0 | 13 |
| 4 | HIV | 130 000 | 50 | 38 | 4 | 8 | 24 |
| 5 | HIV | 9400 | 75 | 25 | 0 | 0 | 4 |
| 6 | HIV | 5100 | 43 | 43 | 7 | 7 | 14 |
| 7 | HIV | 22 000 | 58 | 42 | 0 | 0 | 19 |
| 8 | HIV | 6300 | 25 | 38 | 25 | 12 | 16 |
| 9 | Tx | NA | 75 | 25 | 0 | 0 | 4 |
| 10 | Tx | 30 000 | 54 | 38 | 8 | 0 | 24 |
| 11 | Tx | 6200 | 40 | 40 | 20 | 0 | 10 |
| 12 | LyG/HL | 170 000 | 64 | 27 | 6 | 3 | 77 |
| 13 | Severe primary EBV infection | 280 000 | 27 | 54 | 11 | 8 | 37 |
| 14 | PTLD | 1 300 000 | 36 | 29 | 9 | 16 | 33 |
| 15 | ATL EBV-LPD | 190 000 | 48 | 44 | 4 | 4 | 25 |
| 16 | LyG | 28 000 | 70 | 25 | 0 | 5 | 20 |
| 17 | ICL | 110 000 | 45 | 31 | 21 | 3 | 38 |
| 18 | AA | 64 000 | 69 | 23 | 8 | 0 | 13 |
| 19 | AA | 110 000 | 46 | 21 | 25 | 8 | 24 |
| 20 | AA | 6 500 000 | 36 | 37 | 18 | 9 | 55 |
| 21 | B-cell CAEBV | 15 000 | 65 | 30 | 5 | 0 | 20 |
| 22 | B-cell CAEBV | 30 900 | 31 | 54 | 9 | 6 | 35 |
| 23 | B-cell CAEBV | 310 000 | 54 | 33 | 13 | 0 | 15 |
| 24 | T-cell CAEBV | 54 666/μg | 75 | 24 | 1 | 0 | 88 |
| 25 | T-cell CAEBV | 67 612/μg | 71 | 29 | 0 | 0 | 21 |
| 26 | T-cell CAEBV | 230 000/μg | 56 | 41 | 3 | 0 | 78 |
| 27 | T-cell CAEBV | 4154/μg | 54 | 39 | 7 | 0 | 18 |
| 28 | IM | 1700 | 65 | 29 | 6 | 0 | 17 |
| 29 | IM | 10 651 | 57 | 29 | 14 | 0 | 14 |
| Patient . | Diagnosis . | EBV DNA load/106 cells . | % of range of EBV genomes per cell . | Total EBV+ cells counted . | |||
|---|---|---|---|---|---|---|---|
| 1-10 . | 11-20 . | 21-30 . | > 30 . | ||||
| 1 | HIV | 12 000 | 10 | 30 | 40 | 20 | 10 |
| 2 | HIV | 4300 | 34 | 44 | 22 | 0 | 9 |
| 3 | HIV | 18 000 | 31 | 31 | 38 | 0 | 13 |
| 4 | HIV | 130 000 | 50 | 38 | 4 | 8 | 24 |
| 5 | HIV | 9400 | 75 | 25 | 0 | 0 | 4 |
| 6 | HIV | 5100 | 43 | 43 | 7 | 7 | 14 |
| 7 | HIV | 22 000 | 58 | 42 | 0 | 0 | 19 |
| 8 | HIV | 6300 | 25 | 38 | 25 | 12 | 16 |
| 9 | Tx | NA | 75 | 25 | 0 | 0 | 4 |
| 10 | Tx | 30 000 | 54 | 38 | 8 | 0 | 24 |
| 11 | Tx | 6200 | 40 | 40 | 20 | 0 | 10 |
| 12 | LyG/HL | 170 000 | 64 | 27 | 6 | 3 | 77 |
| 13 | Severe primary EBV infection | 280 000 | 27 | 54 | 11 | 8 | 37 |
| 14 | PTLD | 1 300 000 | 36 | 29 | 9 | 16 | 33 |
| 15 | ATL EBV-LPD | 190 000 | 48 | 44 | 4 | 4 | 25 |
| 16 | LyG | 28 000 | 70 | 25 | 0 | 5 | 20 |
| 17 | ICL | 110 000 | 45 | 31 | 21 | 3 | 38 |
| 18 | AA | 64 000 | 69 | 23 | 8 | 0 | 13 |
| 19 | AA | 110 000 | 46 | 21 | 25 | 8 | 24 |
| 20 | AA | 6 500 000 | 36 | 37 | 18 | 9 | 55 |
| 21 | B-cell CAEBV | 15 000 | 65 | 30 | 5 | 0 | 20 |
| 22 | B-cell CAEBV | 30 900 | 31 | 54 | 9 | 6 | 35 |
| 23 | B-cell CAEBV | 310 000 | 54 | 33 | 13 | 0 | 15 |
| 24 | T-cell CAEBV | 54 666/μg | 75 | 24 | 1 | 0 | 88 |
| 25 | T-cell CAEBV | 67 612/μg | 71 | 29 | 0 | 0 | 21 |
| 26 | T-cell CAEBV | 230 000/μg | 56 | 41 | 3 | 0 | 78 |
| 27 | T-cell CAEBV | 4154/μg | 54 | 39 | 7 | 0 | 18 |
| 28 | IM | 1700 | 65 | 29 | 6 | 0 | 17 |
| 29 | IM | 10 651 | 57 | 29 | 14 | 0 | 14 |
Only patients with HIV (patients 1, 3, and 8) and those receiving antithymocyte globulin for aplastic anemia (patients 19 and 20) had more than 25% of EBV-positive cells with more than 21 EBV genomes per cell.
Analysis of the 7 patients with B- or T-cell CAEBV showed that only one of these patients (patient 22) had more than 30 EBV genomes per cell. Despite the high EBV loads in these patients, 6 of 7 patients had a majority of EBV-positive cells with only 1-10 EBV genomes per cell. Thus, in these patients, the high EBV DNA load was caused by a large number of EBV-infected cells, rather than a high copy number per cell. Similarly, the 2 patients with IM also had a majority of EBV-positive cells with 10 or fewer EBV genomes per cell.
We also analyzed the distribution of the EBV genome copies per cell based on their phenotype. The mean number of EBV genome copies per cell for each of the different cell types showed no correlation with the total EBV DNA load in the blood (Figure 7). A more detailed analysis of the distribution of the number of EBV copies in B cells also showed no correlation with the EBV DNA load in the blood (Table 3; supplemental Figure S4). Taken together, these results suggest that patients with higher EBV DNA loads have a higher number of virus-infected cells, rather than a higher number of EBV genome copies per cell.
Correlation between the EBV DNA load in peripheral blood and the mean number of EBV DNA copies per cell for each cell subpopulation for each patient. Patient numbers are indicated at the bottom of the figure, and the vertical bars for each patient indicate the mean EBV DNA copy number per cell type.
Correlation between the EBV DNA load in peripheral blood and the mean number of EBV DNA copies per cell for each cell subpopulation for each patient. Patient numbers are indicated at the bottom of the figure, and the vertical bars for each patient indicate the mean EBV DNA copy number per cell type.
Discussion
Using the Immuno-FISH procedure, we analyzed 29 patients with different diseases that had high EBV DNA loads in the peripheral blood. All of the patients, except for the 4 T-cell CAEBV patients from Japan, were expected to have a B-cell EBV lymphoproliferative process. In all but 1 of these 25 patients (HIV patient 5), the majority of the EBV-positive cells were B cells; however, there was a wide range (52%-100% of the EBV-positive cells were B cells). Using this technique, EBV was detected in both B and non-B cells in 88% of these patients. Thus, the generally accepted belief that EBV is usually present only in B cells is not the case for patients with elevated EBV DNA loads.
Circulating plasmablast/plasma cells are detected in the blood of healthy persons, although they represent approximately 0.14% of PBMCs.28 Plasmablast/plasma cells have been detected in the blood in patients with virus infections,29 and these cells represent 5.5% of B cells in the peripheral blood of patients with HIV viremia.30 EBV has been detected in plasma cells in human tonsils; approximately 10%-20% of these cells undergo lytic virus replication and most do not complete the full cycle.31 We detected EBV in plasmablasts/plasma cells in the blood of approximately 50% of the patients. Therefore, in patients with lymphoproliferative diseases and high EBV DNA loads, EBV is present in circulating plasmablasts/plasma cells, in addition to B cells.
We detected EBV in peripheral blood T cells in a patient with HIV and in one with Hodgkin lymphoma, as well as in the 4 Japanese patients with T-cell CAEBV. Using cell sorting followed by PCR, EBV was found in circulating T cells in HIV-1–infected children and adolescents with elevated EBV DNA loads.26 EBV was found in CD4+ and CD8+ cells at approximately 10-fold lower EBV copy numbers than in CD19+ cells. In addition to T-cell CAEBV,12,13 EBV has been detected in T cells from patients with EBV-associated hemophagocytic lymphohistiocytosis32 and from patients with T-cell lymphoma.33,34 In contrast to the EBV-infected B cells, we were unable to detect CD21 on EBV-infected T cells. CD21 has been detected on the surface of T-cell lines35 and at low levels on primary T cells.36 Infection of a human T-lymphoblastic cell line resulted in virus internalization and viral gene expression.37,38 It is possible that EBV infected T cells using CD21 in our patients and down-regulated the expression of CD21; alternatively, EBV may have used a different receptor than CD21 to enter the T cells.
We detected EBV in monocytes from 4 patients. EBV infection of peripheral blood monocytes has been described in patients with HIV based on sorting of cells by flow cytometry and EBV DNA PCR.15 EBV has also been reported to infect monocytes in vitro and result in the activation of the viral lytic cycle.39,–41 Our observation that very few monocytes were infected by EBV, and that the viral copy number per cell was very low (ranging from 1 to 3 copies per cell), is consistent with the hypothesis that EBV infection of monocytes might be very transient with lytic replication, resulting in the rapid death of infected cells, or that monocytes have a shorter lifespan than B cells in the circulation.
We found that 69% (20 of 29) of the patients had EBV in circulating non-B, non-T, non-monocyte cells. These cells might represent one or more of several cell types. First, these non-B, non-T, non-monocyte cells might be plasmablasts/plasma cells. Approximately half (9 of 17) of the patients with EBV in non-B, non-T, non-monocyte cells had virus in plasmablasts/plasma cells (CD20−, positive for κ and λ immunoglobulin light chains). Therefore, some of the non-B, non-T, non-monocyte cells may have been plasmablasts/plasma cells that were not stained with antibody to κ and λ immunoglobulin light chains. These cells, however, have a very prominent cytoplasm (Figure 3A), and most of the non-B, non-T, non-monocyte cells did not have a prominent cytoplasm. Second, some of the non-B, non-T, non-monocyte cells might be B cells with very low levels of CD20 that are undetectable by antibody staining. This seems unlikely, based on the sensitivity of Immuno-FISH to detect B cells after sorting for B cells (supplemental Table 1). Furthermore, only 1 of the 29 patients (patient 21) had received anti-CD20 antibody (rituximab), and EBV was still detected in his B cells. Third, EBV could be present in cells other than T, B, NK cells, and monocytes. For example, EBV infects pre-Langerhans cells in the peripheral blood.16 However, we did not detect EBV DNA in pre-Langerhans cells (CD1a+ CD11c+, and CD14−) or in basophils (CD123+) in PBMCs from 4 patients who had EBV in non-B, non-T, non-monocyte cells (patients 12, 15, 16, and 20), using Immuno-FISH (data not shown). Infection of non-B, non-T, non-monocyte cells may be more common in immunocompromised persons. The only groups of patients for whom we did not detect EBV in these cells were patients with T-cell CAEBV and IM; the latter were otherwise healthy persons. In addition, the 1 HIV patient without EBV in non-B, non-T, and non-monocyte cells had the highest CD4 T-cell count (354 cells per μL); the other HIV patients all had CD4 T-cell counts less than 250 cells per μL.
We detected evidence of lytic EBV infection in nearly all of the patients for whom we performed immunofluorescence staining for BZLF1 protein. BZLF1 is the first viral protein expressed during the reactivation of EBV in B cells; it initiates a cascade of viral lytic gene expression, culminating with the death of the infected cells.42 It is interesting to note that although some of the BZLF1-positive cells were CD19/C20+, others were negative when stained with antibodies for B and T cells. These cells could be plasmablast/plasma cells (negative for CD19/CD20), because EBV-positive plasmablast/plasma cells were detected by Immuno-FISH in most of these patients. BZLF1 was previously detected in lymphocytes in tonsils of patients with IM; most of these cells were CD20− and VS38c+ with morphologic features indicative of plasma cells, whereas only rare cells were CD20+.43 The BZLF1 promoter in B cells is activated only after they differentiate into plasma cells.31 In the latter study, BZLF1 was detected almost exclusively in CD20lo plasma cells in human tonsils. Plasma cells in the bone marrow are CD20−, whereas plasma cells in tonsils express low levels of CD20.44
Immuno-FISH allows one to determine both the EBV genome copy number and phenotypic characterization of individual cells in the peripheral blood. This procedure combines the highly sensitive, specific FISH technique with multicolor immunofluorescence using monoclonal antibodies directly conjugated to Alexa dyes. These fluorochromes are very stable at high temperatures and low pH and therefore maintain their activity during the FISH procedure. Moreover, the use of directly conjugated antibodies obviates the problems of the cross-reacting secondary antibodies and permits the detection of multiple surface markers in the same slide.
Other studies have analyzed EBV copy numbers in various cell populations by sorting cells with monoclonal antibodies to cell surface markers and then performing PCR on the sorted populations. Although this technique has been useful,2,4 it has several limitations. First, the sensitivity has been estimated to be approximately 90%2 ; therefore, one cannot detect cell populations containing less than 10% of a given cell phenotype. Second, the specificity of cell sorting followed by PCR amplification has limitations; if very rare EBV PCR-positive cells are detected in sorted cells, this could be a result of a low level of EBV-positive B cells contaminating these sorted cells. Third, the number of EBV genomes per cell cannot be determined. Recently, combined cell-surface staining and in situ hybridization for EBER followed by flow cytometry has been used to identify EBV-positive T-cell subsets in the blood18 ; however, the number of the EBV genomes per cell could not be determined. Fourth, sorting cells requires a large number of cells and may not be feasible in patients with low numbers of PBMCs. Although labor intensive, the Immuno-FISH procedure has improved specificity, compared with the cell sorting and PCR procedure, and allows one to accurately measure the number of EBV genomes per cell. Using this procedure, we were able to determine the phenotype of EBV-infected cells with a specificity of more than 97% (supplemental Table 1; supplemental Figure 1).
Immuno-FISH might be used to better characterize the cell types infected with EBV and viral copy number of individual cells in the peripheral blood of immunocompromised patients. Immuno-FISH does not require tissue biopsies and therefore might be used to noninvasively monitor transplant patients or other immunocompromised patients who have high EBV DNA loads. Although an elevated EBV DNA load is often predictive of EBV PTLD in recipients of T cell–depleted allogeneic hematopoietic cell transplants, EBV DNA loads in non-T cell–depleted allogeneic hematopoietic cell and solid organ transplant recipients may remain persistently elevated without disease and often does not require treatment with potentially toxic agents5 ; currently, there is no noninvasive procedure to predict which of these patients with high EBV DNA loads would develop PTLD. Determination of the number of EBV genomes per cell and the subtype of B (or non-B) cells infected with the virus might provide additional prognostic information for the development of PTLD in patients with high EBV DNA loads. Immuno-FISH should be useful to identify EBV-infected cell populations in the blood of persons with CAEBV whose B, T, or NK cells are infected with the virus. Immuno-FISH might also be useful for studies of other virus infections in the blood.
In conclusion, using Immuno-FISH, we found that EBV is present in B cells as well as in plasmablasts/plasma cells and non-B, non-T, non-monocyte cells in the blood of patients with lymphoproliferative diseases. In patients with high EBV loads who are immunocompromised, the number of genomes per cell and the type of cells infected may provide additional prognostic information for the development of EBV lymphoproliferative diseases. Moreover, the identification of EBV-positive, CD20-negative cells in the blood may have important implications for rituximab therapy in these patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Owen Schwartz, Lily Koo, and Steven Becker for assistance with confocal microscopy; Mark Connor and Charles Brown for advice on the FISH procedure; Gary Fahle for measuring EBV DNA in the blood; Jing Qin for statistical analyses; Susan Moir and Pratip Chattopadhyay for advice on B cells and antibody staining; and Gregg Roby, Gilbu Uzel, and Wyndham Wilson for help in obtaining clinical samples.
This study was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, the National Heart, Lung, and Blood Institute, and the National Cancer Institute.
National Institutes of Health
Authorship
Contribution: S.C. performed the experiments, analyzed the data, and wrote the manuscript; I.S., P.S., H.K., and R.W.C. provided PBMC samples and critically reviewed the manuscript; and J.I.C. designed the study, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jeffrey I. Cohen, Laboratory of Infectious Diseases, National Institutes of Health, 50 South Dr, Building 50, Room 6134, Bethesda, MD 20892; e-mail: jcohen@niaid.nih.gov.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal