Abstract
The Polycomb group (PcG) of proteins is a major mechanism of epigenetic regulation that has been broadly linked to cancer. This system can repress gene expression by chromatin modification and is essential for establishing cell identity. PcG proteins are important for stem cell function and differentiation and have a profound impact during hematopoiesis. In recent years, several published studies have deepened our knowledge of the biology of the PcG in health and disease. In this article, we review the current understanding of the mechanisms of PcG-mediated repression and their relation to DNA methylation, and we discuss the role of the PcG system in hematopoiesis and hematologic malignancies. We suggest that alteration of different PcG members is a frequent event in leukemia and lymphomas that confers the stem cell properties on tumor cells. Thus, drugs targeting Polycomb complexes could be useful for treating patients with these diseases.
Introduction
In the past few years, evidence has built up concerning the critical function of the Polycomb group of proteins (PcG) and other epigenetic regulators for the establishment of cell identity, stem cell function, and differentiation. The PcG is a major mechanism of epigenetic regulation that has been broadly linked to cancer. Taking into account the increasingly extensive literature reflecting recent advances in this field and the abundant data on PcG protein alterations and functions, we review PcG functions in lymphopoiesis and hematopoietic stem cell function as well as their role in the initiation and maintenance of hematologic malignancies. Specifically, we also discuss the use of PcG genes as prognostic or therapeutic markers.
The histone code and epigenetic regulation by Polycomb proteins
Chromatin structure and modifications and, therefore, chromatin regulatory factors are involved in the control of gene expression affecting stemness, differentiation, and proliferation processes and, consequently, tumorigenesis. Among these factors, the PcG and trithorax group (trxG), together with histone demethylases, histone acetylases, and deacetylases, among others, are important sets of proteins that regulate gene activity at the chromatin level.
PcG and trxG proteins were first described in Drosophila,1 where they are responsible for maintaining homeotic gene activity in the appropriate segments during fly development. These systems act by silencing (PcG) or activating (TrxG) gene expression when they bind to specific regions of DNA. In Drosophila, most of the genes regulated by PcG contain consensus sequences called Polycomb-repressed elements (PREs).2 However, although some specific regions in the human genome have been shown to have a similar function than PREs,3 a general DNA sequence specific for Polycomb binding in mammals has not been identified.
PcG proteins form DNA-binding protein complexes, and although the composition of these is variable and dynamic,4,5 2 biochemical and major functional PcG complexes, termed Polycomb-repressive complexes (PRCs), have been described in mammals. One complex is composed of enhancer of zeste homolog 2 (EZH2), suppressor of zeste 12 homolog (SUZ12), and embryonic ectoderm development (EED), which form the nucleus of the PRC2 complex. EZH2 has histone methyltransferase activity and di- and trimethylates lysine 27 of histone H3 (H3K27me2 and H3K27me3). However, PRC2 composition is variable. In fact, some investigators have postulated the existence of PRC3 and PRC4, depending on the partners present in the complex.6,7 These include different EED isoforms, RbAp48, RbAp46, PHD finger protein 1 (PHF1), adipocyte enhances binding protein 2 (AEBP2), sirtuin 1 (SIRT1), histone deacetylase 1 (HDAC1), and HDAC2, among others. The patterns of expression of these various complexes depend on the differentiated status (eg, PRC4 is detected in undifferentiated, but not in differentiated, cells) or the normal/tumoral status of the cell (PRC4 is found in tumoral, but not in normal, differentiated cells).6
The second complex, PRC1, is even more heterogeneous than PRC2, but the core of the complex is formed by RING finger protein 1 (RING1), RNF2 (RING finger protein 2) BMI1 (B lym-phoma Moloney murine leukemia virus integration site 1), MEL18/PCGF2 (melanoma nuclear protein 18/polycomb group ring finger 2), polyhomeotic homolog 1 (PH), nervous system polycomb 1 (NSPC1), MEL18- and BMI1-like ring-finger protein (MBLR), and chromobox homolog (CBX) proteins (Table 1). PRC1 is believed to recognize the PRC2-methyl mark, H3K27me3, that is recruited to the DNA at the appropriate genomic locations. RNF2 in PRC1 complex has histone ubiquitination E3 ligase activity (H2K119ub),8 and this modification is also associated with gene repression.8,9 These complexes could have different compositions with distinct targets, depending on the cellular context, cell type, cell cycle, or differentiation stage, and on the association with other proteins (reviewed in Kerppola10 ). For example, although BMI1 and MEL18 are both homologous to Posterior sex combs (Psc) in Drosophila, their expression seems to be mutually exclusive and confers different specificities on the PRC1 complex.11-13
Common members of PRCs and their alterations in cancer
| Human . | Mouse homolog . | Alteration in cancer . | Tumor type . |
|---|---|---|---|
| PRC1 complex | |||
| BMI1 | Bmi1 | Overexpression/amplification | Acute myeloid leukemia,155 Hodgkin lymphoma,5,102,103,113 B cell non-Hodgkin lymphoma,5,54,112,126 gastrointestinal tumors,5 pituitary adenoma,5 parathyroid adenoma,6 breast cancer,156 glioma,157 neuroblastoma,158 medulloblastoma,159 non-small-cell lung cancer,160 skin cancer161 |
| MEL18/RNF110/PCGF2 | Mel18 | Loss | Cutaneous squamous-cell carcinoma,6 gynecological tumors,5 prostate cancer158 |
| RING1A | Ring1a | Overexpression | Large B-cell lymphoma126 |
| Loss | Clear-cell renal-cell carcinoma and testicular germ-cell tumors5 | ||
| RNF2 | Ring1b | Overexpression | Gastric and colonic tumors,5 melanoma,5 lung cancer,5 gynecological tumors,5 diffuse large B-cell lymphoma,5 follicular lymphoma,5 Burkitt lymphoma,5 Hodgkin lymphoma5,102 |
| HPH1, HPH2, HPH3 | Rae28/Mph1, Phc2, Phc3 | Lack of expression | Acute lymphocytic leukemia162 |
| Overexpression | Large B-cell lymphoma,156 central nervous system tumors6 | ||
| CBX2, CBX4, CBX6, CBX7, CBX8 | M33/Cbx2, Cbx4, Cbx6, Cbx7, Cbx8 | Loss | Pancreatic cancer,159 aggressive thyroid carcinomas160 |
| Overexpression | Follicular lymphoma163 | ||
| PCGF6 | Pcgf6 | Loss | Mantle-cell lymphoma164 |
| RYBP | Rybp | Overexpression | Oligodendroglioma,5 pituitary adenoma,5 T cell lymphoma,5 Hodgkin Lymphoma5,102 |
| PRC2 complex | |||
| EZH2/EZH1 | Ezh2/Ezh1 | Overexpression/amplification | Prostate carcinoma,109,165 breast carcinoma,127,165,166 melanoma,160 bladder cancer,127,167 glioma,168 endometrial carcinoma,165 lymphomas,111,127 colon carcinoma,127 glioblastoma,127 Ewing tumor169 |
| Deletion/mutation | Lymphomas,114 myeloid malignancies117,118 | ||
| Altered localization | Acute myeloid leukemia104 | ||
| SUZ12 | Suz12 | Overexpression/amplification/translocation | Gastrointestinal tumors,108,170 lung tumors,108 thyroid follicular carcinoma,108 pituitary adenoma,108 parathyroid adenoma,108 skin tumors,108 breast and gynecological tumors,108,170 mantle-cell lymphoma,108 chronic myeloid leukemia,119 endometrial stromal tumor119 |
| EED | Eed | Overexpression | Salivary gland adenoid cystic carcinoma,171 Hodgkin lymphoma,103 prostate cancer6 |
| Lack of expression | Prostate cancer172 | ||
| RBBP7 | Rbbp7 | Overexpression | Non-small-cell lung cancer,173 breast cancer174 |
| RBBP4 | Rbbp4 | Down-regulation | Cervical cancer175 |
| PCL3 | Phf19 | Overexpression | Cancers of the colon, skin, lung, rectum, cervix, uterus, and liver 176 |
| Human . | Mouse homolog . | Alteration in cancer . | Tumor type . |
|---|---|---|---|
| PRC1 complex | |||
| BMI1 | Bmi1 | Overexpression/amplification | Acute myeloid leukemia,155 Hodgkin lymphoma,5,102,103,113 B cell non-Hodgkin lymphoma,5,54,112,126 gastrointestinal tumors,5 pituitary adenoma,5 parathyroid adenoma,6 breast cancer,156 glioma,157 neuroblastoma,158 medulloblastoma,159 non-small-cell lung cancer,160 skin cancer161 |
| MEL18/RNF110/PCGF2 | Mel18 | Loss | Cutaneous squamous-cell carcinoma,6 gynecological tumors,5 prostate cancer158 |
| RING1A | Ring1a | Overexpression | Large B-cell lymphoma126 |
| Loss | Clear-cell renal-cell carcinoma and testicular germ-cell tumors5 | ||
| RNF2 | Ring1b | Overexpression | Gastric and colonic tumors,5 melanoma,5 lung cancer,5 gynecological tumors,5 diffuse large B-cell lymphoma,5 follicular lymphoma,5 Burkitt lymphoma,5 Hodgkin lymphoma5,102 |
| HPH1, HPH2, HPH3 | Rae28/Mph1, Phc2, Phc3 | Lack of expression | Acute lymphocytic leukemia162 |
| Overexpression | Large B-cell lymphoma,156 central nervous system tumors6 | ||
| CBX2, CBX4, CBX6, CBX7, CBX8 | M33/Cbx2, Cbx4, Cbx6, Cbx7, Cbx8 | Loss | Pancreatic cancer,159 aggressive thyroid carcinomas160 |
| Overexpression | Follicular lymphoma163 | ||
| PCGF6 | Pcgf6 | Loss | Mantle-cell lymphoma164 |
| RYBP | Rybp | Overexpression | Oligodendroglioma,5 pituitary adenoma,5 T cell lymphoma,5 Hodgkin Lymphoma5,102 |
| PRC2 complex | |||
| EZH2/EZH1 | Ezh2/Ezh1 | Overexpression/amplification | Prostate carcinoma,109,165 breast carcinoma,127,165,166 melanoma,160 bladder cancer,127,167 glioma,168 endometrial carcinoma,165 lymphomas,111,127 colon carcinoma,127 glioblastoma,127 Ewing tumor169 |
| Deletion/mutation | Lymphomas,114 myeloid malignancies117,118 | ||
| Altered localization | Acute myeloid leukemia104 | ||
| SUZ12 | Suz12 | Overexpression/amplification/translocation | Gastrointestinal tumors,108,170 lung tumors,108 thyroid follicular carcinoma,108 pituitary adenoma,108 parathyroid adenoma,108 skin tumors,108 breast and gynecological tumors,108,170 mantle-cell lymphoma,108 chronic myeloid leukemia,119 endometrial stromal tumor119 |
| EED | Eed | Overexpression | Salivary gland adenoid cystic carcinoma,171 Hodgkin lymphoma,103 prostate cancer6 |
| Lack of expression | Prostate cancer172 | ||
| RBBP7 | Rbbp7 | Overexpression | Non-small-cell lung cancer,173 breast cancer174 |
| RBBP4 | Rbbp4 | Down-regulation | Cervical cancer175 |
| PCL3 | Phf19 | Overexpression | Cancers of the colon, skin, lung, rectum, cervix, uterus, and liver 176 |
Bold text indicates members known to be frequently altered in cancer.
In embryonic stem (ES) cells, which have been the subject of most of the studies, a high level of cooccupancy among different PcG proteins (eg, EED, SUZ12, Ring1B, and Ph1) has been described in H3K27me3 promoters. The presence of PRC1 and PRC2 is interrelated, and, as previously mentioned, H3K27me3 has been described as the epigenetic mark that recruits PRC1 to specific promoters.14-16 PRC1 and PRC2 then provoke changes in chromatin structure,17,18 and/or the ubiquitin ligase activity of Ring1B contributes to gene silencing. However, independence from this mark has also been proposed for PRC1 recruitment.19,20 Therefore, the mechanisms for gene repression mediated by PcG proteins depend on PRC1 and PRC2, alone or in association (for a recent review, see Simon and Kingston21 ). These epigenetic modifications of histone proteins and chromatin conformational changes seem to be the main way by which PcG mediates gene silencing. An interrelationship between PcG silencing and DNA hypermethylation has also been suggested for EZH222,23 and has been proposed as being critical to transcriptional silencing. PRC2 can recruit DNMTs to PcG target genes, leading to de novo methylation in these loci. BMI1 is associated with DNMT1 and DMAP1 (DNMT1-associated protein).24 Other studies showing association between PcG targets and methylated cytosine guanine dinucleotide (CpG) islands corroborate the relationship between PcG and DNA methylation25 (see below). However, other authors report the presence of PcG marks (H3K27me3) outside CpG islands in tumoral cells,26 suggesting that there is a mechanism of gene silencing that is independent of promoter DNA methylation in cancer.
Once bound to chromatin, PcG complexes can interfere with other systems. They can inhibit ATP-dependent chromatin remodeling by SWI/SNF,27 interact with components of the transcriptional machinery, as is suggested by the presence of TFIID in the PRC1 complex,28 or block RNA polymerase II association with the target promoter, leading to the inhibition of transcriptional initiation and elongation.29
As mentioned above, the DNA sequences specific for Polycomb binding have not been identified in mammals. Therefore, the question remains as to what mechanisms direct PcG proteins to target promoters in vertebrates. One of those proposed is mediated by PcG-interacting proteins that provide the sequence specificity for PcG binding to target promoters, such as transcription regulators with DNA-binding capacity. This has been described for E2F6, a member of the E2F transcription factor protein family, but unlike other E2F members, this behaves as a transcriptional repressor by interacting with Ring1a and Ring1b, together with YAF2 and DP1.30 BCOR (BCL-6 interacting corepressor), a POZ that is required for germinal center formation,31 is also associated with RING1, RNF2, and Ring1 and YY1 binding protein (RYBP) PcG proteins, among others.32,33 This formation of the BCOR complex with PcG proteins could explain some of the enzymatic activities that can be recruited by BCL6 and, also, how BCL6 may determine some of the targets of PcG proteins. Other transcription regulators, such as OCT4, NANOG, and SOX, interact with PcG proteins from both PRC2 (SUZ12)34 and PRC1 complexes (RNF2)35 in ES cells. These factors could repress differentiation-promoting genes by recruiting PcG proteins to their promoters (see below).34,36
Gene-specific recruitment by ncRNA14,37-39 has been implicated in PcG target repression and specificity. Recently, Kanhere et al40 showed that short RNAs transcribed from Polycomb target genes by RNApolII interact with PRC2, forming a stem-loop structure stabilizing the association of PRC2 with chromatin through SUZ12 and, consequently, repressing the gene expression in cis.
Long ncRNA, such as HOTAIR (Hox transcript antisense intergenic RNA, encoded in the HOXC cluster) interacts with PRC2 and is required for SUZ12 recruitment and trimethylation of H3K27 in HOXD loci.37 Something similar happens with Kcnq1ot1, an ncRNA situated in the intronic region of the Kcnq1 gene. Kcnq1ot1 interacts with chromatin and PRC2 complex in a lineage-specific manner (it seems to function in placenta, but not in fetal liver),38 and this correlates with H3K27me3 enrichment at their target promoters (for review, see Hekimoglu and Ringrose41 ). This mechanism is similar to that producing X-chromosome inactivation and genomic imprinting, mediated by Xist, which targets PRC2 to the inactive X-chromosome (for review, see Chandrasekhar et al42 ).
RNAi machinery can play a major role in transcriptional gene silencing by targeting promoter sequences, among other mechanisms, and this has been associated with PcG silencing. In Drosophila, Argonaute-1 (AGO1; the effector protein in the complexes involved in the RNAi mechanism) and other RNAi components colocalize with PcG bodies, suggesting a role for the RNAi machinery in the chromatin organization of PcG targets.39 In humans, AGO1 is colocalized with EZH2 and TRBP2 proteins in some siRNA-targeted promoters, such as RASFF1A.43
Polycomb functions
A growing number of studies reflect the relevance of PcG in stem cell function, differentiation, development, and cell-cycle control, and consequently in cancer. It has also been implicated in cellular senescence, X-inactivation, genomic imprinting, and actin polymerization (for review, see Rajasekhar and Begemann44 ).
The role of BMI1 in self-renewal of hematopoietic stem cells (HSCs)45 and the fact that CDKN2, a cell-cycle regulator, is the paradigm of polycomb targets, both indicate the involvement of PcG in cell proliferation. There are several lines of evidence for the role of PcG proteins in controlling cell proliferation. Several cell-cycle regulators (both activators and repressors) are found among PcG targets, such as inhibitor of kinase 4/alternate reading frame (INK4a/ARF), MYC, JUN, FOS, CDC25, human telomerase reverse transcriptase, cyclins, cyclin-dependent kinases (CDKs), checkpoint kinase 1 homolog (CHEK1), mitotic arrest deficient–like 1 (MAD2L1), and budding uninhibited by benzimidazoles 3 (BUB3).46-48 Recent studies in Drosophila show that PcG can also regulate cell growth by signaling pathways, such as JAK/STAT.49,50 Changes occur in the expression levels of PcG proteins during hematopoietic differentiation, at least, in part, through the regulation of Hox genes, whose dysregulation leads to alterations in lymphocyte proliferation.51-53 These alterations of PcG members occur in a wide variety of cancers (described below), highlighting the role in controlling cellular proliferation, whereby some PcG genes act as oncogenes (BMI15,54 ), and others function as tumor suppressor genes (MEL18)5,55 (reviewed in Martinez and Cavalli56 ). A special case is Rnf2, which has been shown to restrict proliferation of early myeloid progenitors through the inhibition of cyclin D2 and Cdc6 and, at the same time, to promote the expansion of maturing cell lineage–committed precursors of the myeloid and lymphoid compartments. Moreover, Rnf2 deficiency accelerates lymphomagenesis in the absence of Ink4a.57
Studies of various species have demonstrated the importance of the PcG system not only in embryonic development, but also in adult differentiation and homeostasis.58 PcG regulates genes involved in development and differentiation pathways and are critical regulators during embryogenesis, as is demonstrated by the fact that disruption of several PcG genes is embryonic lethal or leads to postnatal lethality.51-53,59,60
In ES cells, PRC2, and PRC1 are both present in the promoter region of key development regulators. These development regulator genes in mammalian stem cells have 2 epigenetic marks: H3K27me3, a signal of gene silencing, and H3K4me, a mark of gene activation associated with TrxG.61 These 2 marks, with their opposite functions in the same region, result in a bivalent promoter with both negative and positive possibilities and are thought to be important for the repressive or activating function in stem cells, to switch gene expression upon changes in cell status. These bivalent marks have also been found in nonstem cells, such as T cells and human lung fibroblasts,62-65 indicating that in more highly differentiated cells, the dual PcG/TrxG system also enabled fine-scale control of the expression of certain genes with bivalent domains in response to external or internal signals.66,67
It is worth mentioning that ES cells can be generated in the absence of H3K27me3.68 Thus, H3K27me3 seems to mark genes that need to be activated during differentiation, rather than having a direct role on pluripotency at the bivalent loci. As proposed by Bernstein and colleagues,69 during ES cell differentiation, the abundance of bivalent promoters might be correlated with differentiation. However, the possibility that other histone modifications may have a bigger impact in these domains cannot be ruled out.
The localization and expression patterns of PcG genes change during the stages of differentiation.6,12 PcG proteins are displaced from one set of target genes while being directed to another during lineage specification (see below), but the mechanisms are not well understood. Genome-wide mapping by chromatin immunoprecipitation (ChIP) experiments of PcG members in human and murine ES cells has shown that PcG proteins bind and repress several genes involved in the regulation of development: transcription factors involved in regulating early stages in neurogenesis or hematopoiesis (eg, Pax, Lhx) and members of the Sox, Tbx, and Gata families.34,36 Therefore, there is a prevalence of developmental regulators and genes involved in cell-fate decisions among PcG target genes.34,36,70
A large proportion of PcG targets in ES cells are also occupied by the transcription factors, OCT4, SOX2, and NANOG, which are known to be essential for stemness and also indicate that PcG is involved in this process.71 These factors could repress differentiation-promoting genes by recruiting PcG genes to their promoters, thereby maintaining the stem cell capacity of these cells.34,71 PcG complexes are displaced from their target promoters when cells are committed to differentiation.
The role of Polycomb in HSCs
All blood cells derive from a common undifferentiated progenitor, which undergoes subsequent differentiation through a series of binary decisions; the cell must be able both to self-renew and to differentiate into any of the hematopoietic cell lineages. The function of HSCs is regulated by both extrinsic and intrinsic signals. Several systems that are active during embryonic development and act extrinsically on HSCs, such as the Sonic Hedgehog, Notch, and Wnt pathways,76,77 have been shown to induce self-renewal in adults. However, very few effectors are known that are downstream of these external signals. Intrinsic effectors important for HSCs include the HOX genes, Hoxa5, Hoxa9, Hoxa10, Hoxb3, Hoxb4, and Hoxb6.78 For instance, Hoxb4 is a transcription factor that, when overexpressed in HSCs, leads to the expansion of this cellular subset.79 It also seems to protect HSCs from multiple extrinsic signals, such as tumor necrosis factor-α (TNF-α).80 Strikingly, the development of knockout mice for Hoxb4 has shown this gene not to be required for the generation of HSCs or the maintenance of steady-state hematopoiesis. Hoxb4−/− HSCs show only mild defects in proliferation. The conclusion is that either Hoxb4 promotes proliferation of HSC, but not self-renewal, or that Hoxb4 deficiency can be compensated for by neighboring or paralogous Hox genes.81,82 In any case, there must be other intrinsic effectors that are essential to the function and survival of HSCs.
PcG genes play a major role in regulating hematopoietic function.11,45,83-85 It has recently been demonstrated that the levels of BMI1 and MEL18 determine, to a great extent, the capability of HSCs to function as progenitors,13 whereby BMI1 is associated with the enhancement of HSC properties and MEL18 is more closely related to the differentiated phenotypes. These data are consistent with the findings of a previous report showing that BMI1 is expressed in primitive human bone marrow cells, while many other components, such as MEL18, RAE28, and EZH2,12 are increasingly expressed upon differentiation. Moreover, Bmi1-deficient mice have severely impaired HSC self-renewal, and bone marrow progenitors lacking Bmi1 have a restricted proliferative potential,45,52,86 while forced expression of Bmi1 leads to enhanced lymphoproliferation, promotion of HSC self-renewal, and a higher probability of inheriting stemness through cell division.87
Until now, the main mechanism known to regulate HSC capacities by Bmi1 is that acting by regulating the Ink4a/Arf locus. Bmi1−/− HSCs have markedly higher levels of Ink4a and Arf,46 and forced expression of the latter causes a decrease in HSC compartment due to the induction of cell-cycle arrest and p53-mediated apoptosis.45 Because the deletion of both genes partially restores the proliferative capacity of HSCs in a Bmi1−/− background, it seems that Bmi1 protects HSCs from premature loss through the inhibition of Ink4a and Arf,88 although other downstream pathways must mediate the effect of Bmi1 on proliferation.
Alterations in other PcG components also affect hematopoietic function. Overexpression of Ezh2 confers long-term repopulating potential on HSCs, preventing its exhaustion after replicative stress.89 Expression of one null eed allele or 2 hypomorphic eed alleles results in greater lymphoproliferation, developmental blockade during thymocyte differentiation, and a greater risk of developing hematologic tumors.90 On the other hand, targeted deletion of Mel-18 causes severe defects in lymphoid organs, including hypoplasia, while, in bone marrow, most hematopoietic cells are replaced by adipocytes.51 However, the defects in Mel-18−/− mice seem to be due to the impaired response to cytokines, because down-regulation of Mel-18 in bone marrow–derived HSCs promotes their self-renewal, while its forced expression reduces self-renewal capacity of HSCs.11 Similar to Bmi1 and Mel-18, M33-deficient mice also show hypoplasia in the spleen and thymus, and defects in T and B cells.53 Mph1/Rae-28 is another important component of the PcG system that affects proper hematopoietic function. Mph1/Rae-28 mutant embryos die because HSC activity in these animals cannot maintain the hematopoietic system during embryo development and show a progressive impairment in the numbers and proliferative capacity of colony-forming cells, compared with their wild-type littermates. Moreover, HSCs lacking the Mph1/Rae-28 gene cannot reconstitute the bone marrow after the irradiation of mice.91 Likewise, deletion of Rnf2 in the HSC compartment affects HSC function. Rnf2 is able to restrict the proliferation of progenitor cells while promoting the expansion of their maturing progeny, having a dual role in this system.57 All these data suggest a complex regulation of HSC function, based on a delicate equilibrium between enhancing and repressing PcG complexes that, when altered, induces mainly a differentiation blockade and/or impairment of the HSC function.
Polycomb and hematologic malignancies
Human tumors are characterized by a broad spectrum of genetic alterations. It is well documented that progression from normal to tumoral cells also involves epigenetic changes, including extensive DNA methylation at promoter-associated CpG islands, and an aberrant pattern of histone modifications. Chromatin structure is crucial for the regulation of DNA accessibility and thus for the regulation of gene expression. Furthermore, an altered chromatin structure provokes altered gene patterns and genomic instability that can be propagated to daughter cells, causing cellular transformation to a malignant status.92
There is increasing evidence that many tumors depend on the presence of a subpopulation of cancer stem cells (CSCs) with a genetic program that partially resembles that of normal stem cells. In fact, several signaling pathways that are important for tumor development, such as Sonic Hedgehog, Notch, and Wnt, also regulate self-renewal in stem cells (reviewed in Taipale and Beachy93 ), and some embryonic genes are reexpressed in human cancers.94 Several studies have demonstrated the presence of CSCs in tumors, such as acute myeloid leukemia (AML), breast cancer, glioblastoma, and others (for review, see Gupta et al95 ). This is accompanied by a stem cell–like signature at several levels, including gene expression96 and chromatin structure.97
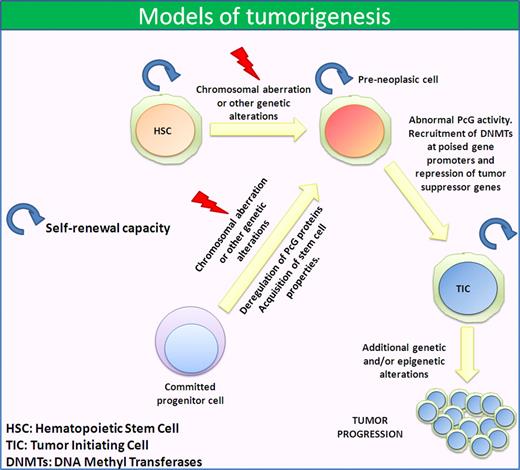
There are several ways in which CSCs can be generated. Tumor-initiating cells with stem cell properties could arise from a stem cell that loses the capacity to regulate its mitotic potential, or a downstream progenitor (or committed progenitor) cell could transiently acquire the ability to self-renew through some molecular alterations, possibly induced by the microenvironment (Figure 1). Both possibilities can be integrated in a single hypothesis through the demonstrated cell plasticity that cancer cells harbor, as suggested by a recent report on melanoma.98
Models of tumorigenesis. Cancer cells can originate from a cell with stem cell properties, such as an HSC, that acquire an altered phenotype through chromosomal aberrations and/or mutations. This leads to an abnormal PcG activity that represses several tumor-suppressor genes promoting cancer development. Another possibility is that the first chromosomal aberration and/or mutation occurs in a committed progenitor cell. In this case, the abnormal PcG activity confers stem cell properties and self-renewal capacity as well as tumorigenic potential.
Models of tumorigenesis. Cancer cells can originate from a cell with stem cell properties, such as an HSC, that acquire an altered phenotype through chromosomal aberrations and/or mutations. This leads to an abnormal PcG activity that represses several tumor-suppressor genes promoting cancer development. Another possibility is that the first chromosomal aberration and/or mutation occurs in a committed progenitor cell. In this case, the abnormal PcG activity confers stem cell properties and self-renewal capacity as well as tumorigenic potential.
PcG proteins have been identified as being important proteins in tumorigenesis due to their potential to repress tumor suppressor genes and regulate genes related to stemness and differentiation.19,34,36,45,70,72,91 Thus, genetic phenomena that alter the expression of PcG members such as BMI1 could be one of the key events that allow the malignant cell to acquire a stem cell phenotype.
Among the most important genes that confer transforming capacity to the cell, Cdkn2a is one of the main targets of Bmi1. In fact, Bmi1 has been identified as an important factor cooperating with Myc-induced lymphomagenesis,99,100 and Bmi1 knockdown has been shown to promote cancer-specific cell death in neuroblastoma.101 However, in some tumor types, such as Hodgkin lymphoma (HL), the regulation of CDKN2A by BMI1 might be disrupted by other mechanisms.102,103 A recent report has found the presence of EZH2 and the H3K27me3 polycomb repressive mark not only at the CDKN2A locus but also at the CDKN2B locus in AML cell lines and patient samples.104 The regulation of the Cdkn2a locus is a mechanism shared by another PcG protein, Cbx7, which is critical for expanding the cellular lifespan and for bypassing senescence by inhibiting the expression of Ink4a/Arf. In contrast to Bmi1, however, Cbx7 is unable to induce telomerase activity, although it still can initiate T-cell lymphomagenesis in mouse models.105,106 Other oncogenic mechanisms have been ascribed to BMI1, such as the inhibition of phosphatase and tensin homolog.107 Furthermore, the effects of many other PcG members are independent of Cdkn2a and are much more diverse.
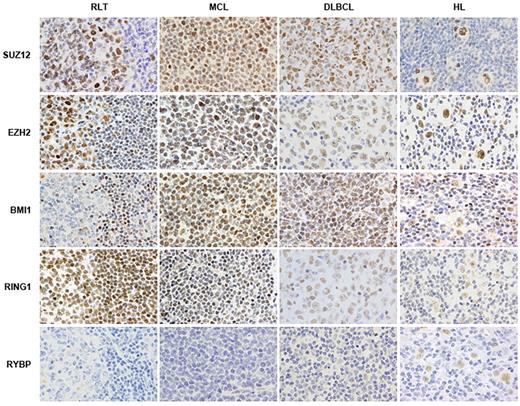
Several studies have shown an extensive relationship between alterations in different PcG members and cancers originating from various tissues (Table 1). Our group and others have found overexpression/down-regulation of PcG members in many tumors, including several lymphomas5,102,108 (Figure 2). Many solid tumors also present alterations in members of the Polycomb system. Prostate cancer is an important example of this, in which EZH2 levels are correlated with aggressiveness of the disease in a mechanism independent of DNA methylation.109 The cancer types studied are diverse, but particular attention has been paid to hematologic malignancies. Several observations support the role of PcG genes in abnormal hematopoiesis through the regulation of HSC self-renewal/proliferation. BMI1 up-regulation is associated with leukemia and mantle cell lymphoma (MCL) and is linked to bad prognosis, and its locus is found to be amplified in MCL.5,54 Furthermore, hematopoietic progenitors of Bmi1-deficient mice are resistant to transformation by the E2a-Pbx1 fusion gene that is frequently present in human acute pre-B lymphoblastic leukemias.110 Bmi1 also seems to be important for the maintenance of the leukemic phenotype (or for the maintenance of leukemia) initiated by the Hoxa9 and Meis1a collaborative oncogenes in a model of AML.86 EZH2 is also up-regulated in MCL111 and anomalously coexpressed with BMI1 in several non-HLs, including small lymphocytic lymphoma, follicular lymphoma (FL), diffuse large B-cell lymphoma (DLBCL), and Burkitt lymphoma (BL),112 as well as in HL itself103,113 (Figure 2). EZH2 is also a target of other genetic abnormalities. For instance, mutations have been described in the catalytic domain of EZH2 in FL and DLBCL of germinal center origin.114 This mutation leads to a reduction of the in vitro enzymatic activity of EZH2, suggesting that, at least in DLBCL and FL of germinal center origin, the mechanism by which EZH2 collaborates to produce malignant transformation is different from those of other types of cancer, such as prostate cancer. Although we do not know for certain, it is possible that the mutations in EZH2 alter not only its enzymatic activity, but also the product or target specificity of the complex. This alteration might be especially important for germinal center–derived B cells, because EZH2 is necessary in these cells for early B-cell development, including rearrangement of the immunoglobulin heavy-chain (IGH) locus.115 Furthermore, in AML, EZH2 seems to be aberrantly localized in the genome,104 and the chromosomal location of EZH2 is a hotspot of genomic aberrations present in myeloid disorders.116 Strikingly, 2 recent studies reported genetic aberrations affecting EZH2, including deletions, mutations, and uniparental disomy, in myeloid malignancies.117,118 All the studied alterations result in the inactivation of EZH2 and loss of H3K27 trimethylation, suggesting that EZH2 may have a dual role as an oncogene or tumor-suppressor gene, depending on cell context. SUZ12 is also the target of genomic aberrations and has been found to be translocated in endometrial stromal tumors.119 The fusion protein is able to restore H3K27me3 after SUZ12 knockdown, inhibits apoptosis, and promotes cell proliferation.120 Our laboratory has shown that SUZ12 is overexpressed in several germinal center (GC)–derived lymphomas, although normal GCs also express SUZ12.108 On the other hand, we found that SUZ12 is also overexpressed in MCL, while the normal mantle zone cells are negative for SUZ12 expression.108 In some cases, this overexpression was accompanied by genetic gains and/or amplifications. The overexpression of SUZ12 seems to be relevant for the tumoral cells, because silencing of SUZ12 in MCL-derived cell lines induces the reexpression of several SUZ12 target genes and increases apoptosis.108 Likewise, the knockdown of SUZ12 in acute promyelocytic leukemia (APL)–derived cells is able to induce granulocytic differentiation. In this leukemia, the fusion gene PML-RARα interacts with the PRC2 complex and directs it to specific target genes121 ; this is a good example of how targeting Polycomb complexes by an oncogene can be of great relevance in tumorigenesis.122 Another example is the WNT pathway, which is altered in several malignancies and constitutively active in chronic myeloid leukemia (CML). In this tumor, during the blastic crisis of the disease, SUZ12 is overexpressed due to the activation of its promoter by some members of the WNT pathway, such as WNT5A and WNT11.123
Immunohistochemical staining for PcG members reveals frequent alterations of these proteins in hematologic malignancies. RLT: reactive lymphoid tissue; MCL: mantle cell lymphoma; DLBCL: diffuse large B-cell lymphoma; HL: Hodgkin lymphoma.
Immunohistochemical staining for PcG members reveals frequent alterations of these proteins in hematologic malignancies. RLT: reactive lymphoid tissue; MCL: mantle cell lymphoma; DLBCL: diffuse large B-cell lymphoma; HL: Hodgkin lymphoma.
EED, another component of the PRC2, exerts a protective influence upon several lymphomagenesis inducers, such as carcinogens, Moloney murine leukemia virus, and irradiation.90,124,125 Intriguingly, mice with a hypomorphic Eed allele show a blockade during thymocyte differentiation at the β-selection checkpoint. These results support the notion that many hematologic malignancies are derived from not fully differentiated cells or cells with either stem cell or progenitor cell properties.
In some lymphomas, such as HL and DLBCL, the expression pattern of PcG members has been studied extensively. In fact, HL is characterized by a unique pattern of expression of PcG proteins, including coexpression of BMI1, MEL18, RING1, human PH1, HPC1, EED, and EZH2.102,103 Moreover, overexpression of RYBP, a PcG member found in Ring1A/Ring1B-containing complexes, is associated with poorer prognosis in HL patients.102 DLBCL samples also overexpress several members of the PRC1 complex, including BMI1, RING1, and HPH1.126
Regulation of PcG in cancer
Direct alteration of PcG members is not the only phenomenon in cancer that affects PcG function. Other PcG-related systems may be altered in cancer. The E2F/Rb pathway, which is frequently deregulated in cancer, may account for the frequent up-regulation of several PcG members, because E2F transcription factors bind and activate many PcG genes.127 This relationship implies that some PcG members are regulated during cell cycle, but there is also evidence that PcG regulates cell cycle.46,90,128-130
MicroRNAs are another type of PcG regulator with aberrations in cancer, and the down-regulation of several of these is a general hallmark of cancer. MiR-101 and miR-26a can down-regulate EZH2 and are known to be down-regulated in bladder cancer and BL, respectively,131,132 linking other epigenetic systems to PcG and cancer. In the case of miR-26a, it has been shown that MYC, which is frequently translocated in BL, can repress miR-26a, leading to the up-regulation of EZH2. Other microRNAs, such as miR-137 and miR-214, regulate differentiation through the control of Ezh2 protein levels.133,134 Bmi1 has also been shown to be regulated by microRNAs. In fact, the down-regulation of miR-200c, miR-203, and miR-183 seems to be important to ensure the expression of Bmi1 in cancer cells and mouse embryonic stem cells.135,136 However, the relationship between Polycomb and microRNAs is even more profound. We and others have found that Polycomb complexes also target several microRNA promoters in cancer and during embryogenesis.108,137,138 Moreover, in recent years, it has been suggested that some microRNAs, such as miR-320, can direct gene silencing through the interaction between AGO1 and Polycomb complexes.139
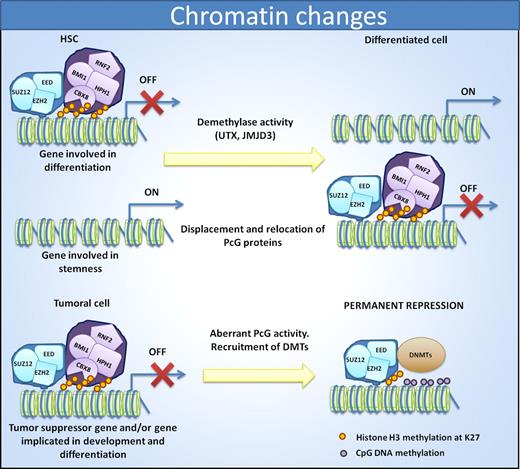
There are some clues that highlight the importance of PcG in cancer even in the absence of any obvious alteration of the PcG components. In recent years, it has been demonstrated that among the genes that suffer anomalous repression in cancer, there is significant enrichment in those targeted by PcG in ES cells.23,140-146 In fact, some investigators have proposed the existence of a Polycomb repression signature in cancer. This signature is constructed on the basis of the identification of PRC2 cancer-occupied genes by ChIP-on-chip, many of which coincide with those targets found in ES cells, and of down-regulated targets in the tumor by gene-expression profiling. This Polycomb fingerprint can predict clinical outcome in several tumors.147 However, it seems that this repression is not brought about uniquely by the direct action of PcG members; the H3K27me3 mark is able to generate the necessary machinery to induce de novo methylation in cancer (Figure 3), because the 2 systems might be connected. EZH2 can directly control DNA methylation, in some models, by serving as a recruitment platform for DNA methyltransferases.22 For instance, many PcG target genes, such as WT1, RARβ, KLF4, inhibitor of DNA binding 4 (ID4), GATA3, CHD5, and SPI1, accumulate DNA methylation at their promoters in cancer.148 This phenomenon occurs in several hematologic malignancies, including primary lymphomas of the central nervous system and DLBCL,141,142,146 FL,143 BL,141,146 acute lymphoblastic leukemia, and CML.144 However, methylation of PcG target genes appears to be an early event in tumorigenesis, because it may be present in early as well as advanced stages of the disease. The fact that EZH2 and the H3K27me3 modification are present at the p15INK4b gene in AML cell lines and human samples, and that this repression can be accompanied by either H3K4me3 activation mark or DNA methylation in a mutually exclusive pattern, probably indicates that DNA methylation induces a more permanent repression, because activation marks also disappear when DNA is methylated.104
Cell identity is reflected by changes in chromatin status. In HSCs, PRC1 and PRC2 complexes help to repress genes involved in differentiation to allow the maintenance of stem cell properties. Differentiation induces displacement of PcG members and relocation to promoters of stemness genes. In cancer, the aberrant PcG activity induces a repression of differentiation promoting genes as well as tumor suppressor genes. In this case, the recruitment of DNMTs induces permanent repression of the genes.
Cell identity is reflected by changes in chromatin status. In HSCs, PRC1 and PRC2 complexes help to repress genes involved in differentiation to allow the maintenance of stem cell properties. Differentiation induces displacement of PcG members and relocation to promoters of stemness genes. In cancer, the aberrant PcG activity induces a repression of differentiation promoting genes as well as tumor suppressor genes. In this case, the recruitment of DNMTs induces permanent repression of the genes.
Conclusions and future perspectives
Other histone modifiers and complexes
The recent arrival on the scene of new players, the histone demethylases (HDMs), in particular, H3K27me3 demethylases UTX and JMJD3, makes the PcG regulation of normal and tumoral cellular processes even more dynamic, complex, and exciting. These HDMs remove the methyl mark, making it possible to derepress genes marked for silencing by PcG complexes. This is evidence of the dynamic regulation of activation/repression of genes involved in the processes described in this review that are controlled by PcG/TrxG complexes.
UTX and JMJD3 have been found in complex with MLL proteins (TrxG members responsible for H3K4 methylation) and proteins associated with them, such as RbBP5. This represents new evidence of the cooperation between H3K4 methylation and H3K27 demethylation.66,149
The first evidence of the role of these enzymes in cancer in general and lymphomas in particular, in the form of mutations of UTX and other HDMs in several types of cancer, has recently been published.150
Implications for therapy
Taken together, the data reviewed here and the new findings from HDMs suggest that alterations in the chromatin modification machinery are a frequent event in cancer and sometimes have prognostic relevance, especially in hematologic malignancies, and that dysregulation of PcG members induces the development of lymphomas and leukemias by blocking the normal differentiation pattern of HSCs and/or conferring stem cell properties on progenitor or differentiated cells.
This has important implications for therapy. Because most chemotherapeutic agents target actively dividing cells, cancer stem cells may be relatively resistant to these kinds of drugs, leading to treatment failure and patient death. Targeting pathways that are especially important for stem cells may offer a better therapeutic window for patient care.
In this regard, there continue to be few drugs developed for use against Polycomb members, despite the importance of this system in cancer cells. Indeed, Tan et al151 reported how DNZep, a drug that disrupts the function of the PRC2 complex, can induce apoptosis in cancer, but not in normal, cells. This is accompanied by the reexpression of genes involved in development and differentiation,152 probably meaning that this kind of treatment induces loss of stemness properties in cancer cells. Some histone deacetylase inhibitors (HDACi), such as LBH589 and LAQ824, can also deplete the PRC2 complex in tumoral cells in AML.153 The authors of this study showed that the inhibition of EZH2 by small interfering RNAs has a synergistically negative effect on the survival of AML cells, favoring the combined use of anti-EZH2 drugs with HDACi for the treatment of AML. In fact, in 2009, the same group published a study in which they combined DNZep with panobinostat, a pan-histone deacetylase inhibitor in AML. The treatment was synergistic and induced apoptosis in AML cells but not in normal CD34+ bone marrow progenitor cells.154 APL patients could also benefit from a PRC2-directed therapy. In this leukemia the PRC2 collaborates with the oncogenic fusion gene PML-RAR-α to repress genes involved in differentiation and to promote tumor development.122 These patients are typically treated with retinoic acid, but depletion of SUZ12 in APL patients reverses the epigenetic marks induced by PML-RAR-α, suggesting that PRC2-targeted therapy could also be used to treat this type of leukemia.
Acknowledgments
We express our gratitude to Dr Miguel A. Vidal for his critical reading of the manuscript.
This work was supported by grants from Ministerio de Educación y Ciencia (SAF2007-65 957-C02-02); Ministerio de Ciencia e Innovacion (SAF2008-03 871); Fondo de Investigaciones Sanitarias (RTICC RD06/0020/0107); and Fundación Científica de la Asociación Española contra el Cáncer, Spain. D.M.-P. was supported by a fellowship from the Ministerio de Educación y Ciencia (AP2005-3972).
Authorship
Contribution: D.M.-P. wrote the manuscript; M.A.P. revised the manuscript; and M.S.-B. wrote and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Margarita Sanchez-Beato, Lymphoma Group, Molecular Pathology Program, Centro Nacional de Investigaciones Oncológicas (CNIO), c/ Melchor Fernández Almagro 3, E-28029 Madrid, Spain; e-mail: msbeato@cnio.es.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal