Abstract
Monocytes are versatile cells that can express different functional programs in response to microenvironmental signals. We show that primary blood monocytes secrete the CXCL12 chemokine, and express the CXCR4 and CXCR7 receptors, leading to an autocrine/paracrine loop that contribute to shape monocyte differentiation to a distinct type of macrophages, with an enhanced expression of CD4, CD14, and CD163, or dendritic cells, with a reduced functional ability to stimulate antigen-specific T-lymphocyte responses. The in vivo relevance of CXCL12 production by mononuclear phagocytes was studied in metastatic melanoma tissues by a thoroughly immunofluorescence phenotyping of CXCL12high expressing cells, which were CD45+, coexpressed the macrophage antigens CD68, CD163, and CD209 and constituted the 60%-90% of tumor-associated macrophages. Microarray analysis of primary monocytes revealed that the vascular endothelial growth factor and the angiogenic chemokine CCL1 mRNA levels were up-regulated in response to CXCL12, leading to enhanced expression of both proteins. In addition, we found that CXCL12 autocrine/paracrine signaling down-regulates the expression of the transcription factor RUNX3 and contributes to maintain the long-term CD4 and CD14 expression in monocytes/macrophages. Together, these results suggest that autocrine CXCL12 production modulates differentiation of monocytes toward a distinct program with proangiogenic and immunosuppressive functions.
Introduction
Monocytes are not fully differentiated cells, derived from the bone marrow, that circulate in blood during 1 to 3 days and enter peripheral tissues to give rise to a heterogeneous lineage of mononuclear phagocytes. Monocytes are highly recruited into foci of active inflammation, but they also enter into healthy tissues as part of the constitutive or steady-state trafficking to become resident tissue macrophages.1 In response to inflammation or microbial products, such as the pro-Th1 stimuli interferon-γ and granulocyte macrophage colony-stimulating factor (GM-CSF) or lipopolysaccharide (LPS), blood monocytes differentiate to inflammatory macrophages (M1) and dendritic cells (DCs). In contrast, in response to pro-Th2/anti-inflammatory stimuli (interleukin-4 [IL-4], IL-10 and M-CSF) monocytes become anti-inflammatory macrophages (M2).2 In vitro, human monocytes can differentiate into DCs in response to GM-CSF and IL-4 or into GM-CSF–derived macrophages (M1), or M-CSF–driven macrophages (M2). In the tumor microenvironment, incoming monocytes are influenced by tumor-derived growth factors, especially M-CSF, IL-10, IL-6, and transforming growth factor-β (TGF-β), which switch monocyte differentiation toward M2 macrophages rather than M1 or DCs.3,4 Thus, monocytes are versatile cells that can express different functional programs in response to environmental signals.
Blood monocytes are recruited into tissues in response to chemoattractants, most of which belong to the chemokine family.5 Chemokines are small (8-14 kDa) secreted proteins that regulate cell trafficking through interactions with a subset of 7—transmembrane, G-protein coupled receptors. The chemokine CXCL12 (stromal cell-derived factor-1, SDF-1) plays a critical role in monocyte extravasation and is both expressed under homeostatic conditions at sites where resident macrophages are present, and highly inducible in some pathologic conditions involving ischemia and/or hypoxia and in proangiogenic environments, such as tumors.6 CXCL12 is expressed by stromal cells in several tissues and organs, including skin, thymus, lymph nodes, lung, liver, and bone marrow. Monocytes express at the plasma membrane the 2 cognate receptors for CXCL12, CXCR4, and CXCR7.7 Binding of CXCL12 to CXCR4 activates multiple downstream pathways that mediate the migration and survival of lymphocytes and monocytes and that regulate gene expression through activation of nuclear factor κB,8 Erk/ELK pathway,9 AP-110 and JAK/STAT activation.11 Functional levels of CXCR4 in monocytes are transiently elevated at the beginning of differentiation, followed by down-regulation in macrophages.12 Recent works indicate that CXCR7 signals through β-arrestin in the absence of G protein activation or by its ability to alter CXCR4-mediated signaling through receptor heterodimerization.13,14
Microenvironmental signals, which operate through the control of gene expression by transcription factors, dictate the terminal differentiation process of monocytes. The RUNX family of context-dependent transcriptional regulators controls proliferation, hematopoiesis, neurogenesis, and lineage-specific gene expression in major developmental pathways.15,16 In the myeloid lineage, RUNX3 mediates TGF-β signaling in DCs and is essential for the generation of Langerhans cells.17 Thus, the transcriptional factor RUNX3 is a good candidate for regulating monocyte differentiation.
The role of chemokine CXCL12 in the immune system has been focused in the ability to orchestrate directional migration to selected tissues of a wide spectrum of cells. However, less is known on the transcriptional events elicited in response to this chemokine. In this work, we have investigated the effect of CXCL12 on monocyte differentiation. Our data indicate that CXCL12, by an autocrine/paracrine mechanism, regulates a distinct program of macrophage differentiation, characterized by the expression of the angiogenic factors vascular endothelial growth factor (VEGF) and CCL1. Interestingly, we found CD163+CD209+CXCR4+ macrophages at tumoral perivascular sites, which also highly expressed CXCL12. As a part of the transcriptional program induced by CXCL12, we show the down-regulation of RUNX3 expression, which maintains CD4 and CD14 expression in mononuclear phagocytes.
Methods
Cells and transfections
Hospital General Universitario Gregorio Marañón (HGUGM) Ethical Committee approved experimentation involving human cells. Peripheral blood mononuclear cells were obtained from healthy donors by Ficoll density gradient centrifugation. Monocytes were isolated with anti-CD14–labeled magnetic microbeads or monocyte isolation kit (Miltenyi Biotec) obtaining an enrichment of 99% and 95%, respectively. Primary cells, monocytic THP-1, and U-937, and lymphoid HSB-2 cell lines were cultured in Teflon dishes in RPMI 10% fetal calf serum (FCS). Monocyte-DC in vitro differentiation was induced with GM-CSF (1000 U/mL) plus IL-4 (1000 U/mL; R&D Systems) and maturation with LPS from Escherichia coli 055:B5 (10 ng/mL; Sigma-Aldrich). Monocytes were differentiated to macrophages with M-CSF (10 ng/mL). THP-1 cells were transfected with 2 μg of small interfering RNA (siRNA) by using diluted X-treme reagent (Roche Applied Science). The silencing efficiency was checked by flow cytometry or Western blot within 24-48 hours, and THP-1 siRNA transfectants were assayed 48 hours posttransfection.
Reagents, antibodies, and siRNAs
The chemokine CXCL12 was from R&D Systems. The CXCR7 antagonist CCX733 was kindly provided by Dr Penfold (Chemocentryx). The CXCR4 antagonist AMD3100 was from Sigma-Aldrich. Antibodies were as follows: RUNX3 (Alexis Biochemicals); GAPDH, FITC-CD163 (Santa Cruz Biotechnology); anti–human melanoma HMB-45 and Factor VIII (DakoCytomation); Tie-2 (Biolegend); FITC-CD14, percP-CD1a, PercP-HLA-DR (anti–MHC-II), PE-CD80, PE-CD83, PE-CD86, FITC-CD209, PE-CXCR4 clone 12G5, and FITC-CD90 (Becton Dickinson); CXCR4 clone 44 717, CXCR4 clone 12G5, CXCR7-PE clone 11G8, and goat anti–human CXCL12 were form R&D Systems; CXCR7 antibody clone 11G8 (Chemocentryx); VE-Cadherin was kindly provided by Dr Sánchez-Madrid (Hospital de la Princesa, Madrid, Spain); rabbit anti–human CXCL12, biotinylated anti–human CXCL12, rabbit anti–human M-CSF, biotinylated anti–human M-CSF (Peprotech). Fluorochrome-conjugated antibodies were from Jackson ImmunoResearch Laboratories. RUNX3 siRNA (ID no. 107390 and ID no. 115509), CXCR7 siRNA (ID no. 109229), and negative control siRNA (AM4611) were from Ambion.
Cell migration
Freshly isolated monocytes (1 × 105 cells) were pretreated or not with CXCR4 (AMD3100; 25 μg/mL) or with CXCR7 (CCX733; 100nM) antagonists for 3 hours or with anti-CXCR4 and/or anti-CXCR7 monoclonal antibody (mAb) at 50 μg/mL for 1 hour in RPMI at 37°C to block recycling receptors. Monocytes were allowed to migrate to CXCL12 (100 ng/mL) for 1 hour and quantified as previously described.18 Silencing of CXCR7 was performed in THP-1 cells, and the effect on CXCL12 migration was evaluated within the 48 hours after transfection. Relative migration to CXCL12 was represented. Assays were run in duplicate.
Antigen-specific T-lymphocyte proliferation assay
Monocytes were derived to DCs, in the absence or presence of CXCL12 (100 ng/mL). At day 6, purified protein derivative (PPD; 50 IU/mL), tetanus toxoid (TT; 10 fl/mL), or no antigen were added, and 4 hours later, cells were treated with LPS (10 ng/mL) for 1 more day to obtain mature DCs. Autologous T lymphocytes were purified (Pan-T cell isolation kit; Miltenyi Biotec) and labeled with carboxyfluorescein succinimidyl ester (CFSE). Pulsed mature DCs were washed, counted, and adjusted at ratios of 1:10 DC/T cells in 300-μL final volumes of medium (RPMI 1640 supplemented with 10% autologous serum). Assays were performed in triplicates. After 6 days of coculture, lymphocyte proliferation was assessed by the CFSE-dilution method.
Microarrays
CD14+ monocytes (2.5 × 106 cells) isolated from 3 different blood donors were cultured in Teflon dishes for 1 hour in RPMI 10% FCS and then, were treated or not with CXCL12 for 6 hours. Total RNA from each condition was extracted and purified using the RNeasy kit (QIAGEN). Labeled RNA was used as hybridization probes on human Codelink Whole Genome Bioarray. All experimental procedures were performed following manufacturer instructions. Microarrays were scanned with a GenePix 4000B (Axon Instruments) scanner. Scanned images and raw data were processed using the Codelink Expression Software. Log2-transformed intensity values were used for gene expression analysis. The ratio of Log2 intensity data corresponding to untreated monocytes versus CXCL12 stimulated monocytes was calculated for each single blood donor (n = 3). All microarray data have been deposited into the Gene Expression Omnibus (GEO) database under accession number is GSE24496.
Real-time PCR and qualitative PCR
Total RNA from 2.5 × 105 cells was extracted with the RNasey kit. Total RNA and cDNAs were quantified with nanodrop (Thermo Fisher Scientific). The GeneAmp Gold RNA was performed using Lightcycler FastStart DNA Master plus SYBR Green I kit in a Capillar Lightcycler (Roche). The reactions were carried out following the manufacturer instructions. The DNA Master SYBR Green I mix contained 4mM MgCl2. For RUNX3 amplification, annealing temperatures at 56°C were used. The specificity of the polymerase chain reaction (PCR) product was determined by melting curve analysis and by agarose gel eletrophoresis. Specific primers were used for RUNX3 forward primer 5′-TCGGAACTGAACCCATTCTC-3′ and reverse 5′-GCCAGAGGATGTCCGTAGT-3′; GAPDH forward primer 5′-GAGTCAACGGATTTGGTCGT-3′ and reverse 5′-GGTGCTAAGCAGTTGGTGGT-3′ as control. Oligonucleotides for CCL1 were designed according to the Roche software for qualitative PCR (qPCR), and RNA was amplified using the Universal Human Probe library (Roche). Conventional RUNX3 PCRs were performed using GC-rich PCR System (Roche), and gel was stained with SYBR Gold (Molecular Probes). Results were normalized according to the expression levels of 18S and GAPDH RNA.
ELISA and flow cytometry
Supernatants from 48 hours cultured cells (1 × 106 cells) were collected and frozen. Human VEGF enzyme-linked immunosorbent assay (ELISA) kit, anti–human CXCL12, and anti–human M-CSF antibodies for capture and detection were from Peprotech. Human IL-10 ELISA kit was from Immunotools. The assays were performed following manufacturer instructions. Samples were run in triplicate.
Direct staining of blood samples with anti–CXCR7-PE or anti–CXCR4-PE, was followed by Facs lysis solution (BD). Detection of membrane CXCR4 and CXCR7 receptors were performed incubating cells for 15 minutes at 4°C with the appropriate antibodies diluted in phosphate-buffered saline with 2% FCS, 0.01% NaN3. Simultaneous permeabilization and fixation was performed with 1.85% formaldehyde 0.2% Triton X-100 in phosphate-buffered saline for 15 minutes. Intracellular CXCL12 and CCL1 were detected by flow cytometric analysis using Cytofix/Cytoperm kit (BD). For RUNX3 intracellular staining cells were fixed with 2% formaldehyde for 10 minutes at 37°C, and permeabilized with ice-cold 90% methanol for 30 minutes at 4°C and frozen. Cells were analyzed on a FACScan cytometer using CellQuest software (BD).
Confocal microscopy and immunohistochemistry
HGUGM Ethical Committee approved the protocols and process involving melanoma samples. Melanoma metastases were immediately frozen, and acetone-fixed 4-μm–thick sections were first incubated with 10% nonimmune goat serum (Zymed). Samples were then incubated for 1 hour at 23°C with the appropriated antibodies or isotype-matched control antibodies. All antibodies were first tested for reactivity on lymphoid tissue. The slides were mounted using an aqueous permanent mounting medium (DAKO). Confocal microscopy images were captured using a Leica SP2-AOBS confocal microscope.
Western blot
Cells (2.5 × 106) were washed and then lysated in sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis buffer (100mM Tris-HCl, pH 6.8, 0.05mM sodium orthovanadate, 1mM EDTA [ethylenediaminetetraacetic acid], 3% SDS, 25mM dithiothreitol, 5% glycerol). Proteins were resolved by SDS polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes that were subsequentially incubated with 0.2 μg/mL anti-Runx3 antibody and horseradish peroxidase-conjugated secondary antibodies. Immunoreactive bands were visualized using enhanced chemiluminescence reagents.
Statistical analysis
Results are given as means ± standard deviation (SD), and statistical significance was determined. The Wilcoxon test was used for nonparametric data analysis. When normal distribution of data were assessed with the Kolmogorov-Smirnov test, means of 2 groups were compared with the Student paired t test. Statistical significance was determined as P values of < .05. SPSS 15.0 software was used for statistical analyses.
Results
CXCR4 and CXCR7 are expressed by blood monocytes and both are necessary for CXCL12-mediated chemotaxis
The chemokine CXCL12 has 2 known receptors, the initially described CXCR4 and the second cognate receptor CXCR7.7 CXCL12-CXCR4 interaction mediates migration of peripheral blood monocytes (PBMos), but the role of CXCR7 in monocyte chemotaxis is yet unexplored. Controversial results and hypothesis have been proposed for CXCR7 function in different migration models and cell types.19,20 Recent works indicate that the role of CXCR7 in cell migration is variable and depends on the particular cell system.21,22 We first analyzed the surface expression of CXCR4 and CXCR7 in whole blood samples by flow cytometry. Compared with the high CXCR4-expression of PBMos, CXCR7-surface expression showed high interindividual variability among blood donors, ranging from high to low (Figure 1A and data not shown). However, a significant level of total CXCR7 was detected after monocyte permeabilization (Figure 1A). In primary lymphocytes and the THP-1 (monocytic) and HSB-2 (lymphoid) cell lines, which were included for comparison, CXCR7 was marginally expressed at the cell surface and mainly detected in a large intracellular pool (Figure 1A and supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We next assessed the effect of receptor blocking with specific mAbs, as well as with receptor antagonists, AMD310023 for CXCR4 and CCX73320 for CXCR7, in the CXCL12-mediated chemotaxis of blood monocytes (Figure 1B). Either the CXCR4 or the CXCR7-blocking mAbs inhibited CXCL12-triggered PBMo chemotaxis by approximately 50%, whereas a combination of both blocking mAbs almost completely abrogated the chemotactic response. In addition, either AMD3100 or CCX733, significantly reduced the CXCL12-induced migratory response, whereas they did not affect cell viability at the doses used (Figure 1B and supplemental Figure 1B). The involvement of CXCR7 in CXCL12-mediated chemotaxis was further analyzed using CXCR7 depletion by siRNA in THP-1 cells (supplemental Figure 1C), which reduced the migratory response compared with control siRNA cells (Figure 1B). Thus, both CXCR4 and CXCR7 receptors are expressed by primary monocytes and contribute to CXCL12-mediated chemotaxis.
Expression of CXCL12 receptors, CXCR4 and CXCR7, in blood monocytes and contribution to CXCL12 mediated-chemotaxis. (A) CXCR4 and CXCR7 immunofluorescence of blood monocytes from 2 different donors (top and middle panels), and the THP-1 monocytic cell line (bottom panel). Unpermeabilized (membrane, left and middle) and permeabilized (total, right) cells were analyzed by flow cytometry. (B) Monocytes were pretreated with the CXCR4 antagonist (AMD3100; 25 μg/mL), with the CXCR7 antagonist (CCX733; 100nM) or with anti-CXCR4/CXCR7 antibodies (50 μg/mL) and allowed to migrate to CXCL12. Untreated (−) cells were used as migration control (100%). CXCL12-induced chemotaxis of CXCR7 siRNA-transfected THP-1 cells was compared with control siRNA-transfected cells. The percentages of migration referred to CXCL12-mediated chemotaxis are shown. Mean ± SD; n = 3; *P < .05, **P < .005, ***P < .0005.
Expression of CXCL12 receptors, CXCR4 and CXCR7, in blood monocytes and contribution to CXCL12 mediated-chemotaxis. (A) CXCR4 and CXCR7 immunofluorescence of blood monocytes from 2 different donors (top and middle panels), and the THP-1 monocytic cell line (bottom panel). Unpermeabilized (membrane, left and middle) and permeabilized (total, right) cells were analyzed by flow cytometry. (B) Monocytes were pretreated with the CXCR4 antagonist (AMD3100; 25 μg/mL), with the CXCR7 antagonist (CCX733; 100nM) or with anti-CXCR4/CXCR7 antibodies (50 μg/mL) and allowed to migrate to CXCL12. Untreated (−) cells were used as migration control (100%). CXCL12-induced chemotaxis of CXCR7 siRNA-transfected THP-1 cells was compared with control siRNA-transfected cells. The percentages of migration referred to CXCL12-mediated chemotaxis are shown. Mean ± SD; n = 3; *P < .05, **P < .005, ***P < .0005.
CXCL12 is constitutively expressed and secreted by monocytes and modifies macrophage differentiation
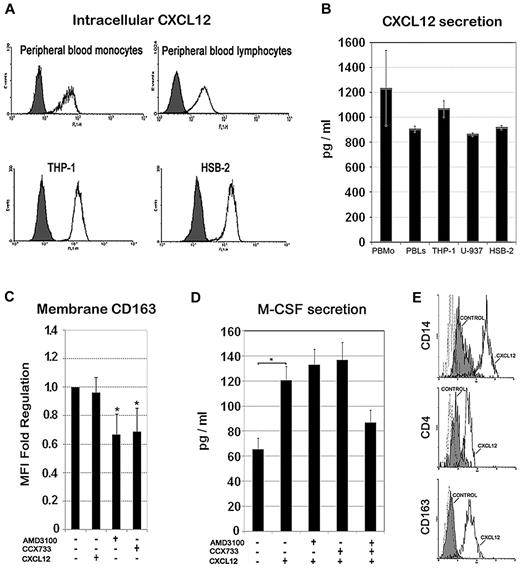
Next, we analyzed by flow cytometry the CXCL12 expression in freshly isolated monocytes and lymphocytes, as well as in both THP-1 and HSB-2 cell lines. In all cases, we detected a large constitutive intracellular pool of CXCL12 (Figure 2A). Furthermore, monocytes (primary, THP-1, and U-937 cells) and lymphocytes (primary and HSB-2 cells) secreted significant amounts of this chemokine to the medium during 48 hours of culture, which was comparable with the amount of CXCL12 secreted by tumor-associated fibroblasts24 (Figure 2B). These results indicate that CXCL12 secretion could serve both to paracrine and autocrine functions in monocytes. CXCL12 ligand binding is known to induce CXCR7 receptor internalization,21 a feature that is consistent with the observed intracellular pool of CXCR7. The above observations led us to hypothesize that the CXCL12 autocrine loop might contribute to the spontaneous differentiation of monocytes toward macrophages that occurs upon in vitro culture, even in the absence of exogenous M-CSF. Therefore, we assessed the effect of CXCR4 and CXCR7 receptor antagonists on the expression of the macrophage scavenger receptor CD163 (Figure 2C). Either AMD3100 or CCX733 significantly reduced CD163 expression of monocytes after 48 hours of culture, suggesting that autocrine CXCL12 production may play a role in the default macrophage differentiation pathway. As M-CSF is a major macrophage differentiation factor, we measured its baseline production under the same experimental conditions or in the presence of exogenous CXCL12 at 100 ng/mL (equivalent to 12.5nM). Figure 2D shows that the autocrine production of M-CSF was significantly increased by exogenous CXCL12, whereas a combination of CXCR4 and CXCR7 receptor antagonists partially blocked this response, suggesting that CXCL12 may induce the differentiation of human monocytes, at least partially, through the regulation of autocrine M-CSF production. Interestingly, addition of exogenous CXCL12 during in vitro M-CSF–induced macrophage differentiation committed a particular macrophage phenotype characterized by higher surface expression of CD4, CD14, and CD163 compared with M-CSF alone, suggesting that CXCL12 might impinge a specific pathway of macrophage differentiation (Figure 2E).
Intracellular expression and secretion of CXCL12 and its role in monocyte to macrophage differentiation. (A) Intracellular expression of CXCL12 in monocytes (primary or THP-1 cell line) or lymphocytes (resting or HSB-2 T cell line) detected by flow cytometry in permeabilized cells. Control isotype (filled histogram) and CXCL12 (empty histogram) are shown in a representative experiment of 3. (B) ELISA quantification of the secreted CXCL12 during 48 hours of culture by peripheral blood monocytes, monocytic cell lines (THP-1 and U-937), and lymphocytes (resting and HSB-2). Mean ± SD; n = 3. (C) CD163 membrane expression on peripheral blood monocytes treated for 48 hours with CXCL12 (100 ng/mL), CXCR4 antagonist (AMD3100; 25 μg/mL), CXCR7 antagonist (CCX733; 100nM) or untreated (−) was analyzed by flow cytometry. Mean fluorescence intensity (MFI) of positive cells was determined and fold change regulation relative to untreated control cells is represented. Mean ± SD; n = 4. (D) M-CSF ELISA quantification of culture supernatants of peripheral blood monocytes treated with AMD3100 and/or CCX733 at the concentrations previously indicated during 48 hours. Mean ± standard error; n = 3; *P < .05. (E) CD14, CD4, and CD163 expression analysis by flow cytometry of M-CSF–derived macrophages (5 days) in the absence (control, shaded profile) or in the presence of CXCL12 (100 ng/mL, black line). A representative experiment of 3 is shown.
Intracellular expression and secretion of CXCL12 and its role in monocyte to macrophage differentiation. (A) Intracellular expression of CXCL12 in monocytes (primary or THP-1 cell line) or lymphocytes (resting or HSB-2 T cell line) detected by flow cytometry in permeabilized cells. Control isotype (filled histogram) and CXCL12 (empty histogram) are shown in a representative experiment of 3. (B) ELISA quantification of the secreted CXCL12 during 48 hours of culture by peripheral blood monocytes, monocytic cell lines (THP-1 and U-937), and lymphocytes (resting and HSB-2). Mean ± SD; n = 3. (C) CD163 membrane expression on peripheral blood monocytes treated for 48 hours with CXCL12 (100 ng/mL), CXCR4 antagonist (AMD3100; 25 μg/mL), CXCR7 antagonist (CCX733; 100nM) or untreated (−) was analyzed by flow cytometry. Mean fluorescence intensity (MFI) of positive cells was determined and fold change regulation relative to untreated control cells is represented. Mean ± SD; n = 4. (D) M-CSF ELISA quantification of culture supernatants of peripheral blood monocytes treated with AMD3100 and/or CCX733 at the concentrations previously indicated during 48 hours. Mean ± standard error; n = 3; *P < .05. (E) CD14, CD4, and CD163 expression analysis by flow cytometry of M-CSF–derived macrophages (5 days) in the absence (control, shaded profile) or in the presence of CXCL12 (100 ng/mL, black line). A representative experiment of 3 is shown.
Differentiation of monocytes to DCs in the presence of CXCL12 decreases the functional ability of DCs to stimulate T-lymphocyte responses
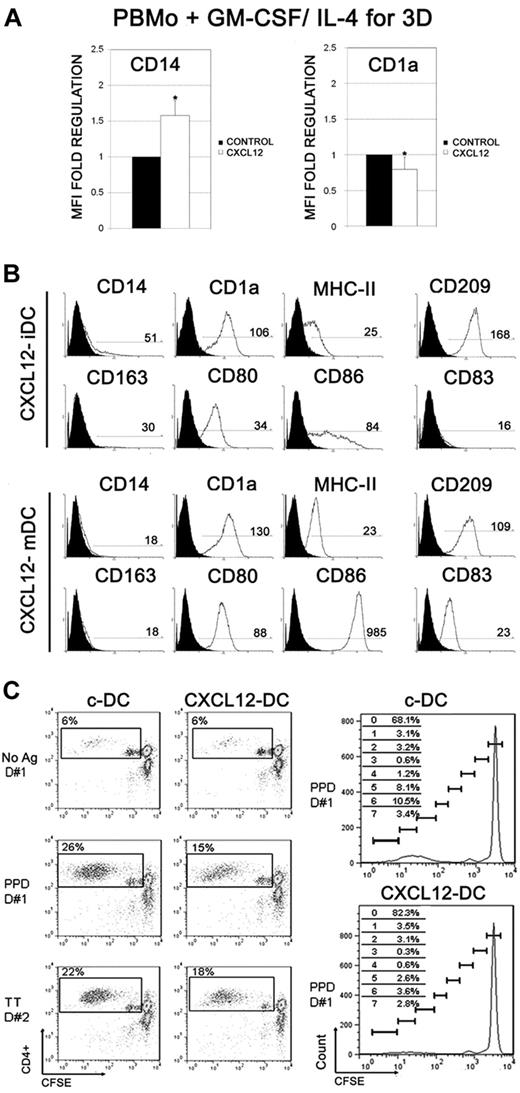
To test the hypothesis that CXCL12 signaling might also modulate monocyte differentiation to DC, PBMos were derived to DCs with GM-CSF and IL-4, in the presence or absence of CXCL12 (CXCL12-DCs and c-DCs, respectively). Monocyte-derived DCs in the presence of CXCL12 displayed delayed down-regulation of CD14 and delayed up-regulation of CD1a at early DC differentiation compared with c-DCs (day 3; Figure 3A; n = 5). However, those differences disappeared during the differentiation process, and both fully differentiated CXCL12-DCs and c-DCs showed similar expression of CD1a, CD209, and MHC-II (immature DCs day 6), and had similar maturation, displaying high expression of CD80, CD83, and CD86 (Figure 3B and supplemental Figure 2; LPS-matured DCs day 8; n = 10). We also analyzed the capacity of c-DCs and CXCL12-DCs to present specific antigens to autologous T lymphocytes. As shown in Figure 3C, CXCL12-DCs had reduced ability to stimulate antigen-specific T-cell proliferation compared with c-DCs, which ranged from approximately 50% inhibition of the PPD response and approximately 25% of TT response. These results suggest that CXCL12 may modulate the ability of DCs to induce antigen-specific T-cell responses. Therefore, despite similar up-regulation of antigen-presenting and costimulatory molecules in both c-DCs and CXCL12-DCs, their immunogenic function may be different.
Effect of CXCL12 on monocyte-DCs differentiation and maturation and immunogenic function. (A) PBMos were treated with GM-CSF and IL-4, in the presence of CXCL12 (100 ng/mL) for 3 days. The expression of CD14 and CD1a was analyzed by flow cytometry. Fold regulation relative to control untreated cells are shown. Mean ± SD; n = 5; *P < .05. (B) PBMos were derived to DCs with GM-CSF and IL-4, in the presence of CXCL12 (100 ng/mL), during the differentiation process. Inmature DCs or LPS-matured DCs were stained with a panel of mAb as indicated. The regions of positive cells were selected using the matched isotype control for each mAb. A representative experiment out of 10 is presented. MFI of positive cells is shown (C) CFSE-labeled autologous T lymphocytes were cocultured with unloaded, PPD- or TT-loaded c-DCs and CXCL12-DCs, at 1:10 DC:T ratio. After 6 days, lymphocytes were harvested, and the percentages of proliferating T cells (CFSElow) were calculated. CXCL12-DCs induced lower CD4+ T lymphocyte proliferation than c-DCs, both in PPD- and TT-specific responses, as shown in 2 representative donors (D#1 and D#2), from 4 individual donors with similar results. Unpulsed c-DCs and CXCL12-DCs served as controls. The frequency of CD4+ T cells in each CFSE-dilution peak for PPD-specific proliferative responses of D#1 is shown in the right panels.
Effect of CXCL12 on monocyte-DCs differentiation and maturation and immunogenic function. (A) PBMos were treated with GM-CSF and IL-4, in the presence of CXCL12 (100 ng/mL) for 3 days. The expression of CD14 and CD1a was analyzed by flow cytometry. Fold regulation relative to control untreated cells are shown. Mean ± SD; n = 5; *P < .05. (B) PBMos were derived to DCs with GM-CSF and IL-4, in the presence of CXCL12 (100 ng/mL), during the differentiation process. Inmature DCs or LPS-matured DCs were stained with a panel of mAb as indicated. The regions of positive cells were selected using the matched isotype control for each mAb. A representative experiment out of 10 is presented. MFI of positive cells is shown (C) CFSE-labeled autologous T lymphocytes were cocultured with unloaded, PPD- or TT-loaded c-DCs and CXCL12-DCs, at 1:10 DC:T ratio. After 6 days, lymphocytes were harvested, and the percentages of proliferating T cells (CFSElow) were calculated. CXCL12-DCs induced lower CD4+ T lymphocyte proliferation than c-DCs, both in PPD- and TT-specific responses, as shown in 2 representative donors (D#1 and D#2), from 4 individual donors with similar results. Unpulsed c-DCs and CXCL12-DCs served as controls. The frequency of CD4+ T cells in each CFSE-dilution peak for PPD-specific proliferative responses of D#1 is shown in the right panels.
CXCL12 is expressed by perivascular macrophages in tumor tissues
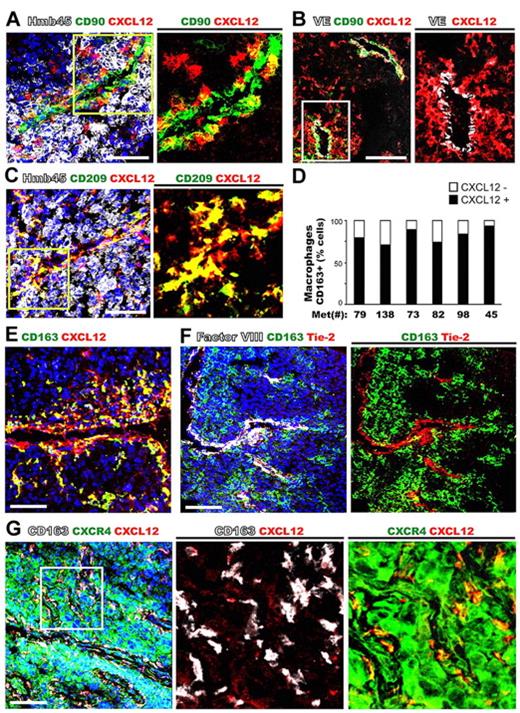
CXCL12 is involved in multiple aspects of tumor progression including the trafficking and metastasis of malignant cells to organs that express high levels of this chemokine, such as lymph nodes, lungs, liver, and bone.25 Furthermore, CXCL12 can be synthesized by tumor cells, fibroblasts, or perycytes within tumor tissues,3 where it may initiate the recruitment of blood monocytes to become cancer-supporting macrophages in a CXCL12-rich microenvironment. Thus, we examined metastatic melanoma samples that are frequently infiltrated by tumor-associated macrophages (TAMs) to analyze their phenotype. Human melanoma tissues were stained with a panel of antibodies that recognize the major cell subsets: melanocytic tumor cells (HMB45), macrophages (CD68, CD163, and CD209), all leukocytes (CD45), mesenchymal cells (CD90 and α-smooth muscle actin [α-SMA]), and endothelial cells (VE-cadherin, factor VIII; Figure 4, supplemental Figure 3, and data not shown). The coexpression of CXCL12 with the different cell subset panel Abs was analyzed by multicolor confocal microscopy in 6 metastatic melanoma samples. Elevated levels of CXCL12 were detected in stromal areas within the tumors and associated with VE-cadherin+ blood vessels, but not within HMB45+ tumor cells (Figure 4A-B). Interestingly, in all the samples studied there were 2 main subsets of highly expressing CXCL12 (CXCL12high) cells; one subset colocalized with CD90+ αSMA+ myofibroblasts, and a second CXCL12high cell subset did not colocalize with fibroblast markers (Figure 4A-B and data not shown). A thoroughly immunofluorescence phenotyping showed that this second subset of CXCL12high cells coexpressed the macrophage antigens CD68, CD163, and CD209 and were often accumulated in the vicinity of vascular structures (Figure 4C-E and data not shown). These CXCL12high cells constituted the 60%-90% of CD163+ TAMs (Figure 4D) and were the predominant subset found in tumor tissues (84.6 ± 7.7 CD163+/CD68+). As shown in Figure 4F, CD163+ TAMs did not coexpress Tie-2, which was expressed by endothelial cells (identified by costaining with Factor VIII), suggesting that CD163+ TAMs were distinct from the minor subpopulation of highly proangiogenic monocytes that express Tie-2.3 In contrast, in normal human skin, dermal macrophages (CD68+CD163+CD209+) were not accumulated at blood vessels and displayed low costaining with CXCL12, which was highly expressed by the basal layer of keratinocytes (supplemental Figure 3). We also analyzed the expression of CXCR4 and CXCR7 receptors by melanoma tissues. As we previously described, most melanoma tumor cells highly express CXCR4,26,27 which was also coexpressed by CXCL12highCD163+ TAMs (Figure 4G). In contrast to the wide CXCR4 expression found in melanoma tissues, CXCR7 was restricted to endothelial cells, and most intratumoral CXCL12highCD163+ TAMs barely expressed this receptor (supplemental Figure 3B-C). These data indicate that CXCL12/CXCR4 autocrine loop may play a primary role in the modulation of macrophage differentiation at tumor tissues, whereas CXCR7 appears to be down-regulated in TAMs. Furthermore, these results also reveal that TAMs comprise an important source of perivascular CXCL12 that could serve as a guidance clue for the recruitment of either blood leukocytes or CXCR4+ tumor cells at perivascular sites.
Most CXCL12+ cells in metastatic melanoma tissues are CD209+ and CD163+ tumor-associated macrophages that display a perivascular location and express CXCR4. Metastatic melanoma tissues were stained with anti-CXCL12 and the following tissue markers: (A) melanocytic cells (HMb45) and myofibroblasts (CD90); (B) endothelial cells (VE-cadherin, VE) and myofibroblasts (CD90); (C) macrophages (CD209) and melanocytic cells (HMb45); and (D) quantification of CXCL12 expression in macrophages CD163+ was given as percentage. Several melanoma metastases were analyzed: #45 lung metastasis; #73, 79, 82, and 138 lymph node metastasis; and #98 skin metastasis. (E) Macrophages (CD163). (F) Macrophages (CD163), endothelial cells (Factor VIII), and TIE-2 positive cells. (G) Macrophages (CD163) and CXCR4+ cells (tumor and macrophages).
Most CXCL12+ cells in metastatic melanoma tissues are CD209+ and CD163+ tumor-associated macrophages that display a perivascular location and express CXCR4. Metastatic melanoma tissues were stained with anti-CXCL12 and the following tissue markers: (A) melanocytic cells (HMb45) and myofibroblasts (CD90); (B) endothelial cells (VE-cadherin, VE) and myofibroblasts (CD90); (C) macrophages (CD209) and melanocytic cells (HMb45); and (D) quantification of CXCL12 expression in macrophages CD163+ was given as percentage. Several melanoma metastases were analyzed: #45 lung metastasis; #73, 79, 82, and 138 lymph node metastasis; and #98 skin metastasis. (E) Macrophages (CD163). (F) Macrophages (CD163), endothelial cells (Factor VIII), and TIE-2 positive cells. (G) Macrophages (CD163) and CXCR4+ cells (tumor and macrophages).
CXCL12 induces the expression of CCL1 and VEGF
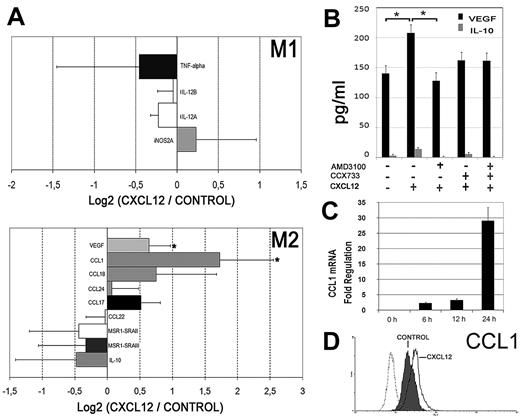
Perivascular CXCL12high expressing cells may ensure the ongoing recruitment of monocytes into tumors, which are then entrapped and subverted by exposition to local high levels of CXCL12. Therefore, the early transcriptional events associated with monocyte exposition to CXCL12 were investigated. Resting human monocytes from 3 separate donors were treated with either CXCL12 or media alone, for 6 hours on Teflon dishes to minimize the effect of adhesion. Microarray analysis was performed using the ratios of the log2-transformed intensity values for each probe. We focused our attention on the expression of macrophage differentiation genes that characterize the M1-M2 (or classic alternative) activation model.1,4 As shown in Figure 5A, CXCL12 stimulation did not significantly modify the mRNA expression of a panel of prototypic M1 genes. CXCL12 had no effect on most M2 genes either, including IL-10 mRNA and protein secretion (Figure 5A-B). Interestingly, among the panel of M2 genes, we identified the up-regulation of the chemokine CCL1 and the endothelial growth factor VEGF, which are proangiogenic factors that characterize a distinct pathway of M2 polarization.4 We confirmed that primary monocytes cultured in the presence of CXCL12 for 48 hours significantly augmented VEGF secretion, and this increase was blocked by the CXCR4 antagonist AMD3100 (Figure 5B). CCL1 mRNA expression was further confirmed by real-time reverse transcription PCR analysis that revealed a time-dependent induction in response to CXCL12 treatment (Figure 5C). Besides, an enhanced pool of intracellular CCL1 protein was detected by flow cytometry in monocytes exposed to CXCL12 (Figure 5D). These results indicate that the important angiogenic factor VEGF and the proangiogenic chemokine CCL1 are selectively up-regulated in response to CXCL12 in human monocytes.
CXCL12 induces the proangiogenic factors VEGF and CCL1. (A) mRNA extracted from PBMos stimulated with CXCL12 (100 ng/mL) or unstimulated (control) was analyzed by microarrays. The ratios of the log2 intensity data of CXCL12-treated cells versus Control for some M1 (top histogram) and M2 (bottom histogram) genes are shown. Mean ± SD; n = 3. (B) ELISA quantification of secreted VEGF (black filled histogram) and IL-10 (gray histogram) from peripheral blood monocytes cultured for 48 hours. Cells were treated with AMD3100 and/or CCX733 as described in Figure 2D. Mean ± SE; n = 3; *P < .05. (C) Time course of CCL1 mRNA expression (qPCR) from PBMos stimulated with CXCL12 (100 ng/mL). Results were normalized to GAPDH levels. Fold regulation relative to control untreated cells is represented. Mean ± SD; n = 3. (D) Flow cytometric analysis of intracellular CCL1 protein expression in freshly isolated PBMos cultured for 12 hours (control, shaded profile) compared with CXCL12-stimulated PBMos (black line). Control isotype (discontinuous line). A representative experiment of 3 is shown.
CXCL12 induces the proangiogenic factors VEGF and CCL1. (A) mRNA extracted from PBMos stimulated with CXCL12 (100 ng/mL) or unstimulated (control) was analyzed by microarrays. The ratios of the log2 intensity data of CXCL12-treated cells versus Control for some M1 (top histogram) and M2 (bottom histogram) genes are shown. Mean ± SD; n = 3. (B) ELISA quantification of secreted VEGF (black filled histogram) and IL-10 (gray histogram) from peripheral blood monocytes cultured for 48 hours. Cells were treated with AMD3100 and/or CCX733 as described in Figure 2D. Mean ± SE; n = 3; *P < .05. (C) Time course of CCL1 mRNA expression (qPCR) from PBMos stimulated with CXCL12 (100 ng/mL). Results were normalized to GAPDH levels. Fold regulation relative to control untreated cells is represented. Mean ± SD; n = 3. (D) Flow cytometric analysis of intracellular CCL1 protein expression in freshly isolated PBMos cultured for 12 hours (control, shaded profile) compared with CXCL12-stimulated PBMos (black line). Control isotype (discontinuous line). A representative experiment of 3 is shown.
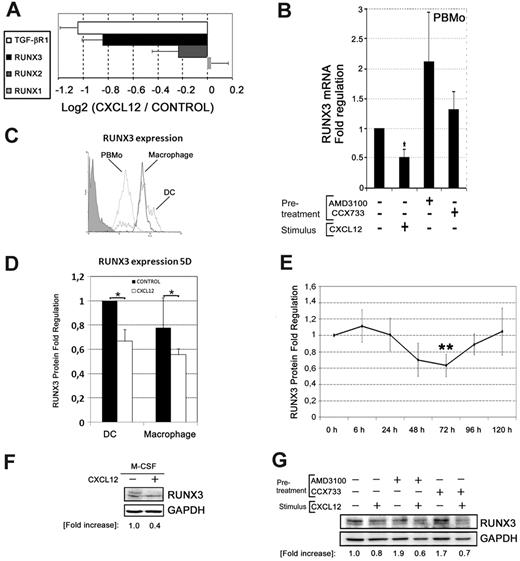
CXCL12 modulates the transcription factor RUNX3 in primary monocytes
To assess the transcriptional basis of CXCL12-induced monocyte differentiation, we decided to focus on the RUNX family of transcription factors because it has been reported that RUNX3 mediates the TGF-β responses in DCs.17 In addition, the expression of PU.1 (Sp1), which controls monocyte development and differentiation, is regulated by RUNX1.28 We found that RUNX1 and RUNX2 transcription did not change upon CXCL12 treatment, whereas RUNX3 and TGF-β receptor appeared to be down-regulated in the microarray data and in the qualitative PCR (Figure 6A and supplemental Figure 4A). qPCR showed that CXCL12 induced a significant down-regulation of RUNX3 mRNA expression both in primary monocytes and in THP-1 cells (Figure 6B and supplemental Figure 4B). Remarkably, treatment of PBMos or THP-1 cells with the CXCR4 antagonist AMD3100 induced RUNX3 expression, indicating that the inhibition of the autocrine/paracine CXCR4-CXCL12 axis was releasing the expression of RUNX3 (Figure 6B and supplemental Figure 4B). A lesser effect in RUNX3 expression was observed by blocking the CXCR7-CXCL12 interaction with the CCX733 antagonist at this early time point (6 hours) in PBMos and THP-1 cells (Figure 6B and supplemental Figure 4B). These results demonstrate that interaction of CXCL12 with CXCR4 is necessary and sufficient to repress early RUNX3 transcription in blood monocytes, and therefore, CXCL12 may down-regulate RUNX3-mediated TGF-β signaling in mononuclear cells.
CXCL12 modulates the transcription factor RUNX3. (A) mRNA extracted from PBMos stimulated with CXCL12 (100 ng/mL) or unstimulated (control) was analyzed by microarrays. CXCL12-induced regulation of the expression of the RUNX family proteins and TGF-β receptor was compared. The ratios of the log2 intensity data of CXCL12-treated cells versus Control for TGF-β/RUNX selected genes are represented. Mean ± SD; n = 3. (B) PBMos were incubated for 6 hours in the presence (+) or absence (−) of CXCL12 (100 ng/mL), or the CXCR4 or CXCR7 antagonists AMD3100 (25 μg/mL) and CCX733 (10nM), respectively. RNA was extracted from cells and subjected to reverse transcriptase reaction and qPCR for RUNX3 and normalized to GAPDH. Fold regulation relative to control untreated cells is shown. Mean ± SD; n = 4; *P < .05. (C) Runx3 expression was analyzed by flow cytometry on PBMos (punctuated line), DCs (gray line), or macrophages (black line), by stimulation for 5 days with GM-CSF plus IL-4 or M-CSF, respectively. Control Isotype was also shown (shaded profile). A representative experiment of 3 is shown. (D) PBMos were differentiated to DCs or macrophages as described in panel C in the absence or presence of CXCL12 (100 ng/mL), during the differentiation process. RUNX3 levels were analyzed by flow cytometry. Fold regulation relative to control untreated (−) cells are shown. Mean ± SD; n = 3; *P < .05. (E) PBMos were cultured in the absence or in the presence of CXCL12 (100 ng/mL) during 5 days. Percentages of RUNX3 positive cells were analyzed by flow cytometry at the indicated times. Fold regulation relative to control untreated cells are shown. Mean ± SD; n = 9; **P < .01. (F) PBMos induced with M-CSF during 2 days were untreated (−) or treated with CXCL12 (100 ng/mL) as indicated. Western blot of total lysates was performed. Results were quantified by densitometry, normalized for differences in loading, and expressed as fold increase compared with unstimulated controls. A representative experiment out of 3 is shown. (G) PBMos were pretreated (+) with AMD3100 (25 μg/mL) or CCX733 (10nM) or untreated (−) as control. Subsequently, cells were stimulated with CXCL12 (100 ng/mL) as indicated. Western blot of total lysates of 48 hours cultured cells was performed. Results were quantified by densitometry, normalized for differences in loading, and expressed as fold increase compared with unstimulated controls. A representative experiment of 3 is shown.
CXCL12 modulates the transcription factor RUNX3. (A) mRNA extracted from PBMos stimulated with CXCL12 (100 ng/mL) or unstimulated (control) was analyzed by microarrays. CXCL12-induced regulation of the expression of the RUNX family proteins and TGF-β receptor was compared. The ratios of the log2 intensity data of CXCL12-treated cells versus Control for TGF-β/RUNX selected genes are represented. Mean ± SD; n = 3. (B) PBMos were incubated for 6 hours in the presence (+) or absence (−) of CXCL12 (100 ng/mL), or the CXCR4 or CXCR7 antagonists AMD3100 (25 μg/mL) and CCX733 (10nM), respectively. RNA was extracted from cells and subjected to reverse transcriptase reaction and qPCR for RUNX3 and normalized to GAPDH. Fold regulation relative to control untreated cells is shown. Mean ± SD; n = 4; *P < .05. (C) Runx3 expression was analyzed by flow cytometry on PBMos (punctuated line), DCs (gray line), or macrophages (black line), by stimulation for 5 days with GM-CSF plus IL-4 or M-CSF, respectively. Control Isotype was also shown (shaded profile). A representative experiment of 3 is shown. (D) PBMos were differentiated to DCs or macrophages as described in panel C in the absence or presence of CXCL12 (100 ng/mL), during the differentiation process. RUNX3 levels were analyzed by flow cytometry. Fold regulation relative to control untreated (−) cells are shown. Mean ± SD; n = 3; *P < .05. (E) PBMos were cultured in the absence or in the presence of CXCL12 (100 ng/mL) during 5 days. Percentages of RUNX3 positive cells were analyzed by flow cytometry at the indicated times. Fold regulation relative to control untreated cells are shown. Mean ± SD; n = 9; **P < .01. (F) PBMos induced with M-CSF during 2 days were untreated (−) or treated with CXCL12 (100 ng/mL) as indicated. Western blot of total lysates was performed. Results were quantified by densitometry, normalized for differences in loading, and expressed as fold increase compared with unstimulated controls. A representative experiment out of 3 is shown. (G) PBMos were pretreated (+) with AMD3100 (25 μg/mL) or CCX733 (10nM) or untreated (−) as control. Subsequently, cells were stimulated with CXCL12 (100 ng/mL) as indicated. Western blot of total lysates of 48 hours cultured cells was performed. Results were quantified by densitometry, normalized for differences in loading, and expressed as fold increase compared with unstimulated controls. A representative experiment of 3 is shown.
Next, we analyzed the regulation of RUNX3 protein levels by flow cytometry during the differentiation of PBMos to either macrophages or DCs in the presence or absence of CXCL12. Figure 6C shows the relatively low RUNX3 expression in PBMos compared with the high RUNX3 expression that occurs during both macrophage and DC differentiation (day 5). Moreover, the presence of CXCL12 during the differentiation process down-regulated RUNX3 protein expression in both macrophages and DCs (day 5; Figure 6D). In addition, we analyzed the effect of CXCL12 on the expression of RUNX3 by PBMos in several kinetics experiments using 24-hour intervals, which showed a significant decrease in RUNX3 protein levels at 72 hours in PBMos treated with CXCL12 compared with unstimulated cells (Figure 6E). The down-regulatory effect of CXCL12 treatment in RUNX3 expression was also detected by Western blot in M-CSF treated monocytes (day 2) and THP-1 cells (Figure 6F and supplemental Figure 4C). Similarly to the effect of exogenous CXCL12, its autocrine secretion (see Figure 2), may contribute to maintain a reduced basal level of RUNX3 expression in PBMos and THP-1 cells. As expected, treatment with CXCR4 or CXCR7 antagonists enhanced RUNX3 protein expression, whereas addition of exogenous CXCL12 (100 ng/mL) diminished RUNX3 protein levels in CXCR4 or CXCR7 antagonist-treated cells via the alternative receptor that remained functional (Figure 6G and supplemental Figure 4D). RUNX3 protein changes in response to CXCL12 or CXCR4 or CXCR7 antagonists were also detected by flow cytometry in the HSB-2 T lymphocytes, which expressed high basal levels of RUNX3 (supplemental Figure 4E). Taken together, these results indicate that CXCL12 down-regulates RUNX3 expression at transcriptional and protein levels in blood monocytes and slows down the up-regulation of RUNX3 that occurs during the initial phases of macrophage and DC differentiation.
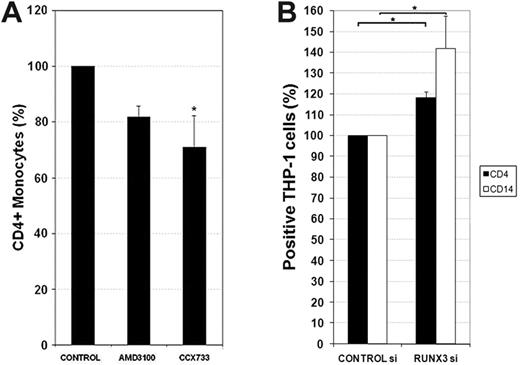
CXCL12-mediated RUNX3 down-regulation maintains the expression of CD4 and CD14 in monocytes
We showed above that M-CSF–derived macrophages in the presence of high doses of CXCL12 maintained higher surface expression of CD4 compared with control macrophages (Figure 2E). RUNX3 silences the expression of CD4 in CD8+ T cells (recently reviewed29 ). As CXCL12 inhibited RUNX3 expression, we next addressed its ability to regulate CD4 expression on blood monocytes. To assess the effect of basal autocrine CXCL12 secretion on CD4 expression, blood monocytes were pretreated with CXCR4 or CXCR7 antagonists and cultured for 48 hours. CCX733 pretreatment significantly decreased the percentage and the fluorescence intensity of CD4+ monocytes (Figure 7A). We next assessed if RUNX3 depletion by siRNA in THP-1 cells could reproduce some of the CXCL12 effects reported above (Figure 2). As expected, enhanced CD4 expression was detected in cells with reduced levels of RUNX3 expression compared with control siRNA (Figure 7B and supplemental Figure 4F). Interestingly, CD14, the most important endotoxin receptor on human monocytes,30 was also significantly augmented by RUNX3 depletion, thus, phenocopying the effects of CXCL12 on CD4 and CD14 expression. The lack of CD163 and CD209 expression on THP-1 cells limited the analysis of the effect of RUNX3 depletion on these molecules (data not shown). These results suggest that CXCL12 maintains the long-term CD4 and CD14 expression in monocytes through RUNX3 down-regulation.
RUNX3 mediates the effects of CXCL12 in CD4 and CD14 expression. (A) PBMos were freshly isolated and cultivated in the presence (+) or absence (−) of AMD3100 (25 μg/mL) or CCX733 (100nM) for 48 hours. Membrane CD4 was analyzed by flow cytometry. Results are expressed as percentages of positive cells. Mean ± SD; n = 4; *P < .05. (B) THP-1 cells were transfected with RUNX3 siRNA or control siRNA. The expression of CD4 and CD14 was evaluated by flow cytometry the third day. Mean ± SD; n = 4; *P < .05.
RUNX3 mediates the effects of CXCL12 in CD4 and CD14 expression. (A) PBMos were freshly isolated and cultivated in the presence (+) or absence (−) of AMD3100 (25 μg/mL) or CCX733 (100nM) for 48 hours. Membrane CD4 was analyzed by flow cytometry. Results are expressed as percentages of positive cells. Mean ± SD; n = 4; *P < .05. (B) THP-1 cells were transfected with RUNX3 siRNA or control siRNA. The expression of CD4 and CD14 was evaluated by flow cytometry the third day. Mean ± SD; n = 4; *P < .05.
Discussion
CXCL12 plays a critical role in monocyte extravasation and is highly expressed at sites where macrophages are present in either physiologic or pathologic situations.31 This study focuses on the role of CXCL12 in shaping the process of monocyte differentiation toward a distinct pathway characterized by proangiogenic and immunosuppressive functions. Our results indicate that in addition to regulate cell migration through both CXCR4 and CXCR7 receptors; CXCL12 enhances the expression of pathogen recognition receptors CD14 and CD163 and induces the secretion of angiogenic factors as VEGF and CCL1. More importantly, the presence of CXCL12 during the differentiation of monocytes to DCs dampers their ability to induce antigen-specific activation of T lymphocytes. Tumors are rich CXCL12 microenvironments, and here we show that CXCL12 is highly expressed by perivascular TAMs within melanoma tissues, supporting an autocrine/paracrine positive feedback loop in the recruitment and differentiation of a subset of monocytes with proangiogenic and immunosuppressive functions, which may promote tumor progression. We provide data indicating that CXCL12 may modulate gene expression and control monocyte differentiation through down-regulation of the transcription factor RUNX3.
Among the multiple processes in which CXCL12 and its cognate receptors, CXCR4 and CXCR7, play a crucial role are the development of embryonic vessels and tumor angiogenesis.6 Accumulating evidence demonstrates that CXCL12 has a major role in the recruitment and retention of CXCR4+ bone marrow–derived cells to the neoangiogenic niches supporting revascularization of ischemic tissue and tumor growth.31 However, the phenotypes of proangiogenic bone marrow–derived cells and their precise role in neovascularization are incompletely defined. Stromal fibroblasts were previously identified as the major source of CXCL12 in human breast carcinomas.24 In addition to fibroblasts, we have identified in metastatic melanoma a large and novel subset of perivascular CXCL12high cells that coexpresses the leukocyte marker CD45 and the macrophage markers CD68, CD163, and CD209. The expression of CD209 has been described on certain tissue macrophages, which are found in noninflamed lymph nodes and at sites of inflammation, but not on DCs.32,33 Perivascular macrophages have been previously reported in other types of human cancer and correlate with increased microvessel density and poor prognosis.34 Perivascular CXCL12 expression has been reported in a model of VEGF-induced adult neovascularization, in which CXCL12 expression is downstream of VEGF and contributes to the perivascular retention of myeloid cells that are essential in the process.35 An important aspect of our work is the proposal that perivascular CXCL12, in addition to participate in the recruitment and retention of blood monocytes, drives a particular pathway of macrophage differentiation, as suggested by the CXCL12-induced up-regulation of important angiogenic factors as VEGF and CCL1. Thus, VEGF appears to be both upstream and downstream of CXCL12, suggesting the existence of a feedback loop in the regulation of both factors. In addition, monocyte recruitment and differentiation into CXCL12high expressing macrophages, which also coexpress CXCR4, may further increase the tissue level of CXCL12 by an autocrine-paracrine loop, further promoting the feedback mechanism for neovascularization associated to chronic inflammation. In fact, low oxygen tension, which is a hallmark of inflammatory diseases and tumor microenvironment, induces CXCR4 and CXCL12 expression through the hypoxia-inducible factor-1.36,37 It has also been shown that hypoxia-inducible factor-1, partly through increases in CXCL12, induces recruitment of bone marrow–derived myeloid CD45+ cells, which provide MMP-9 to promote tumor angiogenesis.38 Identification of the different cell populations and the mechanisms involved in temporally and spatially supporting angiogenesis and/or vasculogeneis in specific types of tumors is currently under intense investigation to define specific treatment strategies. Interestingly, it has been recently reported that the CXCR4 antagonist AMD3100 prevented the influx of myelomonocytes and inhibited the development of tumor vasculature resulting in abrogation of glioblastoma recurrence after irradiation in mice.39
In addition, we found that DCs derived in the presence of CXCL12 are weaker stimulators of antigen-specific T-lymphocyte responses. Mature DCs are traditionally considered to be immunogenic, although there is accumulating evidence that distinct states of DC differentiation and maturation can also induce Ag-specific regulatory T cells.40 Interestingly, it has been recently shown in a mouse model of autoimmune disease that treatment with a CXCL12-immunoglobulin fusion protein suppresses ongoing experimental autoimmune encephalomyelitis, by selecting antigen-specific regulatory T cells.41 Recent findings support that CXCL12 functions as an anti-inflammatory chemokine that may suppress the antigen-specific immune responses.42
Our microarray analysis of CXCL12-treated monocytes revealed that RUNX3, a transcription factor that regulates cell lineage decisions in other cell systems, is down-regulated in the presence of CXCL12. We show at transcriptional and protein levels that the presence of CXCL12 slows down the RUNX3 up-regulation that occurs during monocyte differentiation to either macrophages or DC. Importantly, this is the first report of a signaling pathway that regulates RUNX3 expression. The transcription factor RUNX3 silences CD4 gene expression at the commitment process into CD8 thymocytes.29 Human monocytes, macrophages, and DCs also express CD4, which is known to enhance phagocytosis of apoptotic cells in CD4+ macrophages.43 We show that in monocytes, CXCL12 signaling is important for the maintenance of both CD4 and CD14 expression through the repression of RUNX3 transcription factor. CD14 is part of the lipopolysaccharide “sensing apparatus” of monocytes and is also involved in recognition and phagocytosis of apoptotic cells.30,44 Thus, CXCL12 effect on CD4 and CD14 levels seems to promote the scavenger function of macrophages. In line with our findings, blocking of CXCL12 or CXCR4 with specific antibodies reduced the proportion of CD4+ and CD4+CD8+ thymocytes.45 Furthermore, we have demonstrated that both CXCR4 and CXCR7 receptors are involved in the signaling that inhibits RUNX3 expression at transcriptional and protein levels. Our data also show that both receptors are involved in the migratory response of blood monocytes to CXCL12. These results are in line with recent works that propose that both receptors form functional CXCR4/CXCR7 heterodimers and that CXCR7 is involved in specific CXCL12-mediated signaling.13,14
In summary, we report that as a part of the transcriptional program elicited by CXCL12, this chemokine down-regulates the expression of the transcriptional factor RUNX3. Reduced levels of RUNX3 in turn maintain the expression of CD4 and CD14 by macrophages in the process of differentiation. Other molecules up-regulated by CXCL12 are the proangiogenic factors VEGF and CCL1 and the macrophage scavenger receptor CD163. Importantly, CXCL12 down-modulates the ability of DCs to induce antigen-specific immune responses. Given that CXCL12 is constitutively expressed at sites of macrophage differentiation, as lung or lymph nodes, and is up-regulated during chronic inflammatory processes and cancer, CXCL12 is well-suited to control ongoing monocyte recruitment, tissue retention, and specialized monocytic differentiation under either steady-state or pathologic conditions.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr J. L. Rodríguez-Fernández and Dr C. Cabañas for the valuable contribution to this work; Dr F. Sánchez-Madrid and Dr M. Penfold for antibodies and reactives; Dr M. Ortega, Dr E. Fernández-Ruiz, and Dr M. Alonso (UCAM) for assistance with qPCRs, and J. Villarejo and I. Treviño for expert technical assistance.
This work was supported by grants from the Ministerio de Educación y Ciencia (SAF 2006-08615, 2010-16102, and GEN 2003-20, 649-C06-04 to P.S.-M.) and Ayuda a la investigación científica y técnica 2006 from Fundación Ramón Areces (P.S.-M.), Contrato Juan de la Cierva from Ministerio de Educación y Ciencia and Contrato RETICS RD08/0075 from Ministerio de Ciencia e Innovación to L.S.-M, Contrato de Investigación from Fondo de Investigación Sanitaria/Comunidad de Madrid to R.S and A.E.
Authorship
Contribution: L.S.-M. designed and performed research, analyzed data, made the figures, and wrote the paper; A.E. performed research and analyzed data; R.S. contributed vital analytical tools and made a figure; S.S.-R. and M.A.V. contributed vital analytical tools; and P.S.-M. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Paloma Sánchez-Mateos, Servicio de Inmunología, Hospital Gregorio Marañón, C/Dr Esquerdo 46, 28007 Madrid, Spain; e-mail: rsanchezma.hgugm@salud.madrid.org.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal