Abstract
In classical Hodgkin lymphoma (cHL) the mechanisms underlying primary refractory disease and relapse remain unknown. To gain further insight into cHL pathogenesis and genomic changes linked to treatment response, we studied 53 cHL patients by array comparative genomic hybridization, including 23 patients whose primary treatment failed, using DNA from microdissected HRS cells. Copy number alterations found in more than 20% of cases included gains of 2p, 9p, 16p, 17q, 19q, 20q, and losses of 6q, 11q, and 13q. We identified at high resolution recurrent changes defining minimally gained and lost regions harboring genes involved in nuclear factor κB signaling, such as REL, IKBKB, CD40, and MAP3K14. Gains of chromosome 16p11.2-13.3 were significantly more frequent in pretreatment and relapse biopsies of unresponsive patients and were associated with shortened disease-specific survival (P = .028). In the therapy-resistant HL cell line KMH2, we found genomic gains and overexpression of the multidrug resistance gene ABCC1 mapping to cytoband 16p13.11. We show that doxorubicin exposure to KMH2 induces ABCC1 expression and that siRNA silencing of ABCC1 sensitizes KMH2 cells to doxorubicin toxicity in vitro, suggesting that overexpression of ABCC1 contributes to the drug resistance phenotype found in KMH2.
Introduction
Hodgkin lymphoma (HL) is among the most curable human neoplasms found in adults and accounts for 11% of all malignant lymphomas.1 Advances in treatment have resulted in improved survival with a decrease in mortality of 60% since the early 1970s. Nevertheless, approximately 10% to 20% of advanced-stage patients will die after relapse or progressive disease,2,3 and thus far no reliable prognostic factors are available to predict treatment response for those patients who are not cured by their primary therapy. Although a plethora of biomarkers associated with clinical outcome have been described in classical Hodgkin lymphoma (cHL),4-8 to date none of these factors has influenced clinical practice. After clinical relapse, high-dose chemotherapy regimens with stem cell support (autologous stem cell transplantation) are typically used for younger patients,9,10 whereas novel targeted therapy approaches are lacking. The clinical problem of therapy resistance is further compounded by toxicity of chemotherapy, leading to long-term sequelae for patients achieving long-term remission.11
In cHL, the most common subtype accounting for more than 90% of all HL cases, the paucity of neoplastic Hodgkin Reed-Sternberg (HRS) cells impedes a detailed characterization of the disease. HRS cells typically constitute the minority of cells in the biopsy, often corresponding to less than 1% of cells present in the involved lymph nodes.12 As a result of improvements in laser-capture microdissection and linear nucleic acid amplification techniques, it has become possible to purify HRS cells and study them separately from their surrounding microenvironment.13,14 Despite these advances, however, the mechanisms underlying primary refractory disease and relapse remain largely elusive. In HL cell lines, differences between the gene expression profiles of chemotherapy-resistant and -sensitive cells have been described,15 and in another study protein kinase Cη expression was associated with drug resistance in L428 cells.16 Overall, perhaps related to small case numbers and incomplete clinical data, no investigations of primary HRS cells have been reported yielding definitive target genes that could be linked to a drug resistance phenotype.
The study of copy number changes in microdissected HRS cells with the use of conventional comparative genomic hybridization (CGH) has revealed that cHL shares common chromosomal imbalances. These investigations have defined a characteristic profile of recurrent copy number gains and losses in cHL, including gains of chromosomes 2p, 9p, 16p, and 17q and losses of 13q, 6q, and 11q.13,17 Although these studies were limited by the low resolution of conventional CGH, more recent studies in which the authors used oligonucleotide arrays have provided further insight into copy number imbalances affecting small chromosomal regions.18 Although novel copy number changes were identified, including amplification of STAT6, NOTCH1, and JUNB, the study focused on only 12 HL samples that were unusually rich in HRS cell content.
The constitutive activity of nuclear factor κB (NFκB) transcription factors is a hallmark of HL and is affected by multiple genetic alterations.19 Inactivating somatic mutations of the NFκB signaling inhibitors NFKBIA, NFKBIE, and TNFAIP3 frequently have been described to play a major role in the pathogenesis of HL.20-22 In the aforementioned CGH studies, investigators have identified genomic amplifications of c-REL and BCL3, and in the cases with gains of c-REL, this correlated with increased REL protein expression.23-26
Our study of microdissected HRS cells from 53 cases with cHL identified at high resolution new and recurrent changes defining regions of chromosomal gain or loss harboring potential oncogenes and tumor suppressor genes involved in the pathogenesis of HL, including IKBKB, CD40, MAP3K14, and TNFRSF14. Moreover, we found gains of chromosome 16p to be significantly more frequent in pretreatment and relapse biopsies of patients for which primary therapy failed to eradicate their disease. We provide in vitro data that overexpression of the gene ABCC1 mapping to cytoband 16p13.11 may contribute to the drug-resistance phenotype identified in the cell line KMH2 derived from a patient with relapsed cHL.
Methods
Patient samples
Diagnostic fresh-frozen lymph node specimens from 53 patients with cHL were selected for array CGH (aCGH) analysis diagnosed at the British Columbia Cancer Agency between the years 1989 and 2005. Specimens were selected from the tissue archive according to the following criteria: primary diagnosis of cHL after central review, HIV-negative status, and first-line treatment with systemic chemotherapy ABVD (ie, doxorubicin, bleomycin, vinblastine, and dacarbazine) or a similar regimen with or without radiation therapy if indicated or, in the case of 3 patients with limited-stage disease, wide-field radiation. Pathology review showed predominantly nodular sclerosing subtype with HRS cells of typical morphology and immunophenotype (World Health Organization classification).12 For treatment outcome comparison, we dichotomized the patient cohort into treatment success (n = 30) and failures (n = 23), whereby a patient was judged a treatment failure if the lymphoma progressed at any time after initiation of primary therapy or a treatment success if a patient did not progress or relapse at any time during the follow-up interval. The treatment failure group included 10 biopsies taken at relapse; there were no paired pretreatment/posttreatment samples in this study. Clinical characteristics according to this dichotomy are shown in Table 1. Patients were also risk-stratified according to the International Prognostic Scoring (IPS) system27 : the low-risk group comprised a 0 to 3 score, and the high-risk group comprised a 4 to 7 score. For cases lacking some IPS parameters, incomplete scores were calculated and normalized to a complete set of 7 parameters (Table 1). Ethical approval for this study was obtained from the University of British Columbia-British Columbia Cancer Agency Research Ethics Board (UBC BCCA REB).
Clinical characteristics of 53 patients with classical Hodgkin lymphoma analyzed by the use of array comparative genomic hybridization
| . | All patients . | Treatment success . | Treatment failure . | P . |
|---|---|---|---|---|
| n (%) | 53 | 30 (57) | 23 (43) | |
| Median age, y (range) | 36 | 36 (15-74) | 29 (12-71) | .559 |
| Male sex, % | 66 | 56 | 78 | .100 |
| Histology, % | .831 | |||
| Nodular sclerosis | 82 | 87 | 78 | |
| Mixed cellularity | 8 | 7 | 9 | |
| Lymphocyte rich | 4 | 2 | 4 | |
| Lymphocyte depleted | 0 | 0 | 0 | |
| NOS | 6 | 2 | 9 | |
| Advanced stage, % | 62 | 40 | 91 | < .001* |
| B symptoms, % | 30 | 20 | 43 | .065 |
| IPS 4 or more, high-risk | 17 | 13 | 22 | .419 |
| Mass size, median (range) in cm | 5 | 4 | 6 | .210 |
| 10 or more cm, % | 25 | 23 | 26 | 1.000 |
| Treatment | .249 | |||
| ABVD type ± radiation, % | 94 | 90 | 100 | |
| Extended-field radiation alone, % | 6 | 10 | 0 |
| . | All patients . | Treatment success . | Treatment failure . | P . |
|---|---|---|---|---|
| n (%) | 53 | 30 (57) | 23 (43) | |
| Median age, y (range) | 36 | 36 (15-74) | 29 (12-71) | .559 |
| Male sex, % | 66 | 56 | 78 | .100 |
| Histology, % | .831 | |||
| Nodular sclerosis | 82 | 87 | 78 | |
| Mixed cellularity | 8 | 7 | 9 | |
| Lymphocyte rich | 4 | 2 | 4 | |
| Lymphocyte depleted | 0 | 0 | 0 | |
| NOS | 6 | 2 | 9 | |
| Advanced stage, % | 62 | 40 | 91 | < .001* |
| B symptoms, % | 30 | 20 | 43 | .065 |
| IPS 4 or more, high-risk | 17 | 13 | 22 | .419 |
| Mass size, median (range) in cm | 5 | 4 | 6 | .210 |
| 10 or more cm, % | 25 | 23 | 26 | 1.000 |
| Treatment | .249 | |||
| ABVD type ± radiation, % | 94 | 90 | 100 | |
| Extended-field radiation alone, % | 6 | 10 | 0 |
ABVD indicates doxorubicin, bleomycin, vinblastine, and dacarbazine; IPS, International Prognostic Scoring; and NOS, not otherwise specified.
Statistically significant.
Laser microdissection, DNA extraction, and whole-genome amplification
For a detailed description, see the supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In brief, enrichment of HRS cells was performed by the use of laser microdissection with a Zeiss Axioplan 2 microscope equipped with Molecular Machines Industries Technology. Genomic DNA extraction of the pooled 500 to 1000 HRS cells per case was performed according to standard procedures with the Puregene Cell & Tissue Kit by Gentra Systems, following the manufacturer's protocol. We then performed whole-genome amplification (WGA) using the GenomePlex Whole Genome Amplification kit according to the standard protocol (Sigma-Aldrich) to obtain greater than 200 ng of amplified product per case for array CGH. For quality control purposes, 50 ng of amplified DNA was analyzed by multiplex polymerase chain reaction (PCR; QIAGEN) as previously described.28
Array comparative genomic hybridization
The submegabase resolution tiling array contains 26 819 BAC clones spotted in duplicate and covers more than 95% of the human genome,29 producing high-resolution profiles with a functional resolution of 50 kb.30 aCGH was performed as previously described.31 For details on DNA labeling, target hybridization, scoring of the array CGH raw data, and computational analysis, see the supplemental Methods. We distinguished 3 states of copy number only: (1) loss, (2) neutral, and (3) gain. As normal controls we also included array CGH profiles of microdissected germinal centers derived from 5 benign tonsils.
Fluorescence in situ hybridization and fluorescence immunophenotyping and interphase cytogenetics as a tool for investigation of neoplasms
Fluorescence immunophenotyping and interphase cytogenetics as a tool for investigation of neoplasms (FICTION) was used to validate aCGH findings in CD30+ HRS cells of corresponding frozen HL cell suspensions as described previously.25 We used the following in-house BAC fluorescence in situ hybridization (FISH) probes: RP11-462C07 (16p13.11), RP11-790E15 (16q12.1), and D16Z3 alpha satellite (Vysis). For further details on probe preparation, hybridization and scoring, see the supplemental Methods.
Cell culture, siRNA interference, and doxorubicin treatment
The HL cell lines KMH2 and L428 were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ; http://www.dsmz.de/), and cultures were grown according to the standard conditions. KMH2 cells (DSMZ ACC 8) were established from the pleural effusion of a mixed cellularity subtype HL patient and L428 cells (DSMZ ACC 197) from the pleural effusion of a nodular sclerosis subtype HL patient. Cells of low passage number were transiently transfected in duplicate with 100 pmol each of Silencer Select siRNA (ABI) directed against ABCC1 or Silencer Select control siRNA (nonsilencing control). RNA interference was achieved by multiple (3×) nucleofections with Amaxa (Lonza) nucleofector technology (see supplemental Figure 1 for detailed schedule) according to the manufacturer's instructions. Transfection efficiency was assessed by quantitative reverse transcription (RT)–PCR (see “Quantitative RT-PCR”).
Cytotoxicity experiments of KMH2 and L428 were performed in quadruplicate 4-mL cultures (6-well plates). Doxorubicin working solutions (2×) were prepared from stock solution (2 mg/mL; Novopharm Ltd) in supplemented RPMI-1640 medium under ultraviolet protection directly before treatment. Long-term cultures of resistant KMH2 clones (28 days) were carried out with the use of final doxorubicin concentrations of 0.025 μg/mL after 7 days of initial drug treatment at 0.25 μg/mL. After 7 days of doxorubicin treatment at different dose points, we found 19% of cells viable (Trypan blue staining) at 0.25 μg/mL doxorubicin, whereas viability was less than 0.01% at 0.5 μg/mL and 1 μg/mL (supplemental Figure 2). We accordingly considered doxorubicin doses of 0.5 μg/mL or greater as lethal and doses 0.25 μg/mL or less as sublethal.
Quantitative RT-PCR
RNA from cell cultures was extracted by the use of Allprep extraction kits (QIAGEN) after mechanical homogenization of the cell pellets in RLT buffer. Quantitative RT-PCR was performed with the use of an Applied Biosystems (ABI) 7900HT real-time PCR system with TaqMan RNA-to-CT 1-Step Kits. For ABCC1 mRNA detection, reactions were carried out in triplicate in 384-well plates with the use of inventoried TaqMan probes (Hs00219905_m1 ABCC1) and endogenous control probes (Hu-GAPDH FAM-MGB, Hu-ACTB FAM-MGB). ABCC1 mRNA levels of KMH2 and L428 were calculated relative to the endogenous control and compared with mRNA levels of CD77+ germinal center B cells that are considered the cell of origin in HL.32 CD77+ cells were enriched from 5 benign tonsillar cell disaggregates by MACS technology (Miltenyi Biotec) as previously described.33
Proliferation assay in doxorubicin-treated cell cultures
Cells in 100-μL suspensions from each culture were seeded into 96-well plates and incubated with 10 μL of WST-1 reagent (Roche Applied Science) for 2 to 4 hours.34 Cell numbers correlated directly with the quantity of mitochondrial dehydrogenases available to cleave WST-1. The spectrophotometric absorbance of the sample against the background control was measured at a wavelength of 440 nm on a microplate reader (Varioskan; Thermo Fisher Scientific Inc). Proliferation of cells treated with doxorubicin was calculated relative to untreated controls.
Statistical analysis
For time-to-event analyses, the primary end point was progression-free survival (PFS) and disease-specific survival (DSS). PFS was defined as the time from initial diagnosis to progression at any time, relapse from complete response, or initiation of new previously unplanned treatment. DSS was recorded as the time to death from disease. Cox proportional hazard models and time to event analyses, including log-rank test with the Kaplan-Meier method, were performed with SPSS Software Version 11.0.0. Univariate and multivariate analysis (Cox proportional-hazards models) were used to assess the prognostic significance of regional copy number changes independent of the IPS. Group comparisons were performed by χ2 and Student t tests (2-tailed distribution). In cell culture experiments, KMH2 cells with ABCC1 siRNA interference were directly compared with their matched nonsilencing controls by paired Student t tests. P values less than .05 were reported as significant.
Results
Clinical data
The clinical characteristics of the cohort comprising 53 selected patients with cHL are summarized in Table 1. The majority of patients had a diagnosis of nodular sclerosis, and most presented with advanced-stage disease. Median age at initial diagnosis was 36 years. Effectively all patients received systemic chemotherapy and radiation therapy if indicated; only 3 patients (6%) received limited-field radiation as their sole treatment modality. For clinical group comparisons, the cohort was dichotomized into 30 patients with treatment success versus 23 patients with treatment failure. The treatment failure group included 10 patients with lymph node biopsies sampled at relapse. The complete cohort was enriched for all available treatment failure cases that had fresh-frozen tissue available. For this selected cohort, the median follow-up time was 5.38 years for living patients, and the 10-year overall survival was 61.1%. Expectedly, the treatment failure group consisted of significantly more patients diagnosed with advanced-stage disease (P < .001). All other clinical parameters were equally distributed between the 2 treatment outcome groups. In the treatment failure group, all 23 patients received systemic polychemotherapy. According to the time point of progression or relapse, we distinguished 6 patients with primary refractory disease (progression during treatment), 6 patients with early relapses (relapse within 6 months after treatment), and 11 patients with late relapses (relapse later than 6 months after treatment).
Copy number alterations in microdissected HRS cells
To identify copy number changes in HRS cells we individually picked 500 to 1000 HRS cells per case by microdissection. WGA of the extracted DNA yielded on average 1154 ng of amplified product (range, 207-6092 ng), allowing for comparative hybridization in every case (input 200 ng). All 53 cases passed quality control by multiplex PCR for standard fragment sizes (300-bp fragment present). Analysis of the complete cohort revealed 451 total imbalances, averaging 8.5 (median, 6; range, 1-33) imbalances per case. Of the 451 imbalances, 275 were classified as gains and 176 as losses favoring a preponderance of copy number gains. A detailed list of the individual imbalances per case can be found in supplemental Table 2 (“Regions”). The median segment length was 23 817 kb, ranging from minimally 159 kb to 157.9 Mb. Accordingly, called regions contained on average 227 BAC clones (range, 2-1402).
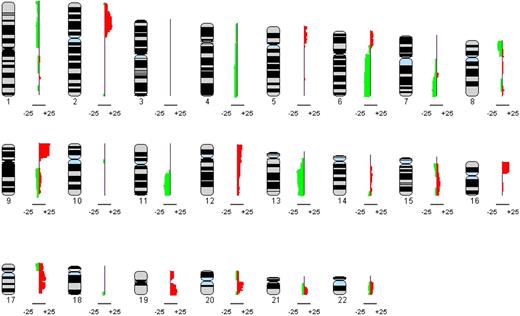
Each aCGH profile was annotated individually and then jointly analyzed to generate a genome-wide copy number profile of all 53 cHL cases studied. Figure 1 shows the composite profile ideogram of all imbalances affecting the 22 autosomes. The most frequently altered regions (>20% of cases) involved gains of chromosomes 2p, 9p, 16p, 17q, 19q, and 20q and losses of chromosomes 6q, 11q, and 13q. For details, see Table 2. We did not observe any copy number changes in the 5 germinal center profiles that served as normal controls. The uncalled raw array CGH data for the 53 patients and the 5 normal controls are included in the supplemental Data (compressed raw data file).
Recurrent imbalances found in HRS cells of 53 cHL samples are shown. The composite frequency plot summarizes the relative frequencies of chromosomal gains as red bars to the right and losses as green bars to the left aligned to each of the 22 autosomes.
Recurrent imbalances found in HRS cells of 53 cHL samples are shown. The composite frequency plot summarizes the relative frequencies of chromosomal gains as red bars to the right and losses as green bars to the left aligned to each of the 22 autosomes.
Chromosomal region with copy number alterations in more than 20% of cases
| Chromosome . | Cytoband . | Peak relative frequency, % . | Length, Mb . |
|---|---|---|---|
| Losses | |||
| 6 | q21-27 | 21 | 63 |
| 11 | q22.2-23.3 | 23 | 16 |
| 13 | q14.13-31.3 | 25 | 45 |
| 13 | q33.2-34 | 21 | 10 |
| Gains | |||
| 2 | p13.3-23.2 | 28 | 41 |
| 9 | p13.1-24.3 | 40 | 39 |
| 16 | p11.2-13.3 | 25 | 29 |
| 17 | q11.2-21.32 | 25 | 16 |
| 17 | q22-24.2 | 23 | 16 |
| 19 | q13.11-13.43 | 25 | 25 |
| 20 | q11.1-11.23 | 23 | 9 |
| 20 | q12-q13.12 | 21 | 3 |
| 20 | q13.13-13.2 | 21 | 1 |
| Chromosome . | Cytoband . | Peak relative frequency, % . | Length, Mb . |
|---|---|---|---|
| Losses | |||
| 6 | q21-27 | 21 | 63 |
| 11 | q22.2-23.3 | 23 | 16 |
| 13 | q14.13-31.3 | 25 | 45 |
| 13 | q33.2-34 | 21 | 10 |
| Gains | |||
| 2 | p13.3-23.2 | 28 | 41 |
| 9 | p13.1-24.3 | 40 | 39 |
| 16 | p11.2-13.3 | 25 | 29 |
| 17 | q11.2-21.32 | 25 | 16 |
| 17 | q22-24.2 | 23 | 16 |
| 19 | q13.11-13.43 | 25 | 25 |
| 20 | q11.1-11.23 | 23 | 9 |
| 20 | q12-q13.12 | 21 | 3 |
| 20 | q13.13-13.2 | 21 | 1 |
We next focused on minimally lost (MLR) or gained (MGR) consensus regions (<5 Mb) across all samples that were copy-number changed in at least 3 individual cases and identified6 MLRs and 14 MGRs, respectively (Table 3; supplemental Table 1). The size of these regions spanned 599 kb (minimum) to 4.8 Mb (maximum). The most frequent cytobands involved in gains were 2p15-16.1, 9p21.1, 9p24.1-24.3, 17q21.31-32, 20q13.11-13.12, and 20q13.2, and the most frequent cytobands involved in losses were 6q23.2, 11q22.3, and 13q14.3-21.1. To identify commonly targeted gene groups and pathways within these regions we performed gene enrichment analysis using all annotated genes aligning to the identified MLRs and MGRs. Most strikingly, we identified several NFκB signaling pathway genes to be overrepresented (P = .023). NFκB pathway genes involved in gains included REL, CD40, IKBKB, BMP4, MAP3K14, and ITGB3, and in losses included TNFRSF14 and PRKCZ (Table 3). Focusing on chromosome 6q deletions, we also found the TNFAIP3 locus (encoding A20) frequently lost (21%; see Table 2). Interestingly, in 1 case (study case 17) we found a whole 6q-arm loss with an additional log-ratio shift, indicating a likely homozygous deletion of a 1.03 Mb segment encompassing TNFAIP3 (supplemental Figure 3).
Minimally lost or gained regions less than 5 Mb in 3 or more cases (data based on the National Center for Biotechnology Information Built 36.1 human assembly), sorted by relative frequency
| Chromsome . | Cytoband . | Length (< 5 Mb) . | Relative frequency . | Potential target genes . |
|---|---|---|---|---|
| Losses | ||||
| 13 | 13q14.3-21.1 | 4797519 | 0.25 | LECT1 |
| 6 | 6q23.2 | 683447 | 0.23 | CTGF |
| 11 | 11q22.3 | 2826040 | 0.23 | ATM |
| 7 | 7q34-35 | 3510852 | 0.17 | ZYX |
| 1 | 1p36.31-33 | 4408633 | 0.13 | TNFRSF14,* PRKCZ,* TP73 |
| 14 | 14q32.31-32.33 | 4714542 | 0.08 | HSP90, TNFAIP2 |
| Gains | ||||
| 9 | 9p24.1-24.3 | 4749650 | 0.40 | JAK2 |
| 9 | 9p21.1 | 757283 | 0.38 | ACO1 |
| 2 | 2p15-16.1 | 4804518 | 0.28 | REL* |
| 17 | 17q21.31-32 | 3894954 | 0.25 | HOXB1-B9, TBX21, MAP3K14,* ITGB3* |
| 20 | 20q13.11-13.12 | 3489871 | 0.21 | CD40* |
| 20 | 20q13.2 | 1442350 | 0.21 | TSHZ2, ZNF217 |
| 5 | 5p13.2 | 2597057 | 0.11 | IL7R |
| 8 | 8q24.21 | 1389530 | 0.09 | MYC |
| 12 | 12q24.31-24.32 | 1789670 | 0.09 | UBC |
| 14 | 14q22.1-22.2 | 3377578 | 0.09 | PTGDR, BMP4* |
| 1 | 1q32.1 | 3976681 | 0.08 | PTPN7 |
| 8 | 8p11.21 | 599180 | 0.08 | IKBKB (IKK-2)* |
| 1 | 1q23.2-23.3 | 1877267 | 0.06 | CD48 (SLAMF1), SLAMF2 |
| 5 | 5q31.2-31.3 | 4339424 | 0.06 | CD14, ETF1, WNT8A |
| Chromsome . | Cytoband . | Length (< 5 Mb) . | Relative frequency . | Potential target genes . |
|---|---|---|---|---|
| Losses | ||||
| 13 | 13q14.3-21.1 | 4797519 | 0.25 | LECT1 |
| 6 | 6q23.2 | 683447 | 0.23 | CTGF |
| 11 | 11q22.3 | 2826040 | 0.23 | ATM |
| 7 | 7q34-35 | 3510852 | 0.17 | ZYX |
| 1 | 1p36.31-33 | 4408633 | 0.13 | TNFRSF14,* PRKCZ,* TP73 |
| 14 | 14q32.31-32.33 | 4714542 | 0.08 | HSP90, TNFAIP2 |
| Gains | ||||
| 9 | 9p24.1-24.3 | 4749650 | 0.40 | JAK2 |
| 9 | 9p21.1 | 757283 | 0.38 | ACO1 |
| 2 | 2p15-16.1 | 4804518 | 0.28 | REL* |
| 17 | 17q21.31-32 | 3894954 | 0.25 | HOXB1-B9, TBX21, MAP3K14,* ITGB3* |
| 20 | 20q13.11-13.12 | 3489871 | 0.21 | CD40* |
| 20 | 20q13.2 | 1442350 | 0.21 | TSHZ2, ZNF217 |
| 5 | 5p13.2 | 2597057 | 0.11 | IL7R |
| 8 | 8q24.21 | 1389530 | 0.09 | MYC |
| 12 | 12q24.31-24.32 | 1789670 | 0.09 | UBC |
| 14 | 14q22.1-22.2 | 3377578 | 0.09 | PTGDR, BMP4* |
| 1 | 1q32.1 | 3976681 | 0.08 | PTPN7 |
| 8 | 8p11.21 | 599180 | 0.08 | IKBKB (IKK-2)* |
| 1 | 1q23.2-23.3 | 1877267 | 0.06 | CD48 (SLAMF1), SLAMF2 |
| 5 | 5q31.2-31.3 | 4339424 | 0.06 | CD14, ETF1, WNT8A |
A selection of genes with biological plausibility is shown. Genomic coordinates for each region are given in the supplemental Methods.
Nuclear factor κB signaling pathway genes by ingenuity analysis.
Cluster analysis and secondary pathways
Using high-resolution array CGH, application of a robust computational analysis, and a comparably large cHL cohort of predominantly nodular sclerosis subtype, we were able to define sample clusters characterized by common patterns of imbalances (supplemental Figure 4). Using the chromosomal imbalances that occurred in more than 10% of cases, we applied the k-medoids algorithm (Hamming distance metric) to all 53 cases. This algorithm identified 4 distinct clusters: (1) gain of 2p, loss of 6p, and gains of 16p, 17p/q, and 19p/q; (2) gains of 2p and loss of 6p, but no gains of chromosomes 16, 17, or 19; (3) gains of 9p; and (4) an indefinable group with various alterations but predominantly no 9p gains (1 exception). None of these groups were significantly enriched with primary treatment failures or relapse biopsies.
Associations of copy number alterations with primary treatment outcome
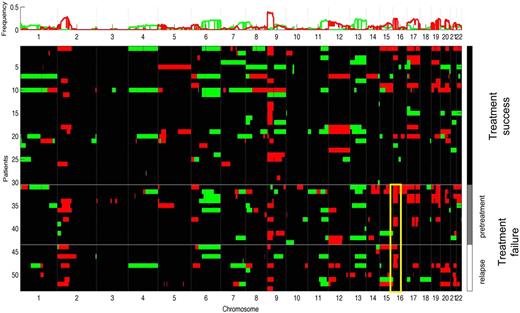
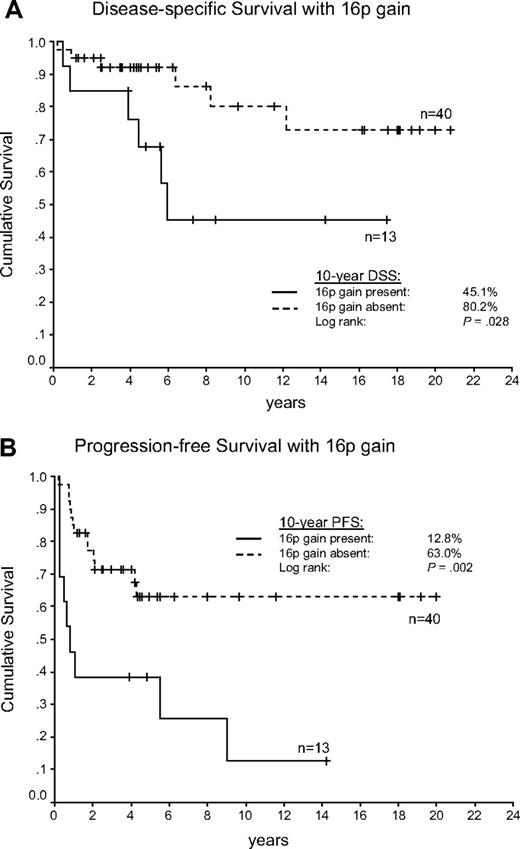
An increased number of imbalances per case was not significantly correlated with adverse outcome (mean in treatment failure vs treatment success, 10.5 vs 7.8, P = .265). However, genome-wide group comparison identified gain of 16p12.1-13.3 to be more frequent in the treatment failure group (Figure 2). Comparing the occurrence of the aberrations in treatment failures versus successes by the χ2 statistic, 16p gains proved to be significant (failures vs successes, 43% vs 10%, P = .005). This statistically significant association could still be observed by the use of pretreatment biopsies only (failures vs successes, 46% vs 10%, P = .007). According to the time point of disease progression or relapse, we found the greatest frequency of 16p gains in primary refractory patients (83.3%), compared with 33.3% of patients with early relapses and 25% with late relapses. Supplemental Figure 5 shows the frequency of gains of chromosome 16 in both outcome groups according to a Hidden Markov Model algorithm (Continuous Master Model [CMM]). The respective minimally gained region on chromosome 16p encompassed 24.5 Mb defined by the 13 cases with 16p gains (Figure 3). Univariate analysis showed that 16p gains were associated with shortened PFS (P = .002) and DSS (P = .028) compared with cases without 16p gain (Figure 4A-B). In univariate analysis, the IPS was associated with inferior DSS when dichotomizing the patient cohort into high-risk (4-7) and low-risk patients (0-3; P = .006). However, when a multivariate Cox-regression model for DSS was used, 16p gain proved to be an adverse prognostic factor independent of the IPS (P = .048). In this series we did not find any other correlations of copy number changes with treatment outcome.
Sample clustering according to treatment outcome reveals overrepresentation of 16p gains in the treatment failure group (yellow box). Red indicates chromosomal gains; green, chromosomal losses. Black boxes next to cluster labels indicate treatment failure cases. (Top) cumulative frequency of imbalances.
Sample clustering according to treatment outcome reveals overrepresentation of 16p gains in the treatment failure group (yellow box). Red indicates chromosomal gains; green, chromosomal losses. Black boxes next to cluster labels indicate treatment failure cases. (Top) cumulative frequency of imbalances.
Minimally gained region on chromosome 16p (chr16: 1 480 000 - 26 000 000) is shown demarcated by the black bars. On the left side a representative profile with 16p gain is demonstrated (study case 19) showing overrepresentation of chromosomal material in the tumor DNA as a shift (at the BAC level) to the right from the neutral line. FISH validation of the finding is shown in the right panel with the use of probes derived from BAC clones: top left, study case 1 (interphase); bottom left, study case 19 (interphase); top right, KMH2 (interphase); and bottom right (metaphase). The red signal spans the ABCC1 locus on 16p; as references, a locus on 16q and a centromere 16 probe were used. FICTION: CD30 stain in light blue. Relative signal number gains of the ABCC1 locus are demonstrated.
Minimally gained region on chromosome 16p (chr16: 1 480 000 - 26 000 000) is shown demarcated by the black bars. On the left side a representative profile with 16p gain is demonstrated (study case 19) showing overrepresentation of chromosomal material in the tumor DNA as a shift (at the BAC level) to the right from the neutral line. FISH validation of the finding is shown in the right panel with the use of probes derived from BAC clones: top left, study case 1 (interphase); bottom left, study case 19 (interphase); top right, KMH2 (interphase); and bottom right (metaphase). The red signal spans the ABCC1 locus on 16p; as references, a locus on 16q and a centromere 16 probe were used. FICTION: CD30 stain in light blue. Relative signal number gains of the ABCC1 locus are demonstrated.
Survival in the aCGH cohort comprising 53 patients with cHL according to 16p chromosomal gains. (A) Disease-specific survival (DSS). (B) Progression-free survival (PFS).
Survival in the aCGH cohort comprising 53 patients with cHL according to 16p chromosomal gains. (A) Disease-specific survival (DSS). (B) Progression-free survival (PFS).
To validate the identified gains of 16p we selected 2 cases with available archival lymph node single cell suspensions and performed FICTION using locus-specific in-house BAC probes for cytoband 16p13.11. Representative images of the aCGH ideograms and the corresponding most common signal constellations in nuclei of CD30+ HRS cells are shown in Figure 3. In both cases signal gains of the 16p probe (red), compared with the reference probes on chromosome 16, confirmed the array CGH findings.
The HL cell line KMH2 harbors genomic amplification of the ABCC1 locus on chromosome 16 and overexpresses ABCC1
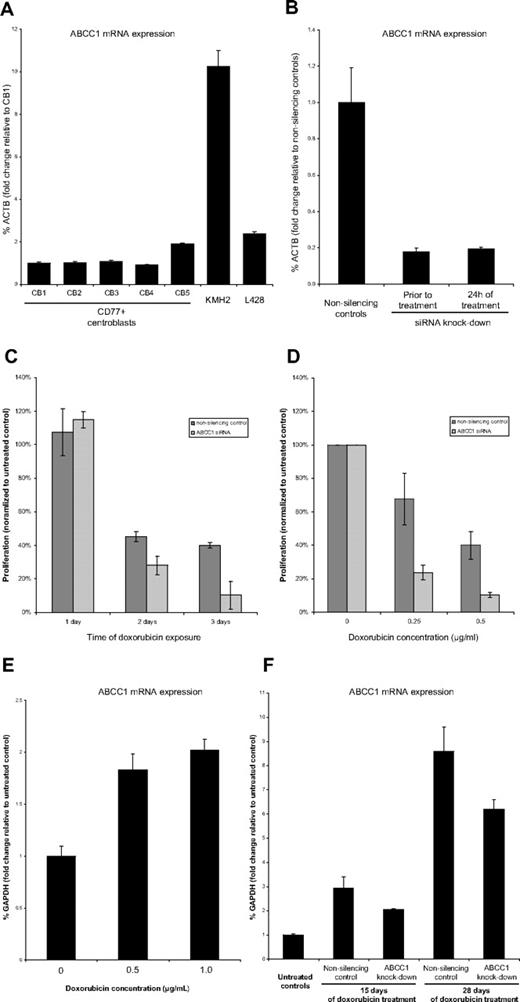
The correlation of chromosome 16p gain with treatment outcome suggested a possible role of individual genes harbored in this region associated with therapy resistance and clinical relapse. Most strikingly, the amplicon on chromosome 16 encompassed the gene location of the multidrug resistance gene ABCC1 (MRP1) at cytoband 16p13.11 that has been previously described to be overexpressed in various solid tumors,35-37 and mapped to a prominent peak in the CMM analysis (supplemental Figure 5). We selected the HL cell lines KMH2 and L428, which were both derived from patients with multiple relapsed cHL, for functional study of ABCC1. Using FISH, we could confirm that KMH2 showed gains of the ABCC1 locus, involving duplication on a marker chromosome consistent with findings by others who used multicolor-FISH24 and aCGH38 (Figure 3). Accordingly, mRNA levels of ABCC1 were 10-fold increased in KMH2 compared with CD77+ germinal center B cells (Figure 5A). In contrast, L428 did not show genomic gain of ABCC1 (supplemental Figure 6) and expressed ABCC1 at much lower levels (2-fold increased compared with CD77+ cell; Figure 5A).
ABCC1 RNA interference and induction in KMH2 cells. (A) ABCC1 mRNA is overexpressed in KMH2 compared with CD77+ centroblasts. (B) siRNA silencing shows effective and continuing knockdown of ABCC1 in KMH2 before treatment and after 24 hours of treatment. (C) Proliferation assay (WST-1) under doxorubicin treatment (0.5 μg/mL) shows increased toxicity in ABCC1 siRNA-silenced KMH2 cells compared with nonsilencing controls (each normalized to proliferation of cells not treated with doxorubicin). (D) Proliferation of siRNA-silenced KMH2 cells on day 3 is significantly decreased by the use of different doxorubicin doses (each normalized to proliferation of cells not treated with doxorubicin). (E) ABCC1 expression in KMH2 is early induced after 2 days of doxorubicin treatment. (F) Sublethal doxorubicin doses (0.25 μg/mL) produce resistant KMH2 clones highly overexpressing ABCC1 compared with doxorubicin untreated controls in long-term culture.
ABCC1 RNA interference and induction in KMH2 cells. (A) ABCC1 mRNA is overexpressed in KMH2 compared with CD77+ centroblasts. (B) siRNA silencing shows effective and continuing knockdown of ABCC1 in KMH2 before treatment and after 24 hours of treatment. (C) Proliferation assay (WST-1) under doxorubicin treatment (0.5 μg/mL) shows increased toxicity in ABCC1 siRNA-silenced KMH2 cells compared with nonsilencing controls (each normalized to proliferation of cells not treated with doxorubicin). (D) Proliferation of siRNA-silenced KMH2 cells on day 3 is significantly decreased by the use of different doxorubicin doses (each normalized to proliferation of cells not treated with doxorubicin). (E) ABCC1 expression in KMH2 is early induced after 2 days of doxorubicin treatment. (F) Sublethal doxorubicin doses (0.25 μg/mL) produce resistant KMH2 clones highly overexpressing ABCC1 compared with doxorubicin untreated controls in long-term culture.
ABCC1 siRNA silencing sensitizes KMH2 to treatment with doxorubicin in vitro
To test the dependency of drug resistance on overexpression of ABCC1 in HL cell lines we studied the proliferation in KMH2 and L428 ABCC1 knockdown derivatives using siRNA interference compared with their respective nonsilencing controls. The mean doubling time of wild-type KMH2 and L428 cells was 48 hours and 36 hours, respectively. The median lethal dose (LD50) of doxorubicin after 48 hours of exposure was 0.7 μg/mL for KMH2 and 0.6 μg/mL for L428. After repeated (3×) transient siRNA interference (supplemental Figure 1), we achieved an effective and persistent knockdown of ABCC1 before and during doxorubicin treatment without significant toxicity (mRNA reduction to 20% in KMH2 and 16% in L428) in both cell lines (Figure 5B; supplemental Figure 7). We therefore selected 24, 48, and 72 hours as measurement time points. In KMH2 cells, proliferation was suppressed to a greater extend in the ABCC1 knockdown cultures compared with nonsilencing controls with increasing time of exposure to doxorubicin (Figure 5C) and with increasing doses of doxorubicin (Figure 5D); in particular, after 3 days of doxorubicin treatment KMH2 cultures with siRNA interference compared with their nonsilencing controls displayed a significantly lower proliferation rate at 0.25 μg/mL (24% vs 68%, P = .030) and 0.5 μg/mL doxorubicin concentrations (10% vs 40%, P = .017). By contrast, no significant differences in proliferation could be observed in L428 ABCC1 knockdown cultures (supplemental Figure 8A-B). Overall, these data show that overexpression of ABCC1 contributes to the drug resistance phenotype of KMH2.
ABCC1 expression is early induced in KMH2 by doxorubicin treatment and in long-term culture during sublethal doxorubicin exposure
To test the hypothesis of whether ABCC1-dependent drug-resistance develops during treatment we measured ABCC1 induction in KMH2 cultures using lethal and sublethal doxorubicin doses. Lethal doses of doxorubicin (0.5 μg/mL and 1 μg/mL) over the course of 48 hours showed that ABCC1 was induced early compared with untreated controls (Figure 5E). Furthermore, in long-term cultures of KMH2 exposed to sublethal doxorubicin doses (0.25 μg/mL) we selected a small proportion (19%) of resistant cells (supplemental Figure 2) that proliferated in maintenance selection media (0.025 μg/mL). These cells strongly overexpressed ABCC1 compared with untreated controls after 15 days and even more after 28 days of doxorubicin treatment (Figure 5F). These effects were visible in both cell cultures with and without transient RNA interference; however, in ABCC1 knock-down cultures the ABCC1 gene induction was less prominent.
Discussion
Our study revealed novel recurrent chromosomal imbalances in cHL in a large number of cases, including 23 samples from patients who progressed following primary therapy. Importantly, we identified gains of chromosome 16p to be associated with treatment failure and accordingly with shortened PFS and DSS. Furthermore, we validated the recurrence of 16p gains in 2 primary lymph node samples and also found gains of 16p in the cHL cell line KMH2 by FISH.
To date, detailed investigations of the molecular processes that characterize treatment failure in cHL have not been described. Interestingly, we observed 16p gains predominantly in therapy-refractory disease, a finding that is consistent with a primary drug resistance in HRS cells with this imbalance. To further explore whether the multidrug resistance gene ABCC1 mapping to the 16p amplicon plays a role in relapsing HL, we chose KMH2 as an in vitro model system because this therapy-resistant cell line harbors gains of 16p and overexpresses ABCC1 as shown in our study and by others.38 We could show that ABCC1 siRNA silencing sensitizes KMH2 to doxorubicin killing, demonstrating that overexpression of ABCC1 contributes to drug resistance in this cell line. In contrast, we did not observe the same effect of siRNA interference in L428, a cHL cell line that did not harbor 16p gains and does not overexpress ABCC1. Our finding that ABCC1 expression is increased by doxorubicin under long-term exposure also suggests that drug resistance might further develop during treatment. This increase in expression could be the result of selection for ABCC1-overexpressing cells that were already present in the untreated KMH2 cultures or to induction of ABCC1 expression in cells that previously did not express ABCC1 at these levels. The finding of ABCC1 gene induction is in agreement with investigations in a daunorubicin-resistant cell line derived from promyelocytic HL60 cells that overexpressed ABCC1 by more than 10-fold compared with the parental daunorubicin-sensitive cell line.37
In vincristine-resistant human ovarian cancer cell lines, overexpression of ABCC1 was linked to the de novo development of 16p gain involving the ABCC1 locus.35 Briefly, it has been well established that cellular efflux pumps of the ATP binding cassette (ABC) transporter family, in particular ABCB1 (MDR1/P-GP), ABCC1 (MRP1), and ABCG2 (BCRP), play important roles in the development of multidrug-resistance in human cancers.39 In a prospective study of primary neuroblastoma, high levels of ABCC1 expression correlated with poor clinical outcome,36 and in early-stage breast cancer patients treated with adjuvant chemotherapy, MRP1 expression predicted shorter relapse-free survival.40 Furthermore, CGH study of chemotherapy-resistant breast cancer tumors after neoadjuvant chemotherapy revealed genomic gain of the gene loci of ABCB1, ABCC1, and ABCG2.41 Overexpression of ABCC1 has also been implicated in clinical outcome prediction of neoadjuvant chemotherapy in nonsmall cell lung cancer.42
Overall, our aCGH data and functional analyses in KMH2 point to a contribution of the multidrug resistance gene ABCC1 for therapy resistance in HL; however, further study is needed to demonstrate overexpression of ABCC1 and other cellular efflux pumps in primary tissue samples.
In this study, we have applied whole-genome tiling path BAC aCGH, with a greater than 200-kb resolution for the detection of copy number alterations and a reported tolerance of up to 70% contamination by nontumor cells.43 This technology enabled us to define minimally gained and lost regions in HL with high accuracy because of a sufficient number of recurrent observations and submegabase resolution; however, because of the necessity of HRS enrichment (by laser microdissection) and WGA, the observed dynamic range of intensity ratios on many arrays was limited so that we only distinguished between copy number gains, neutral copy number, and copy number losses. In defining these small regions, supported by 3 or more cases, we were able to identify chromosomal regions that interestingly were enriched for genes prominently involved in NFκB signaling. Of these, amplification of the REL gene locus has been previously described25 and, although not part of a minimally lost region, we also found recurrent loss of the TNFAIP3 locus on chromosome 6q23.3 as shown by others.22
In addition, we found the CD40 locus on chromosome 20q13.12 gained in more than 20% of cases establishing a link of genomic gain to overexpression of this gene. Overexpression of this member of the TNF receptor family (TNFRSF5) is reported in the vast majority of cHL cases.44 CD40 down-stream signaling is prominently involved in T-cell–mediated B-cell activation and is a major contributor of both the canonical and noncanonical NFκB signaling pathways.19 The region containing IκB kinase complex member IKKβ (8p11.21) was gained in our series, and IKKβ is involved in phosphorylation of IκBα and of IκBϵ, marking them for ubiquitylation, proteasomal degradation, and ultimately leading to nuclear translocation of the NFκB heterodimers.45 Recurrent genomic gain of the IKBKB locus had not been previously described in cHL. In agreement with previous work,17 our study also confirmed the frequent presence of chromosome 17q gains, and our data resolved a minimally gained region of 3.9 Mb containing MAP3K14 as a coamplified gene. MAP3K14 encodes NIK (NF-kappa beta-inducing kinase), a molecule that is central to the noncanonical NFκB pathway activation and integrates upstream signaling from a variety of surface receptors such as TACI (TNFRSF13B), BCMA (TNFRSF17), and LTβ receptor.46 NIK has been recently described to be overexpressed in HRS cells and therefore our data strongly suggests MAP3K14 as a possible target gene in the 17q amplicon.47 Deletions and copy-number neutral LOH of 1p36 have been frequently found in follicular lymphoma31,48 and in other malignancies.49 Our study similarly identifies a minimally lost region in cHL spanning 4.4 Mb. This region contains HVEM (Herpesvirus entry mediator, encoded by TNFRSF14), which has recently been described as a negative regulator of NFκB signaling in B cells after BTLA ligation, suggesting a possible role of TNFRSF14 as a critical gene in this region.50
In the present study, we have shown that the combination of laser microdissection with subsequent WGA and high-resolution aCGH provides a robust and sensitive platform for detecting chromosomal imbalances in microdissected HRS cells. The authors of 2 other studies13,17 have successfully used these techniques in conjunction with aCGH. However, these 2 studies used low-resolution platforms. In a more recent study18 the authors reported only a few cases with selection of HL cases having high HRS cell content, thus avoiding the need for WGA. Despite this selection bias toward cases rich in HRS cells, the authors identified novel imbalances and could validate previous observations without the potential bias by WGA. Reassuringly, our findings are consistent with previous studies and identified the same pattern of large-scale recurrent imbalances and cluster analysis of the identified imbalances in each case revealed a certain pattern of these very common imbalances, with gains of 2p and loss of 6q often traveling together, and 9p gains defining their own cluster. However, relative frequencies varied in comparison to other studies. Although Chui et al found gains of 17q in up to 70% of cases and Joos et al found gains of 2p in 54% of cases,13 in our study the most frequently gained chromosomal region was 9p24.1-24.3 at 40%. These differences are most likely the result of differences in sample size, selection, and the stringency of segmental copy number calls.
In summary, we identified novel recurrent imbalances in cHL and found a correlation of 16p chromosomal gains with PFS and DSS. Our functional data using KMH2 furthermore showed that overexpression of ABCC1 contributes to the drug resistance phenotype in this HL cell line, suggesting that further study of this gene family in primary HL samples linked to treatment failure is warranted.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Center for Translational and Applied Genomics (CTAG) and Pat Allard for excellent technical support.
This work is supported by a postdoctoral fellowship of the Deutsche Forschungsgemeinschaft (DFG) to C.S., the Cancer Research Society (CRS) to C.S., the Michael Smith Foundation for Health Research (MSFHR) to C.S., and the Lymphoma Research Foundation (LRF) to C.S. Operational funds were available through the Canadian Institutes of Health Research (CIHR), grant no. 178536 to R.D.G.
Authorship
Contribution: C.S. designed the research, performed experiments, analyzed results, and wrote the manuscript; A.T., L.B., and M.B. performed experiments; S.P.P. and P.F. analyzed results; J.M.C. provided clinical data; D.E.H. designed the research and analyzed the results; and R.D.G. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Randy Gascoyne, Department of Pathology & Advanced Therapeutics, British Columbia Cancer Agency and the BC Cancer Research Centre, 675 West 10th Ave, Rm 5-113, Vancouver, BC V5Z 1L3, Canada; e-mail: rgascoyn@bccancer.bc.ca.