Abstract
Recent studies of WT1 mutations in acute myeloid leukemia (AML) mostly report an association with unfavorable clinical outcome. We screened 842 patients treated on 3 consecutive pediatric AML trials for WT1 zinc-finger mutations. Eighty-five mutations were detected in 70 of 842 patients (8.3%). Mutations occurred predominantly in exon 7 (n = 74) but were also found in exons 8 (n = 5) and 9 (n = 6). Normal karyotype was observed in 35.3% of WT1mut patients, whereas 27.5% WT1mut patients harbored favorable risk cytogenetics. Patients with or without mutations had similar rates of complete remission after one course of induction chemotherapy. Overall survival (OS) for patients with WT1 mutations was 41% versus 54% for those without mutations (P = .016). Corresponding event-free survival (EFS) was also significantly worse for those with WT1 mutations (28% vs 42%; P = .01). However, FLT3/ITD was present in 36% of the WT1mut cohort; WT1mut patients without FLT3/ITD had similar OS (56% vs 56%, respectively; P = .8) and EFS (35% and 44%, respectively; P = .34) to patients who were wild type for both mutations. In current risk stratification schemes incorporating cytogenetics and FLT3/ITD status, the presence of WT1 mutations has no independent prognostic significance in predicting outcome in pediatric AML. The clinical trials are registered at www.clinicaltrials.gov as #NCT00002798 and #NCT00070174.
Introduction
Acute myeloid leukemia (AML) is a clinically and genetically heterogeneous disease that constitutes 15% to 20% of childhood leukemias. In recent years, recurring mutations in several genes with biologic and prognostic implications have been characterized in AML, particularly within the normal karyotype and/or intermediate-risk cytogenetic subset. Those genes in which mutations may affect disease classification and risk stratification schemes include FLT3, NPM, NRAS, MLL, and CEBPA.1,2 Several series in adult AML have added mutations in the Wilms tumor 1 (WT1) gene to this list.3–7
The WT1 gene, located on chromosome 11p13, encodes a zinc-finger protein that exists in multiple isoforms and functions as a transcription factor.8 WT1 is expressed primarily in tissues of the developing genitourinary and hematopoietic systems, and mutations in WT1 occur in both syndrome-associated and sporadic cases of nephroblastoma (Wilms tumor), the most common childhood renal malignancy.9 WT1 is also expressed in CD34+ hematopoietic progenitors and is overexpressed in a subset of acute leukemias.10 An AML-associated somatic WT1 mutation was first reported in the remaining allele of a patient with the WAGR contiguous gene-deletion syndrome in whom a secondary leukemia developed.11
The WT1 protein consists of a proline-glutamine–rich N-terminal transcriptional regulatory domain (exons 1-6), as well as 4 C-terminal zinc finger domains (exons 7-10) that facilitate DNA binding.8 Exons 5 and 9 are subjected to alternative splicing, yielding 4 different splice isoforms. Posttranslational modifications and alternate start codons lead to additional protein products, furthering the complexity. The WT1 protein may act as a transcriptional activator or a transcriptional repressor, depending on the level of expression, the specific isoform, and the cellular context.12 AML-associated mutations of WT1 have been reported almost exclusively in the zinc-finger domains, resulting in a protein predicted to be incapable of binding DNA. The most commonly reported of these mutations are frameshift mutations of exon 7,4–7 leading to a premature stop codon and a truncated protein lacking the C-terminal zinc fingers. In exon 9, missense mutations predominate; these types of mutations have been shown to interrupt DNA binding capacity by affecting amino acid residues either directly involved in DNA binding or essential to the structure of the zinc-finger motif.13
Recent studies from the Cancer and Leukemia Group B (CALGB),4 Medical Research Council (MRC),5 and AML Study Group (AMLSG)7 have reported WT1 zinc-finger mutations in approximately 10% of adults with cytogenetically normal AML. Among these studies, there are subtle differences in the prognostic significance of harboring such a mutation. The MRC reported that patients with WT1 mutations had a significantly lower complete remission (CR) rate after induction chemotherapy, whereas the CALGB and AMLSG found no statistically significant difference between the CR rates of those with or without mutations. Both the CALGB and MRC studies found the presence of a WT1 mutation to be an independent predictor of inferior overall survival (OS) and disease-free survival (DFS); in contrast, the AMLSG study found that WT1 mutations lacked independent prognostic significance. Notably, only the AMLSG study performed explorative subset analysis, examining outcome measures separately in subsets defined by combined WT1 and FLT3/ITD status. Hollink et al14 reported WT1 mutations in 35 of 298 (12%) pediatric patients with de novo AML, the largest pediatric series to date. These patients were treated on AML-BFM-SG/DCOG or LAME protocols, and WT1 mutations independently predicted poorer survival and greater risk of relapse.
In our study, we examined pediatric patients enrolled on 3 consecutive Children's Cancer Group (CCG) and Children's Oncology Group (COG) trials, providing the largest cohort of patients with AML screened for WT1 mutations to date. Herein, we present a comprehensive evaluation of the prevalence and prognostic significance of WT1 mutations, in the context of other validated biologic, cytogenetic, and molecular risk factors, in a large cohort of pediatric patients with de novo AML.
Methods
Patient samples
Pediatric patients with newly diagnosed de novo AML enrolled in 3 consecutive pediatric AML protocols, CCG-2941, CCG-2961, or COG-AAML03P1, were eligible for this study. Details of these studies have been previously described.15,16 Of the 1328 patients treated on the 3 consecutive studies, 842 diagnostic specimens were available and were obtained from the COG AML Reference Laboratory. Approval by the institutional review board was obtained before mutation analysis, and this study was approved by the COG Myeloid Disease Biology Committee. Informed consent for study protocol treatment and tissue sample evaluation was obtained in accordance with the Declaration of Helsinki.
Mutation screening
Genomic DNA was extracted from the diagnostic marrow specimens with the Puregene protocol (Gentra Systems Inc). Polymerase chain reaction (PCR) amplification of the zinc-finger domains of WT1 (exons 7-10) was performed with the use of 4 primer pairs (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). For exons 7 through 9, the PCR reaction was carried out in a mixture containing Failsafe PCR Premix (Epicentre Biotechonologies) Buffer A (exons 7 and 9) or Buffer F (exon 8), 1.25 U of Platinum Taq DNA Polymerase (Invitrogen), 5 pmol of each primer, and 10 ng of genomic DNA. For fragment-length analysis screening, forward primers were labeled with 6-FAM dye. Thermocycler conditions were as follows: 95°C for 5 minutes; 40 cycles at 95°C for 45 seconds, 59°C for 30 seconds, and 72° for 45 seconds; and a final extension step at 72°C for 7 minutes. Fluorescently labeled fragments were separated on an ABI 377xl automated DNA sequencer, and analysis was performed with the use of Genemapper software (Applied Biosystems). Mutations were confirmed by direct sequencing; for samples containing more than one WT1 mutation, cloning was performed with the use of the TOPO-TA Cloning Kit (Invitrogen), and direct sequencing was performed on plasmid DNA obtained from multiple isolated clones.
For exon 10, the PCR reaction was carried out in 10× PCR Buffer (Invitrogen) containing 1.5 μL of 50mM MgCl2, 0.5 μL of 10mM deoxynucleoside triphosphates, 1.25 U of Platinum Taq DNA Polymerase, 5 pmol of each primer, and 10 ng of genomic DNA. The following conditions were used for PCR amplification: 94°C for 5 minutes; 35 cycles at 94°C for 30 seconds, 65°C for 30 seconds, and 72° for 30 seconds; and a final extension step at 72°C for 7 minutes. Direct sequencing was carried out with the use of the Big Dye Terminator v3.1 Cycle Sequencing Reaction (Applied Biosystems) and run on an ABI 3730xL DNA analyzer.
Screening for FLT3/ITD, NPM, and CEBPA mutations was performed as previously described.17–19
Statistical methods
The Kaplan-Meier method was used to estimate OS, event-free survival (EFS), and DFS. OS was defined as the time from study entry to death. EFS was defined as the time from study entry until failure to achieve remission by the end of 2 courses of therapy, relapse, or death. DFS was defined as the time from end of course 1 for patients in CR (bone marrow aspirate containing < 5% blasts by morphology and no evidence of extramedullary disease) until relapse or death. Estimates of relapse risk (RR) were obtained by the method of cumulative incidence that accounts for competing events. RR was defined as the time from end of course 1 for patients in CR to relapse or death due to progressive disease whereby deaths from nonprogressive disease were considered competing events. The significance of predictor variables was tested with the log-rank statistic for OS, EFS, and DFS and with Gray statistic for RR. Children who also received a stem cell transplant while on study were censored at the time of transplantation for all analyses, unless otherwise indicated. Children lost to follow-up were censored at their date of last known contact or 6 months before the cutoff date of analyses being April 14, 2005, for patients on CCG-2941, November 6, 2009, for patients on CCG-2961, and November 16, 2009, for patients on COG-AAML03P1. The significance of observed differences in proportions was tested using the χ2 test and Fisher exact test when data were sparse. The Mann-Whitney test was used to determine the significance between differences in medians. Cox proportional hazard models were used to estimate hazard ratios (HRs) for univariate and multivariate analyses for OS, DFS, and relapse-free survival (RFS). RFS was defined as the time from end of course 1 for patients in CR to relapse or death due to progressive disease, whereby deaths from nonprogressive disease were censored.
Results
Patient population
From September 1995 to November 2005, 1328 pediatric patients with AML were treated on CCG-2941, CCG-2961, and COG-AAML03P1, 842 (63%) of whom had diagnostic bone marrow specimens available for analysis. Demographics, laboratory and clinical characteristics, and outcome for those with and without available specimens were compared. Patients without available diagnostic specimens (N = 486) had similar clinical outcomes with 5-year OS of 55% (± 5%) compared with 53% (± 4%) for those who were analyzed (P = .261). Induction CR rates and EFS from study entry were also similar. The study population differed from those not tested mainly in regard to age and diagnostic white blood cell (WBC) count, whereby those not tested were younger (median age, 6.9 vs 10.4 years; P < .001) and had lower median diagnostic WBC counts (15.1 vs 21.8 × 109/L [15 100 vs 21 800/μ/L]; P = .002). In addition, the population that was not tested had a higher proportion of patients with megakaryocytic leukemia (10.7% vs 4.4%; P < .001).
WT1 mutation analysis
Initial mutation screening of the exons encoding all 4 zinc-fingers (exons 7-10) was performed by direct sequencing on 100 samples from CCG-2961. Frameshift mutations were detected in exon 7 (n = 6) and exon 8 (n = 1). Although a known single nucleotide polymorphism (SNP; c.1293A>G) was detected in exon 7 in 21 patients, no other exon 7 point mutations were detected in this initial 100-patient screen. Point mutations (n = 2) were detected in exon 9, and no mutations were detected in exon 10. Thus, in the remaining 742 available diagnostic specimens from patients enrolled on CCG-2941, CCG-2961, or COG-AAML03P1, fragment-length analysis was used to screen exons 7 and 8, whereas direct sequencing was performed on exon 9, and exon 10 was not screened further. Mutations detected by fragment-length analysis were subjected to confirmation by direct sequencing.
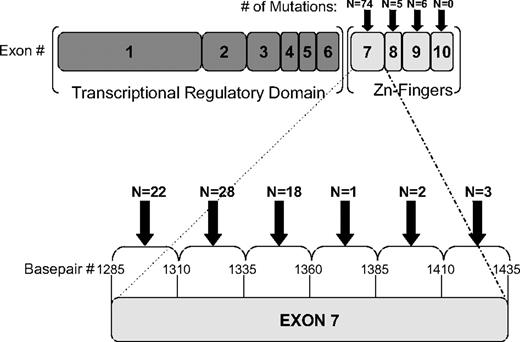
Overall, 85 WT1 zinc-finger mutations were detected in 70 of the 842 patients (8.3%) screened (supplemental Table 2). Mutations clustered overwhelmingly in exon 7 (74 mutations in 59 patients; Figure 1), but they were also detected in exon 8 (n = 5) and exon 9 (n = 6). Nearly all exon 7 mutations were frameshift mutations leading to a stop codon, including insertions (n = 62) ranging between 1 and 31 base pairs and deletions (n = 9) ranging between 1 and 38 base pairs. Also detected were 3 single base pair substitution, missense mutations. Most exon 7 mutations occurred within 2 “mutational hotspot” regions: between base pairs 1295 and 1309 (n = 22) or between base pairs 1323 and 1340 (n = 33). For patients in whom 2 mutated peaks were detected on fragment-length analysis, or in whom 2 mutations were detected on confirmatory sequencing, TOPO-TA cloning (Invitrogen) was performed. Plasmid DNA from multiple isolated clones was then directly sequenced. In this manner 15 of the 70 patients (21.4%) with mutations were shown to harbor biallelic WT1 mutations in exon 7: 4 patients had homozygous frameshift mutations, 8 patients had 2 different frameshift mutations, and 3 patients had both a frameshift and a missense mutation.
Location of WT1 zinc-finger mutations. Graphical depiction of the 85 mutations detected in 70 of 842 pediatric patients with AML screened. Mutations clustered in the N-terminal portion of exon 7 but were also detected in exons 8 and 9.
Location of WT1 zinc-finger mutations. Graphical depiction of the 85 mutations detected in 70 of 842 pediatric patients with AML screened. Mutations clustered in the N-terminal portion of exon 7 but were also detected in exons 8 and 9.
Eleven mutations were detected outside of exon 7, including 5 frameshift mutations in exon 8 and 6 missense mutations in exon 9. None of these 11 patients carried concomitant exon 7 mutations. Exon 8 frameshift mutations included insertions (n = 4) ranging from 2 to 23 base pairs, and one deletion of 22 base pairs, all of which were predicted to lead to a premature stop codon. Exon 9 mutations included substitutions at 3 amino acid residues known to be affected by constitutional WT1 mutations in Denys-Drash syndrome (DDS), a congenital anomaly syndrome consisting of the triad of ambiguous genitalia, congenital nephrotic syndrome, and predisposition to Wilms tumor. These amino acid changes included H397A (n = 1), D396N (n = 2), R394P (n = 2), and R394W (n = 1). Such DDS-associated exon 9 point mutations have been shown to decrease the DNA-binding affinity of the WT1 protein.20
Characteristics of the study population
Demographic, laboratory, and clinical characteristics of patients with or without WT1 mutations were compared (Table 1). There were no significant differences in sex, race, median diagnostic blast percentage, or median diagnostic WBC count between patients with and patients without WT1 mutations. However, WT1 mutations were less common in the youngest patients with AML aged from birth to 2 years; this age group accounted for 7.1% of patients with mutations, as opposed to 25% of those without (P = .001). Such mutations were also less common in patients with French-American-British class M5 (these accounted for 6% of patients with WT1 mutations and 18.7% of patients without; P = .014).
Characteristics of patients with or without WT1 mutations
| . | WT1 mutant . | WT1 wild-type . | P . |
|---|---|---|---|
| Study | |||
| CCG-2941 | 1 (1.4) | 38 (4.9) | .243 |
| CCG-2961 | 46 (65.7) | 495 (64.1) | .891 |
| AAML03P1 | 23 (32.9) | 239 (31.0) | .846 |
| Sex | |||
| Male | 37 (52.9) | 411 (53.2) | .949 |
| Female | 33 (47.1) | 361 (46.8) | |
| Age, y | |||
| Median (range) | 11.5 (0.85-18.3) | 10.1 (0.01-21.63) | .031* |
| 0-2 | 5 (7.1) | 193 (25.0) | .001* |
| 3-10 | 25 (35.7) | 225 (29.1) | .310 |
| 11-21 | 40 (57.1) | 354 (5.9) | .092 |
| Race | |||
| White | 47 (67.1) | 492 (65.1) | .829 |
| Black | 12 (17.1) | 85 (11.2) | .203 |
| Hispanic | 8 (1.4) | 125 (16.5) | .346 |
| Asian | 2 (2.9) | 26 (3.4) | .999 |
| Other | 1 (1.4) | 28 (3.7) | .503 |
| Unknown | 0 | 16 | |
| WBC count, × 103/μL, median (range) | 35 (1.2-3260) | 20.5 (0.3-860) | .145 |
| BM blasts, % | 75.5 (5-100) | 70 (0-100) | .122 |
| Platelet count, ×103/μL, median (range) | 54 (4-800) | 47.5 (2-46 000) | .252 |
| Hemoglobin level, g/dL, median (range) | 8.2 (3.1-13.7) | 8.3 (0.4-38.6) | .935 |
| FAB classification | |||
| M0 | 3 (4.5) | 32 (4.4) | .999 |
| M1 | 11 (16.4) | 119 (16.3) | .875 |
| M2 | 22 (32.8) | 202 (27.7) | .457 |
| M4 | 23 (34.3) | 191 (26.2) | .199 |
| M5 | 4 (6.0) | 136 (18.7) | .014* |
| M6 | 2 (3.0) | 15 (2.1) | .648 |
| M7 | 2 (3.0) | 33 (4.5) | .760 |
| Other/no data, n | 3 | 44 | |
| Cytogenetics | |||
| Normal | 18 (35.3) | 108 (20.0) | .017* |
| t(8;21) | 5 (9.8) | 87 (16.1) | .327 |
| inv(16) | 9 (17.6) | 62 (11.5) | .283 |
| Abnormal 11 | 4 (7.8) | 125 (23.1) | .019* |
| t(6;9)(p23;q34) | 3 (5.9) | 9 (1.7) | .076 |
| −7/7q− | 2 (3.9) | 18 (3.3) | .687 |
| −5/5q− | 0 (0.0) | 7 (1.3) | .999 |
| +8 | 3 (5.9) | 41 (7.6) | .999 |
| +21 | 0 (0.0) | 4 (0.7) | .999 |
| Pseudodiploid | 4 (7.8) | 31 (5.7) | .531 |
| Hyperdiploid | 2 (3.9) | 8 (1.5) | .210 |
| Hypodiploid | 0 (0.0) | 5 (0.9) | .999 |
| Other | 1 (2.0) | 36 (6.7) | .356 |
| Unknown, n | 19 | 231 | |
| FLT3/ITD status | |||
| ITD+ | 25 (35.7) | 76 (9.8) | < .001* |
| ITD− | 45 (64.3) | 696 (90.2) | |
| Missing, n | 0 | 0 | |
| FLT3 PM status | |||
| FLT3 PM+ | 5 (7.1) | 47 (6.1) | .611 |
| FLT3 PM− | 65 (92.9) | 725 (93.9) | |
| Missing, n | 0 | 0 | |
| CEBPA status | |||
| CEBPA mutant | 3 (4.5) | 28 (3.9) | .743 |
| CEBPA WT | 64 (95.5) | 688 (96.1) | |
| Missing, n | 3 | 56 | |
| NPM status | |||
| NPM mutant | 3 (4.3) | 43 (6.4) | .792 |
| NPM WT | 67 (95.7) | 632 (93.6) | |
| Missing, n | 0 | 97 | |
| Course 1 response | |||
| CR | 49 (72.1) | 603 (80.6) | .127 |
| Not in CR | 19 (27.9) | 145 (19.4) | |
| Unevaluable, n | 27 | 24 | |
| Course 2 response | |||
| CR | 44 (69.8) | 547 (77.6) | .355 |
| Not in CR | 19 (30.2) | 158 (22.4) | |
| Unevaluable, n | 7 | 67 |
| . | WT1 mutant . | WT1 wild-type . | P . |
|---|---|---|---|
| Study | |||
| CCG-2941 | 1 (1.4) | 38 (4.9) | .243 |
| CCG-2961 | 46 (65.7) | 495 (64.1) | .891 |
| AAML03P1 | 23 (32.9) | 239 (31.0) | .846 |
| Sex | |||
| Male | 37 (52.9) | 411 (53.2) | .949 |
| Female | 33 (47.1) | 361 (46.8) | |
| Age, y | |||
| Median (range) | 11.5 (0.85-18.3) | 10.1 (0.01-21.63) | .031* |
| 0-2 | 5 (7.1) | 193 (25.0) | .001* |
| 3-10 | 25 (35.7) | 225 (29.1) | .310 |
| 11-21 | 40 (57.1) | 354 (5.9) | .092 |
| Race | |||
| White | 47 (67.1) | 492 (65.1) | .829 |
| Black | 12 (17.1) | 85 (11.2) | .203 |
| Hispanic | 8 (1.4) | 125 (16.5) | .346 |
| Asian | 2 (2.9) | 26 (3.4) | .999 |
| Other | 1 (1.4) | 28 (3.7) | .503 |
| Unknown | 0 | 16 | |
| WBC count, × 103/μL, median (range) | 35 (1.2-3260) | 20.5 (0.3-860) | .145 |
| BM blasts, % | 75.5 (5-100) | 70 (0-100) | .122 |
| Platelet count, ×103/μL, median (range) | 54 (4-800) | 47.5 (2-46 000) | .252 |
| Hemoglobin level, g/dL, median (range) | 8.2 (3.1-13.7) | 8.3 (0.4-38.6) | .935 |
| FAB classification | |||
| M0 | 3 (4.5) | 32 (4.4) | .999 |
| M1 | 11 (16.4) | 119 (16.3) | .875 |
| M2 | 22 (32.8) | 202 (27.7) | .457 |
| M4 | 23 (34.3) | 191 (26.2) | .199 |
| M5 | 4 (6.0) | 136 (18.7) | .014* |
| M6 | 2 (3.0) | 15 (2.1) | .648 |
| M7 | 2 (3.0) | 33 (4.5) | .760 |
| Other/no data, n | 3 | 44 | |
| Cytogenetics | |||
| Normal | 18 (35.3) | 108 (20.0) | .017* |
| t(8;21) | 5 (9.8) | 87 (16.1) | .327 |
| inv(16) | 9 (17.6) | 62 (11.5) | .283 |
| Abnormal 11 | 4 (7.8) | 125 (23.1) | .019* |
| t(6;9)(p23;q34) | 3 (5.9) | 9 (1.7) | .076 |
| −7/7q− | 2 (3.9) | 18 (3.3) | .687 |
| −5/5q− | 0 (0.0) | 7 (1.3) | .999 |
| +8 | 3 (5.9) | 41 (7.6) | .999 |
| +21 | 0 (0.0) | 4 (0.7) | .999 |
| Pseudodiploid | 4 (7.8) | 31 (5.7) | .531 |
| Hyperdiploid | 2 (3.9) | 8 (1.5) | .210 |
| Hypodiploid | 0 (0.0) | 5 (0.9) | .999 |
| Other | 1 (2.0) | 36 (6.7) | .356 |
| Unknown, n | 19 | 231 | |
| FLT3/ITD status | |||
| ITD+ | 25 (35.7) | 76 (9.8) | < .001* |
| ITD− | 45 (64.3) | 696 (90.2) | |
| Missing, n | 0 | 0 | |
| FLT3 PM status | |||
| FLT3 PM+ | 5 (7.1) | 47 (6.1) | .611 |
| FLT3 PM− | 65 (92.9) | 725 (93.9) | |
| Missing, n | 0 | 0 | |
| CEBPA status | |||
| CEBPA mutant | 3 (4.5) | 28 (3.9) | .743 |
| CEBPA WT | 64 (95.5) | 688 (96.1) | |
| Missing, n | 3 | 56 | |
| NPM status | |||
| NPM mutant | 3 (4.3) | 43 (6.4) | .792 |
| NPM WT | 67 (95.7) | 632 (93.6) | |
| Missing, n | 0 | 97 | |
| Course 1 response | |||
| CR | 49 (72.1) | 603 (80.6) | .127 |
| Not in CR | 19 (27.9) | 145 (19.4) | |
| Unevaluable, n | 27 | 24 | |
| Course 2 response | |||
| CR | 44 (69.8) | 547 (77.6) | .355 |
| Not in CR | 19 (30.2) | 158 (22.4) | |
| Unevaluable, n | 7 | 67 |
Data are reported as n (%) except where otherwise noted. BM indicates bone marrow; FAB, French-American-British; PM, point mutation; and WT, wild-type.
Statistically significant (P = .05).
We also evaluated associations between WT1 mutations and cytogenetic and molecular alterations (Table 1). In terms of cytogenetics, WT1 mutations were found most frequently in the normal karyotype subset (35.3% of WT1mut patients had normal karyotype compared with 20.0% of those without WT1 mutations; P = .017). A substantial number (27.5%) of WT1mut patients with known cytogenetics were also found in the “favorable-risk,” core-binding factor (CBF-AML) cytogenetic subgroup. This was primarily because of a higher incidence of inv(16), which was present in 17.6% of WT1mut patients, but only to 11.5% of patients without WT1 mutations (P = .283); in addition, 9.8% of patients with WT1 mutations had t(8;21). An overlap between CBF translocations and WT1 mutations has not been previously reported. To verify our results, we confirmed the presence of the WT1 mutation, by fragment-length analysis, and the translocation, by reverse transcription–PCR, in diagnostic RNA from all patients with both the mutation and the cytogenetic abnormality; RNA testing confirmed the coexistence of both abnormalities in each patient. Two patients (3.9%) with WT1 mutations were classified as cytogenetically high risk because of the presence of monosomy 7.
Regarding other molecular alterations, there was also a substantial overlap between WT1 mutations and FLT3/ITD, ie, 35.7% of those carrying a WT1 mutation were FLT3/ITD positive as opposed to 9.8% of patients without WT1 mutations (P < .001). In addition, 11q23 alterations were rare in patients with WT1 mutations (7.8% vs 23.1%; P = .019) compared with patients with wild-type WT1. CEBPA mutations were found in 4.5% of those with WT1 mutations; NPM mutations occurred in 4.3%, and FLT3 point mutations occurred in 7.1%. These percentages did not differ significantly from the prevalence of these mutations in patients without WT1 mutations.
Clinical outcome and prognostic effect of WT1 mutations
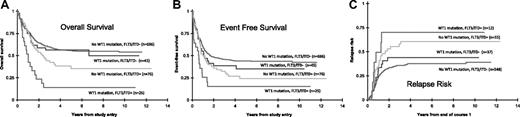
The CR rate was determined for all patients after the first course of induction therapy. Patients with WT1 mutations had a lower rate of CR (72.1%) compared with those without mutations (80.6%), but this difference was not statistically significant (P = .127). Clinical outcome data were examined for the 842 patients with known WT1 mutation status (Figure 2). Actuarial OS from study entry for patients with WT1 mutations was 41% plus or minus 13% versus 54% plus or minus 4% for those without WT1 mutations; (HR = 1.52; P = .017). Corresponding EFS was also significantly worse for those with WT1 mutations (28% ± 12% vs 42% ± 4%; HR = 1.50; P = .011). Of the 652 patients who achieved an initial CR, RR at 5 years from remission was higher for patients with WT1 mutations (51% ± 16%) than for patients without WT1 mutations (40% ± 4%; P = .136) but not significantly different. Corresponding DFS from CR in patients with and without WT1 mutations was 38% plus or minus 15% and 50% plus or minus 5%, respectively (HR = 1.39; P = .119).
Clinical significance of WT1 mutations in pediatric AML. Kaplan-Meier estimates show that patients with WT1 mutations have shorter (A) OS and (B) EFS from study entry than patients without these mutations. (C) Cumulative incidence of relapse was also worse for patients with WT1 mutations.
Clinical significance of WT1 mutations in pediatric AML. Kaplan-Meier estimates show that patients with WT1 mutations have shorter (A) OS and (B) EFS from study entry than patients without these mutations. (C) Cumulative incidence of relapse was also worse for patients with WT1 mutations.
Differences in outcome measures between patients with and without WT1 mutations were similar when analyses were restricted to patients having normal cytogenetics: (OS: 20% ± 22% vs 44% ± 11%, P = .024; EFS: 32% ± 24% vs 57% ± 11%, P = .012; RR: 40% ± 31% vs 44% ± 12%, P = .905; DFS: 31% ± 30% vs 46% ± 12%, P = .181). However, when outcome analyses were restricted to standard-risk patients (excluding those patients risk-stratified by virtue of favorable cytogenetics, unfavorable cytogenetics, CEBPA mutations, or FLT3/ITD), no significant differences were found between patients with and without WT1 mutations: (OS: 53% ± 24% vs 47% ± 6%, P = .935; EFS: 30% ± 21% vs 37% ± 6%, P = .334; RR: 58% ± 29% vs 45% ± 7%, P = .313; DFS: 33% ± 27% vs 46% ± 7%, P = .349).
Of the 842 patients in this study, 128 total patients (15%) underwent allogeneic hematopoietic stem cell transplantation (allo-HSCT) from a matched sibling donor after the third course of chemotherapy (intensification 1) for patients on AAML03P1 and after the second course of induction for patients on CCG-2941 and CCG-2961. For patients in CR by the end of induction 2 or intensification 1, we directly compared the outcome of WT1 mutation–positive patients who received an allo-HSC transplant (n = 9) with that of patients who were treated with chemotherapy only (n = 33). In patients with WT1 mutations, RR (cumulative incidence) at 5 years from the end of course 2 was 22% plus or minus 28% for the HSC transplant recipients versus 53% plus or minus 18% for patients who were treated with chemotherapy only (P = .159). Corresponding OS at 5 years from the end of course 2 was 78% plus or minus 28% versus 64% plus or minus 17% (P = .321) for HSC transplant recipients and chemotherapy-only recipients, respectively. Patients with WT1 mutations did appear to benefit from allo-HSCT, but differences were not statistically significant. Of note, however, of the 9 patients with WT1 mutations who underwent HSCT, only 1 patient was “high-risk” by virtue of FLT3/ITD with high allelic ratio. Within the subset of 128 patients who received a HSC transplant, we also compared the outcome of patients with WT1 mutations (n = 9) with patients without WT1 mutations (n = 119) and found that WT1 status did not have any significant effect on outcome after HSCT in our study (RR from transplantation, 22% ± 28% for WT1-mutated patients vs 14% ± 7% for WT1 wild-type patients [P = .546]; corresponding 5-year OS, 78% ± 28% for patients with WT1 mutations vs 72% ± 9% for patients without [P = .642]).
Given the substantial overlap between WTI and FLT3/ITD mutations, we examined outcome measures separately for WT1mut/ITD+ patients compared with WT1mu+/ITD− patients (Table 2). Only 12 of 23 patients (48%) with WT1mut/ITD+ achieved CR after induction 1, as opposed to 37 of 45 patients (82.2%) with WT1mut/ITD− (P = .020). Patients with both WT1 and FLT3/ITD mutations had an extremely dismal prognosis (OS, 15% ± 15%; EFS, 15% ± 16%), yet patients with WT1 mutations who were ITD negative had similar outcomes to patients who were WT1 wild-type and ITD negative (Table 2; Figure 3). OS from study entry was 56% plus or minus 16% for the WT1mut/ITD− patients compared with 56% plus or minus 4% for patients with neither mutation (P = .80). Corresponding EFS was 35% plus or minus 16% compared with 44% plus or minus 4%, respectively (P = .336), and 5-year RR from CR was similar between the 2 groups (44% ± 18% vs 38% ± 5%; P = .456). Thus, WT1mut/ITD+ patients had shorter survival due to both a higher rate of induction failure and a higher rate of disease recurrence, whereas WT1mut/ITD− patients had a similar outcome to patients who were wild type for both mutations. The prognostic significance of having a WT1 mutation was also determined in patients with a normal karyotype who were FLT3/ITD negative. No significant differences were found in the OS (51% ± 32% and 58% ± 12%, respectively; P = .31) or in the corresponding EFS values (31% ± 29% and 44% ± 12%, respectively; P = .408).
Clinical outcomes of patients stratified by WT1 and FLT3/ITD status
| . | WT1 wild-type . | WT1 mutant . | P . |
|---|---|---|---|
| CR rate at end of course 1 | |||
| FLT3/ITD neg, % | 81.5 | 82.2 | .932 |
| FLT3/ITD pos, % | 72.4 | 52.2 | .119 |
| P | .078 | .020 | |
| OS | |||
| FLT3/ITD neg, %, mean ± SD (n) | 56 ± 4 (696) | 56 ± 16 (45) | .8 |
| FLT3/ITD pos, %, mean ± SD (n) | 35 ± 13 (76) | 15 ± 15 (25) | .004 |
| P | .026 | .001 | |
| EFS | |||
| FLT3/ITD neg, %, mean ± SD (n) | 44 ± 4 (696) | 35 ± 16 (45) | .336 |
| FLT3/ITD pos, %, mean ± SD (n) | 24 ± 12 (76) | 15 ± 16 (25) | .036 |
| P | .017 | .019 | |
| RR | |||
| FLT3/ITD neg, %, mean ± SD (n) | 38 ± 5 (548) | 44 ± 18 (37) | .456 |
| FLT3/ITD pos, %, mean ± SD (n) | 61 ± 15 (55) | 70 ± 30 (12) | .403 |
| P | .005 | .115 | |
| FLT3/ITD, allelic ratio | .131 | ||
| Low, n (%) | 25 (33) | 4 (16) | |
| High, n (%) | 51 (67) | 21 (84) |
| . | WT1 wild-type . | WT1 mutant . | P . |
|---|---|---|---|
| CR rate at end of course 1 | |||
| FLT3/ITD neg, % | 81.5 | 82.2 | .932 |
| FLT3/ITD pos, % | 72.4 | 52.2 | .119 |
| P | .078 | .020 | |
| OS | |||
| FLT3/ITD neg, %, mean ± SD (n) | 56 ± 4 (696) | 56 ± 16 (45) | .8 |
| FLT3/ITD pos, %, mean ± SD (n) | 35 ± 13 (76) | 15 ± 15 (25) | .004 |
| P | .026 | .001 | |
| EFS | |||
| FLT3/ITD neg, %, mean ± SD (n) | 44 ± 4 (696) | 35 ± 16 (45) | .336 |
| FLT3/ITD pos, %, mean ± SD (n) | 24 ± 12 (76) | 15 ± 16 (25) | .036 |
| P | .017 | .019 | |
| RR | |||
| FLT3/ITD neg, %, mean ± SD (n) | 38 ± 5 (548) | 44 ± 18 (37) | .456 |
| FLT3/ITD pos, %, mean ± SD (n) | 61 ± 15 (55) | 70 ± 30 (12) | .403 |
| P | .005 | .115 | |
| FLT3/ITD, allelic ratio | .131 | ||
| Low, n (%) | 25 (33) | 4 (16) | |
| High, n (%) | 51 (67) | 21 (84) |
Outcomes stratified by combined WT1 and FLT3/ITD mutation status. When stratifying patients by combined FLT3/ITD and WT1 status, patients with both mutations had the worst (A) OS, (B) EFS, and (C) RR. Of the 2 mutations, FLT3/ITD imparts a stronger negative effective on survival outcome.
Outcomes stratified by combined WT1 and FLT3/ITD mutation status. When stratifying patients by combined FLT3/ITD and WT1 status, patients with both mutations had the worst (A) OS, (B) EFS, and (C) RR. Of the 2 mutations, FLT3/ITD imparts a stronger negative effective on survival outcome.
Conversely, the presence of FLT3/ITD remained a significant marker of unfavorable prognosis, even in the patients without WT1 mutations (Table 2; Figure 3). WT1−/ITD+ patients had significantly lower OS (35% ± 13% vs 56% ± 4%; P = .026) and EFS (24% ± 12% vs 44% ± 4%; P = .017) than did patients who were wild type for both mutations. Corresponding RR was 61% plus or minus 15% for WT1−/ITD+ patients. As expected, patients with FLT3/ITD with a high allelic ratio (AR > 0.4)21 had an even worse outcome. This remained true in the cohort of patients negative for WT1 mutations, whereby patients with FLT3/ITD with high AR had an OS of 29% plus or minus 16%, EFS of 17% plus or minus 12%, and an RR of 77% plus or minus 15%.
Subset analysis was then performed for patients with CBF-AML. Patients with both WT1mut and CBF-AML had similar OS and EFS compared with patients with CBF-AML and wild-type WT1 (OS: 77% ± 23% vs 74% ± 8%, P = .972; EFS: 60% ± 28% vs 58% ± 9%, P = .885). DFS (65% ± 29% vs 60% ± 9%) and RR (28% ± 27% vs 27% ± 9%) were also not significantly different, suggesting that the favorable prognostic effect of the CBF translocation “trumps” the prognostic effect of the WT1 mutation.
Outcomes for those with “double,” biallelic WT1 mutations were compared with outcomes for patients with single WT1 mutations. Although numbers were too small to detect significant differences, patients with biallelic mutations appeared to have worse outcome than patients with single mutations (OS: 33% ± 24% vs 43% ± 15%, HR = 1.22, P = .587; EFS: 20% ± 21% vs 31% ± 14%, HR = 1.48, P = .249).
Prognostic factors
We performed Cox regression analyses to evaluate the status of the following mutations as predictors of OS and RFS in separate univariate models: WT1, NPM, CEBPA, and FLT3/ITD. In univariate analysis, the presence of FLT3/ITD was the strongest predictor of decreased survival (HR = 1.77; P < .001) and increased risk of relapse (HR = 1.87; P < .001). WT1 mutations also predicted poor outcome in the univariate model (lower OS: HR = 1.52, P = .017; lower RFS: HR = 1.49, P = .092). CEBPA mutations predicted improved OS (HR = 0.43; P = .027) and RFS (HR = 0.42; P = .052), whereas NPM mutations were not prognostic in these analyses.
Cox regression analyses were then performed to evaluate WT1 mutation status as a predictor of EFS and RR alongside the following known prognostic groups: high diagnostic WBC count, high-risk cytogenetic/molecular features (defined as −5/del(5q), −7, or presence of FLT3/ITD with AR > 0.4), as well as favorable-risk cytogenetic/molecular features (CBF-AML or presence of CEBPA mutation). These factors were analyzed as predictors of EFS and RR in univariate and multivariate models (Table 3). In the univariate model, the presence of a WT1 mutation was a significant prognostic factor for decreased EFS with an HR of 1.50 (P = .011). In separate univariate models, high-risk cytogenetic/molecular features (HR = 1.79; P < .001), and diagnostic WBC count greater than 50 × 109/L (HR = 1.32; P < .001) were associated with worse EFS. Patients with favorable-risk cytogenetic/molecular features had superior OS (HR = 0.51; P < .001) and RFS (HR = 0.47; P < .001). In a multivariate model that included the above-mentioned prognostic factors, the presence of WT1 mutations loses independent prognostic significance for EFS (P = .105) and RFS (P = .261), whereas WBC count greater than 50 × 109/L, the presence of high-risk cytogenetic or molecular features, or the presence of favorable-risk cytogenetic or molecular features all remained as independent prognostic factors for EFS. Only WBC count greater than 50 × 109/L did not remain significant in a multivariate model for RFS.
Cox regression analysis of WT1 mutations and other specific prognostic factors
| . | EFS from study entry . | RFS from end of course 1 . | ||||||
|---|---|---|---|---|---|---|---|---|
| N . | HR . | 95% CI . | P . | N . | HR . | 95% CI . | P . | |
| Univariate Cox analyses | ||||||||
| WT1 mutation present (vs absent) | 70 | 1.50 | 1.10-2.05 | .011 | 49 | 1.49 | 0.94-2.36 | .092 |
| High-risk cytogenetic/molecular (vs standard) | 123 | 1.79 | 1.4-2.28 | < .001 | 79 | 1.94 | 1.39-2.72 | < .001 |
| Favorable-risk cytogenetic/molecular (vs standard) | 249 | 0.51 | 0.40-0.66 | < .001 | 227 | 0.47 | 0.34-0.65 | < .001 |
| WBC count greater than 50 × 109/L (vs less than 50 × 109/L | 388 | 1.32 | 1.13-1.56 | < .001 | 283 | 1.26 | 1.00-1.58 | .047 |
| Multivariate Cox analyses | ||||||||
| WT1 mutation present (vs absent) | 59 | 1.33 | 0.94-1.88 | .105 | 41 | 1.33 | 0.81-2.20 | .261 |
| High-risk cytogenetic/molecular (vs standard) | 98 | 1.52 | 1.14-2.04 | .004 | 63 | 1.93 | 1.29-2.89 | .002 |
| Favorable-risk cytogenetic/molecular (vs standard) | 189 | 0.50 | 0.38-0.66 | < .001 | 171 | 0.48 | 0.32-0.70 | < .001 |
| WBC count greater than 50 × 109/L (vs less than 50 × 109/L) | 212 | 1.35 | 1.08-1.69 | .010 | 151 | 1.26 | 0.91-1.74 | .158 |
| . | EFS from study entry . | RFS from end of course 1 . | ||||||
|---|---|---|---|---|---|---|---|---|
| N . | HR . | 95% CI . | P . | N . | HR . | 95% CI . | P . | |
| Univariate Cox analyses | ||||||||
| WT1 mutation present (vs absent) | 70 | 1.50 | 1.10-2.05 | .011 | 49 | 1.49 | 0.94-2.36 | .092 |
| High-risk cytogenetic/molecular (vs standard) | 123 | 1.79 | 1.4-2.28 | < .001 | 79 | 1.94 | 1.39-2.72 | < .001 |
| Favorable-risk cytogenetic/molecular (vs standard) | 249 | 0.51 | 0.40-0.66 | < .001 | 227 | 0.47 | 0.34-0.65 | < .001 |
| WBC count greater than 50 × 109/L (vs less than 50 × 109/L | 388 | 1.32 | 1.13-1.56 | < .001 | 283 | 1.26 | 1.00-1.58 | .047 |
| Multivariate Cox analyses | ||||||||
| WT1 mutation present (vs absent) | 59 | 1.33 | 0.94-1.88 | .105 | 41 | 1.33 | 0.81-2.20 | .261 |
| High-risk cytogenetic/molecular (vs standard) | 98 | 1.52 | 1.14-2.04 | .004 | 63 | 1.93 | 1.29-2.89 | .002 |
| Favorable-risk cytogenetic/molecular (vs standard) | 189 | 0.50 | 0.38-0.66 | < .001 | 171 | 0.48 | 0.32-0.70 | < .001 |
| WBC count greater than 50 × 109/L (vs less than 50 × 109/L) | 212 | 1.35 | 1.08-1.69 | .010 | 151 | 1.26 | 0.91-1.74 | .158 |
Discussion
In this retrospective study, we report that mutations in the zinc-finger domains of the WT1 gene were detected in 70 of 842 patients with pediatric AML, corresponding to 8.3% of our study population, and that these mutations were significantly associated with unfavorable outcome in univariate, but not multivariate, analysis. This is the largest study of WT1 mutations in AML to date. We demonstrate that WT1 mutations are associated with shorter OS and EFS as well as higher risk of relapse. However, we found a substantial overlap between WT1 mutations and other established prognostic markers already in clinical use, notably FLT3/ITD and CBF translocations. The presence of WT1 mutations as well as CBF translocations was confirmed at the RNA level in these patients, as this is an association that has not been previously reported.
We found that WT1 mutation status lacks independent prognostic significance in multivariate analysis, including other established prognostic markers. This is in contrast to adult studies reported by the CALGB4 and the MRC.5 We also recently reported that KIT mutations lack independent prognostic significance in pediatric CBF-AML,22 as opposed to their widely reported association with unfavorable outcome in adult CBF-AML. These differences in the prognostic value of molecular markers in pediatric versus adult AML may reflect underlying differences in disease biology or differences in the interaction of this biology with pediatric versus adult AML treatment schemas. Routine mutation screening of the WT1 gene at diagnosis, for the purpose of risk stratification in future pediatric trials, is not warranted on the basis of the results of our study.
The overlap between WT1 mutations and FLT3/ITD has both biologic as well as prognostic implications. Subset analysis identifies the WT1mut/FLT3/ITD+ group as particularly high risk, suggesting the chemoresistance and resultant effect on outcome conferred by these mutations is potentiated when they occur together. These patients have dismal survival outcomes due to both failure of induction chemotherapy as well as higher rates of recurrence, features known to be associated with FLT3/ITD+ disease. These patients are already stratified into the maximally intensive therapy arm on the basis of FLT3 status alone on current COG protocols. Notably, when excluding the FLT3/ITD+ subgroup in our study, patients with WT1 mutations in the absence of FLT3/ITD had similar outcomes to patients who had neither mutation. Although the prior pediatric AML study reported independent prognostic significance for WT1 mutations, Hollink et al14 also failed to detect a statistically significant difference in OS between WT1mut and WT1 wild-type patients when restricting analysis to FLT3/ITD− patients. In contrast with the prior pediatric study, our study finds that FLT3/ITD remains a significant predictor of poor outcome even in patients without WT1 mutations. The relative sizes of the studies may account for this difference, because our study may have been large enough to detect a difference that was not statistically significant in the Dutch study. In addition, had allelic ratio been used to stratify patients into a high-risk FLT3/ITD group, Hollink et al14 may have been able to demonstrate a more pronounced difference in outcome that was based on ITD status.
When considering only the exons comprising the zinc-finger domains, WT1 mutations were found in approximately 10% of patients in 3 preceding adult AML studies.4,5,7 Note that these 3 studies restricted their cohorts to normal-karyotype AML. The mutation incidence of 8.7% in our study is higher than the 5% reported in unselected adult patients with AML by the Acute Leukemia French Association6 but less than the 12% incidence reported in the prior pediatric study.14 This discrepancy may be explained in part by the fact that Hollink et al14 reported rare missense mutations in exon 7; although we discovered 3 such mutations on confirmatory sequencing of patients with length mutations in the opposite allele, our method of screening by fragment-length analysis in exon 7 would not detect patients who harbored only missense mutations without concomitant insertions or deletions.
Most of the WT1 mutations in our study were found in exon 7, as has been previously described. We also detected 5 novel frameshift mutations in exon 8. The frameshift mutations in exons 7 and 8 are predicted to result in a truncated protein lacking a significant portion of the zinc-finger domains. In exon 9, nearly all of the previously described mutations are single base pair substitutions; we detected 4 such missense mutations. Five of the exon 9 mutations in our study replaced either the arginine at residue 394 or the aspartic acid at residue 396 with a different amino acid. Because R394 and D396 are the residues most commonly affected by germline WT1 mutations in DDS, functional studies have been performed which show mutations at these codons abrogate the WT1 protein's DNA-binding capacity.23 The remaining exon 9 mutation in our cohort was H397A, an amino acid residue also implicated in some cases of DDS. Replacing histidine with a different amino acid would disrupt the precise cysteine-histidine spacing that is crucial to the structure of the zinc-finger. The zinc-finger domains in WT1 are responsible for DNA binding, nuclear localization, and protein interaction. Mutations leading to loss, or alteration, of the zinc-fingers may lead to the loss of these integral functions.
All of the mutations we detected in the zinc-finger domains of WT1 are predicted to either abolish or diminish the function of the WT1 protein. Inactivation of both alleles would be predicted to completely eliminate normal WT1 function. We detected mutations affecting both alleles in 15 of the 70 patients with WT1 mutations; in each case of “double” mutation, both mutations were found in exon 7. The true incidence of biallelic double mutations may be higher than reported in our study, because mutations in C-terminal exons, which we did not screen, have been reported rarely in conjunction with exon 7 mutations.14 Four of the biallelic mutations we detected appear to be homozygous by direct sequencing; previous work has shown that segmental uniparental disomy may be a mechanism of acquiring loss of heterozygosity of a mutated WT1 gene in AML.24 We did not detect any double mutations involving exon 9. There are data to suggest that exon 9 missense mutations may behave in a dominant-negative manner,23 in which case biallelic inactivation would not be required for complete loss of function.
The exact effect of the loss of function of WT1 on either the development or progression of leukemia is unknown. The traditional model of molecular-genetic cooperativity in myeloid leukemogenesis posits that “class II” events, which impair differentiation, must be coupled with “class I” events, which confer a proliferative advantage.25 In our study, WT1 mutations show significant overlap with CBF translocations, which are classic class II events, as well as with the class I mutation FLT3/ITD, so the role of WT1 mutations in the stepwise evolution of leukemia is uncertain. In addition, although WT1 mutations in AML appear to result in loss of WT1 function, marked overexpression of wild-type WT1 is a common finding in AML.26–28 Further, in one study, patients with pediatric AML with WT1 exon 7 mutations had significantly higher levels of WT1 expression than patients with wild-type WT1.28 This apparent contradiction, in which a single gene might function as both an oncogene as well as a tumor suppressor, may stem from the ability of the WT1 protein to function either as a transcriptional activator or repressor, depending on a multitude of factors.12 There is much still to be learned about the biology of WT1 in AML; further insight into the roles that this fascinating gene plays in leukemogenesis may eventually pave the way for targeted therapies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the AML Reference Laboratories of the COG for providing diagnostic specimens. We thank the patients and families who consented to the use of biologic specimens in these trials.
This work was supported by the American Society of Clinical Oncology Young Investigator Award and the University of Washington Child Health Research Center New Investigator Award (P.A.H.), the National Institutes of Health (grants R01 CA114563 and R21 CA10262-01; S.M.), and the Children's Oncology Group Chair (grant NIH U10 CA98543).
National Institutes of Health
Authorship
Contribution: P.A.H. and R.Z. designed and performed research, analyzed data, and wrote the manuscript; T.A.A. served as senior statistician, performed statistical analyses, and edited the manuscript; R.B.G. performed statistical analyses and edited the manuscript; K.L.M. performed research and edited the manuscript; J.A.P., D.L.S., N.A.H., S.C.R., B.H., J.L.F., and B.L. analyzed data and edited the manuscript; and S.M. designed the research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the Children's Oncology Group appears in the supplemental Appendix.
Correspondence: Soheil Meshinchi, Fred Hutchinson Cancer Research Center, Clinical Research Division, D5-380, 1100 Fairview Ave N., Seattle, WA 98103; e-mail: smeshinc@fhcrc.org.
References
Author notes
P.A.H. and R.Z. contributed equally to this study.