Abstract
The transcription factor signal transducer and activator of transcription 5 (STAT5) fulfills essential roles in self-renewal in mouse and human hematopoietic stem cells (HSCs), and its persistent activation contributes to leukemic transformation, although little molecular insight into the underlying mechanisms has been obtained. In the present study, we show that STAT5 can impose long-term expansion exclusively on human HSCs, not on progenitors. This was associated with an enhanced cobblestone formation under bone marrow stromal cells of STAT5-transduced HSCs. Hypoxia-induced factor 2α (HIF2α) was identified as a STAT5 target gene in HSCs, and chromatin immunoprecipitation studies revealed STAT5 binding to a site 344 base pairs upstream of the start codon of HIF2α. Lentiviral RNA interference (RNAi)–mediated down-modulation of HIF2α impaired STAT5-induced long-term expansion and HSC frequencies, whereas differentiation was not affected. Glucose uptake was elevated in STAT5-activated HSCs, and several genes associated with glucose metabolism were up-regulated by STAT5 in an HIF2α-dependent manner. Our studies indicate that pathways normally activated under hypoxia might be used by STAT5 under higher oxygen conditions to maintain and/or impose HSC self-renewal properties.
Introduction
Hematopoiesis is a dynamic process in which self-renewal, proliferation, lineage commitment, differentiation, and apoptosis are tightly controlled. Signal transducer and activator of transcription 5 (STAT5) is a transcription factor that fulfills important roles in many of these processes.1,2 STAT5 is widely expressed throughout the hematopoietic system, both in stem and progenitor cells and in committed erythroid, myeloid, and lymphoid cells, and can be activated by different cytokines.1-3 In most cases, JAK tyrosine kinase activity mediates STAT5 tyrosine phosphorylation, although the tyrosine kinase receptor family can also induce STAT5 phosphorylation in a JAK-independent manner. After cytokine-induced phosphorylation, STAT5 dimerizes and translocates to the nucleus, where it induces the expression of target genes.3,4
Loss-of-function experiments have revealed critical roles for STAT5 in the hematopoietic stem/progenitor compartment. Stat5abΔN/ΔN mice that express a truncated form of STAT5 have been used to assess stem-cell function in the absence of wt (wild-type) STAT5 signaling. These mice were characterized by normal HSC numbers, and stem cells isolated from the bone marrow or fetal liver were capable of engrafting irradiated recipients5 ; however, the competitive repopulating capacity of Stat5abΔN/ΔN HSCs was severely impaired.5-9 Experiments with Stat5ab−/− mice that completely lack expression of STAT5 revealed that STAT5 was required for the development of HSCs, lymphocytes, and erythrocytes, whereas myelopoiesis was not affected in these animals.10 It was demonstrated recently by Wang et al that conditional deletion of Stat5 in a Mx1-Cre mouse model decreases the pool size, survival, and quiescence of HSCs, indicating a role of STAT5 in the maintenance of HSC quiescence under physiologic conditions.11 In human cord blood CD34+ cells, down-modulation of STAT5 resulted in decreased colony-forming cell (CFC) numbers and long-term culture-initiating cell (LTC-IC) frequencies that coincided with a reduction in long-term expansion, whereas cell differentiation was not affected.12
The most direct evidence for STAT5 acting as an oncogene arises from murine bone marrow transplantation studies in which constitutively activated STAT5 (S711F) mutants were overexpressed. Lethally irradiated recipients receiving activated STAT5-transduced bone marrow died within 6 weeks after transplantation of a multilineage leukemia.13 It was demonstrated that a tryptophan residue in the N-terminal region of STAT5 is required for tetramerization of STAT5 dimers, and tetramer-deficient STAT5 mutants were unable to induce leukemia in mice.13 Another activating mutant of STAT5, STAT5A(1*6), which contains 2 point mutations (H299R and S711F),14 was also shown to induce a myeloproliferative disorder,15 but only when the most primitive CD34−Lin−Sca1+c-Kit+ population was transduced.16 In contrast, human stem and progenitor cells overexpressing constitutively activated STAT5A imposed long-term self-renewal but without malignant transformation.17 However, constitutive activation of STAT5 has been observed in the majority of acute myeloid leukemia (AML) cases.18-20 This might be a characteristic feature of upstream activated kinases such as FLT3, KIT, or JAK2,21-23 or it could be due to autocrine growth factor production.18 Targeting STAT5 in primary AML CD34+ cells resulted in impaired long-term expansion and a decline in the formation of leukemic cobblestone-area forming cells (CAFCs).12
Although it is clear that STAT5 fulfills essential roles in both normal hematopoiesis and in the development or maintenance of leukemia, little is known about the molecular mechanisms that are involved. By making use of an inducible model in which constitutive STAT5 activity can be introduced in specific compartments of the hematopoietic system,24,25 our current data show that STAT5 can impose a long-term proliferative advantage on the CD34+/CD38− HSC population, but not on progenitors. Gene-expression profiling in STAT5-transduced HSC and progenitor cells and in STAT5-activated, GATA1–down-modulated cells led to the identification of 32 GATA1-independent STAT5 target genes in the HSC population that could be linked to the STAT5-induced long-term proliferation and self-renewal phenotype. One of the identified genes was hypoxia-induced factor 2α (HIF2α, also known as EPAS1), and functional studies revealed that STAT5-induced expansion of HSCs and enhanced progenitor- and stem-cell frequencies were mediated, at least in part, via HIF2α up-regulation, whereas differentiation was not affected.
Methods
Cell cultures and cell lines
Neonatal cord blood was obtained from healthy full-term pregnancies after informed consent in accordance with the Declaration of Helsinki from the obstetrics departments of the University Medical Center Groningen (UMCG) and the Martini Hospital Groningen, Groningen, The Netherlands. All protocols were approved by the Medical Ethical Committee of the UMCG. After separation of mononuclear cells with lymphocyte separation medium (PAA Laboratories), cord blood CD34+ cells were isolated with the Mini-MACS separation system (Miltenyi Biotec). Cells for MS5 coculture and LTC-IC assays were cultured in α-modified essential medium (Fisher Scientific) supplemented with heat-inactivated 12.5% fetal calf serum and heat-inactivated 12.5% horse serum (both from HyClone Laboratories); 2mM glutamine, penicillin, and streptomycin (all from PAA Laboratories); 57.2μM β-mercaptoethanol (Merck Sharp & Dohme); and 1μM hydrocortisone (Sigma-Aldrich). This medium was designated as Gartner medium. UT7-GM cells were grown in Iscove modified Eagle medium, 20% fetal calf serum, and 10 ng/mL of GM-CSF (Genetics Institute). Before experiments, the cells were deprived of GM-CSF overnight and then stimulated with 10 U/mL of erythropoietin (Eprex; Janssen-Cilag).
CFC and LTC-IC assays
CFC assays were performed as described previously.17 Retrovirally transduced CD34+ cord blood cells were used for LTC-IC limiting dilution assays in the range of 5-1000 cells per well in a 96-well plate using Gartner medium. Methylcellulose (StemCell Technologies) supplemented with 20 ng/mL of interleukin-3, 20 ng/mL of interleukin-6 (both from Gist-Brocades), 20 ng/mL of G-CSF (Rhone-Poulenc Rorer), 20 ng/mL of c-kit ligand (Amgen), and 6 U/mL of erythropoietin were added at week 5. Two weeks later, wells containing CFCs were scored as positive and the LTC-IC frequency was calculated using extreme limiting dilution analysis (ELDA) software.26
Retroviral/lentiviral production and transduction
For lentiviral transduction, we used a pLVUT lentiviral vector24 in which the short hairpin against human HIF2α (GCTGACTCTTTGCTCTAAT) was cloned into the ClaI EcoR1 restriction site. For the control, a short hairpin RNA targeting firefly luciferase was used. Viral particles for lentiviral transduction were produced by cotransfection of 293T cells with 0.7 μg of pcDNA3-VSVg-REV, 3 μg of pCMV D8.91, and 3 μg of pLVUT-luciferase RNA interference (RNAi) or pLVUT-HIF2α-RNAi. Lentiviral supernatants were collected after 24 hours and stored at −80°C. Stable PG13 STAT5A(wt)–estrogen receptor (STAT5A-ER)25 retroviral producers were cultured in Dulbecco modified Eagle Medium (Fisher Scientific) supplemented with heat-inactivated 10% fetal calf serum (HyClone Laboratories) and penicillin/streptomycin (PAA Laboratories). For retroviral transduction, viral particles were harvested from PG13 cultures after 8-12 hours of incubation in hematopoietic progenitor growth medium. Supernatants were collected before the transduction rounds and passed through 0.45-mm filters (Sigma-Aldrich). Cord blood CD34+ cells were cultured for 48 hours in hematopoietic progenitor cell growth medium supplemented with stem cell factor (100 ng/mL), Flt3 ligand (100 ng/mL; Amgen), and thrombopoietin (100 ng/mL; Kirin) and transduced on retronectin-coated plates (Lucron Bioproducts) in 3 consecutive rounds of 8 and 12 hours with lentiviral or retroviral supernatant supplemented with the same cytokines and 4 μg/mL of Polybrene. After transduction, transduced green fluorescent protein–positive, truncated nerve growth factor receptor–positive, or double-positive cells were sorted on a MoFlo sorter (Dako Cytomation).
Flow cytometric analysis and cell sorting
Antibodies used for FACS analysis and for cell sorting were: CD34, CD38, CD45RA, CD123, CD15, CD71 CD235a, CD11b, and CD14 (BD Biosciences). Cell sorting of HSCs and progenitor fractions from CD34+ cord blood cells was performed on the basis of the combinatorial expression of cell-surface antigens. HSCs were determined as CD34+CD38low, common myeloid progenitors (CMPs) as CD34+CD38+CD123+CD45RA−, granulocyte-macrophage progenitors (GMPs) as CD34+CD38+CD123+CD45RA+, and megakaryocyte-erythroid progenitors (MEPs) as CD34+CD38+CD 123−CD45RA−. Sorting of cells was performed on the MoFlo sorter.
Immunoblotting and cytospins
May-Grünwald-Giemsa (MGG) staining was used to visualize cells on cytospins. Images were taken with an Olympus BX50 microscope using a 63×, 1.3 numeric aperture oil objective. For HIF2α Western blot analysis, the primary antibody was from R&D Systems and was used in a 1:1000 dilution. Antibodies against STAT5 (C17) were obtained from Santa Cruz Biotechnology; anti–phosphotyrosine 496-STAT5 was obtained from BD Biosciences. Fluorescence-labeled secondary antibodies were purchased from Invitrogen and were used in a 1:2000 dilution. Blots were scanned on an Odyssey infrared scanner (Li-Cor Biosciences).
mRNA analysis
Total RNA was isolated according to the manufacturer's recommendations from sorted cord-blood CD34+ cells using an RNeasy kit from QIAGEN. For reverse transcription, 200 ng of total RNA was used and cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad). cDNA was diluted 20-fold, 4 μL was amplified using the iQ SYBR Green Supermix (Bio-Rad) in a MyIQ thermocycler (Bio-Rad), and mRNA expression levels were quantified using MyIQ software (Bio-Rad). Hypoxanthine phosphoribosyltransferase, RPL27, and RPL30 expression levels were used to normalize samples and to calculate relative expression levels. Genome-wide expression analysis was performed on BeadChip arrays (Sentrix Human-6 46000-probe sets; Illumina). One microgram of mRNA was combined from 3 independent transduction experiments and used for the labeling reactions. Hybridization with the arrays was performed according to the manufacturer's instructions. Data were analyzed using the BeadStudio v3 gene expression module (Illumina) and Genespring (Agilent). The MIAME-compliant microarray data are available at http://www.ncbi.nlm.nih.gov/geo/ under accession number GSE26775.
Glucose-uptake analysis
2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG; Invitrogen N13195) was added to transduced cells at a final concentration of 10μM per 100 000 cells for 3 hours. Thereafter, cells were washed with phosphate-buffered saline, stained with CD34-APC and CD38-PerCP5.5 for 20 minutes, washed with phosphate-buffered saline again, and analyzed by FACS.
ChIP
ChIP was used to identify STAT5 binding to the HIF2α and cytokine-inducible SH2-containing protein (CISH) promoters, essentially according to established protocols. Detailed information and primer sequences can be found in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Statistical analysis
All values are expressed as means ± SE. The Student t test was used for all other comparisons. Differences were considered statistically significant at P < .05.
Results
Overexpression of STAT5 imposes a long-term proliferative advantage on HSCs but not on progenitors
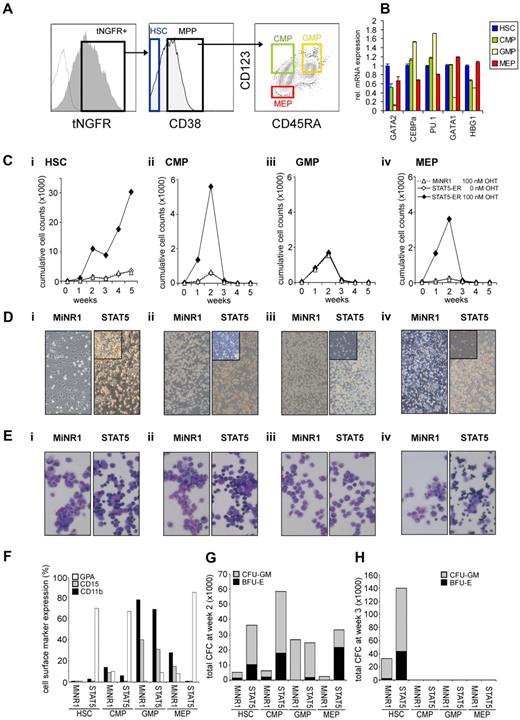
To gain further insight into whether STAT5-imposed long-term expansion is exclusively restricted to the HSC population or if it can also be imposed on progenitor cells, we overexpressed a 4-hydroxytamoxifen (4-OHT)–inducible STAT5 retroviral vector in human cord blood CD34+ cells.25 Cord blood CD34+ cells were transduced with the MiNR1 empty vector or with the STAT5A-ER retroviral vector, after which cells were sorted into HSC and progenitor fractions using the following cell-surface markers: CD34+/CD38low (HSCs), CD38+CD34+CD45RA−CD123+ (CMPs), CD38+CD34+CD45RA+CD123+ (GMPs), and CD38+CD34+CD45RA−CD123− (MEPs; Figure 1A). Upon in vitro culturing of human cord blood CD34+ cells for 3-5 days in serum-free hematopoietic progenitor growth medium supplemented with cytokines, the expression levels of CD38 decreased slightly and the expression levels of CD123 increased somewhat, which warrants caution in terms of our FACS phenotype–defined stem and progenitor subpopulations. Nevertheless, LTC-IC assays using day-5 cultured cord blood cells revealed that the majority of HSCs still resided within the CD34+/CD38− FACS-defined HSC population (Schuringa et al17 and data not shown). Furthermore, the purity of the sort was determined by CFC assays, as reported previously,27 and MEPs and GMPs were sorted to > 95% homogeneity. Quantitative polymerase chain reaction (Q-PCR) analysis was performed on myeloid and erythroid-specific genes, and these studies further confirmed good separation of the HSC and multipotent progenitor (MPP) populations (Figure 1B). In particular, CEBPα and PU.1 myeloid genes were detected in the CMP and GMP fraction, whereas there was a high expression of GATA-1 in the MEP fraction. From each sorted cell population, 4 × 104 cells were plated in long-term MS5 stromal cocultures in the presence or absence of 4-OHT. Cultures were demi-depopulated weekly, and expansion and differentiation of suspension cells were evaluated. As shown in Figure 1C, activation of STAT5 in HSCs resulted in a strong proliferative advantage over a period of 5 weeks. By the end of week 5, the cocultures initiated with STAT5-transduced HSCs displayed a 10-fold increase in cumulative expansion compared with MiNR1-transduced HSCs (Figure 1C). This increase in expansion was associated with the formation of CAFCs under the MS5 stroma that appeared within 1 week after plating and continued to be present throughout the 5-week culture period (Figure 1D), and secondary cultures could be established from these week-5 CAFCs (data not shown17,25 ). In contrast, only a transient proliferative advantage was observed in the cocultures initiated with STAT5-transduced CMP and MEP populations, and none of these progenitor cocultures was able to expand for longer than 3 weeks (Figure 1C). Interestingly, STAT5 overexpression did not induce increased expansion in GMPs (Figure 1C). In contrast to what was observed in HSCs transduced with STAT5, CAFCs were not observed in any of the progenitor subpopulations transduced with STAT5 (Figure 1D). The differentiation profile in the cultures was determined from the suspension cells by MGG staining and FACS analysis. Overexpression of STAT5 in HSC, CMP, and MEP compartments resulted in increased erythroid differentiation, whereby erythroblasts (in HSCs and CMPs) and orthochromatic normoblasts with distinct pyknotic nuclei (in MEPs) could be observed in MGG-stained cytospins at week 1 (Figure 1E). No erythroid differentiation was observed in STAT5-transduced GMPs (Figure 1E). FACS measurements were in agreement with these morphologic data and indicated increased percentages of GPA-positive cells in the STAT5-transduced HSC, CMP, and MEP fractions, whereas CD15 and CD11b percentages were decreased (Figure 1F). Expression of these cell-surface markers was not changed significantly in cocultures initiated with GMPs transduced with STAT5 (Figure 1F). The presence of progenitors in the cocultures was evaluated weekly by performing CFC assays with suspension cells. At week 2, a strong increase in CFC frequency was observed in HSCs, CMPs, and MEPs transduced with STAT5 (Figure 1G). At week 3, no CFCs were observed in any of the progenitor cultures, in agreement with expansion data indicating that after week 3, the cocultures initiated with transduced progenitor populations were exhausted (Figure 1G). Only cocultures initiated with HSCs were able to generate CFCs at weeks 3-5, when activation of STAT5 resulted in a strong increase in the number of CFCs (Figure 1F and data not shown).
STAT5 imposes a long-term proliferative advantage on human HSCs but not progenitors in MS5 stromal cocultures. (A) Human cord blood CD34+ cells were transduced with control and STAT5A-ER retroviral vectors, and tNFGR-positive cells were sorted into HSC, CMP, GMP, and MEP populations. (B) RNA was extracted from each sorted population and used for real-time Q-PCR analysis. (C) Sorted HSCs (i), CMPs (ii), GMPs (iii), and MEPs (iv) were plated on MS5 stromal cells, stimulated with 4-OHT as indicated, and the cultures were demi-depopulated weekly for analysis. Weekly cumulative cell counts are shown for a representative experiment of 3 independent experiments. (D) Representative images from MS5 cocultures at week 2 as described in panel C. (E) MGG-stained cytospins from week-1 suspension cells from cocultures as described in panel C. (F) FACS analysis on week-1 suspension cells from MS5 cocultures as described in panel C. (G-H) Suspension cells from MS5 cocultures as described in panel C were analyzed for progenitor content by CFC assay. At week 2 (G) and week 3 (H), 2500 cells from each coculture were plated in a CFC assay in methylcellulose, and colonies were evaluated 2 weeks after plating. Total CFC numbers are shown. CFU-GM, colony-forming unit–granulocyte-macrophage; BFU-E, burst-forming unit–erythroid.
STAT5 imposes a long-term proliferative advantage on human HSCs but not progenitors in MS5 stromal cocultures. (A) Human cord blood CD34+ cells were transduced with control and STAT5A-ER retroviral vectors, and tNFGR-positive cells were sorted into HSC, CMP, GMP, and MEP populations. (B) RNA was extracted from each sorted population and used for real-time Q-PCR analysis. (C) Sorted HSCs (i), CMPs (ii), GMPs (iii), and MEPs (iv) were plated on MS5 stromal cells, stimulated with 4-OHT as indicated, and the cultures were demi-depopulated weekly for analysis. Weekly cumulative cell counts are shown for a representative experiment of 3 independent experiments. (D) Representative images from MS5 cocultures at week 2 as described in panel C. (E) MGG-stained cytospins from week-1 suspension cells from cocultures as described in panel C. (F) FACS analysis on week-1 suspension cells from MS5 cocultures as described in panel C. (G-H) Suspension cells from MS5 cocultures as described in panel C were analyzed for progenitor content by CFC assay. At week 2 (G) and week 3 (H), 2500 cells from each coculture were plated in a CFC assay in methylcellulose, and colonies were evaluated 2 weeks after plating. Total CFC numbers are shown. CFU-GM, colony-forming unit–granulocyte-macrophage; BFU-E, burst-forming unit–erythroid.
Identification of STAT5 target genes in the HSC, CMP, GMP, and MEP populations
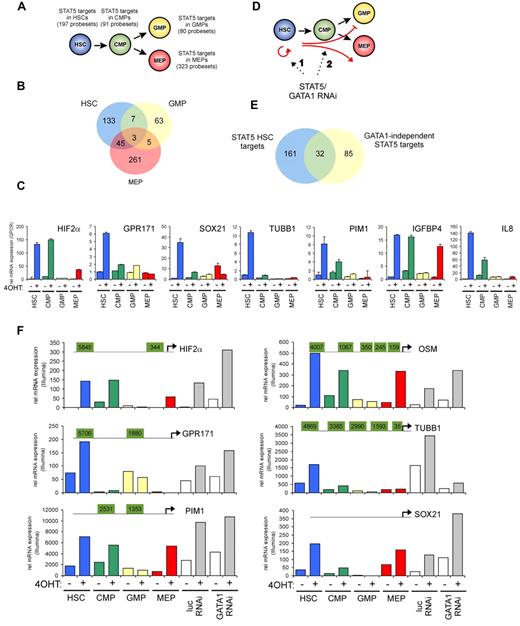
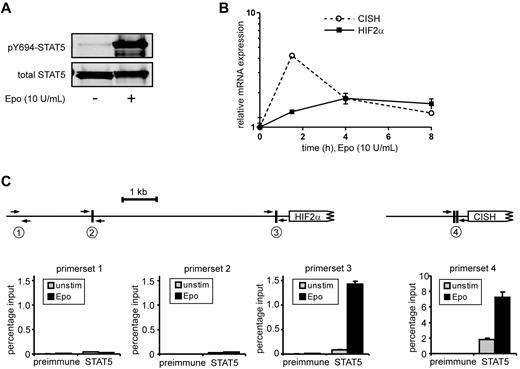
To identify STAT5 target genes in HSCs and progenitor populations, a genome-wide gene-expression profiling was performed using Illumina BeadChip arrays. Cord blood CD34+ cells were transduced with the STAT5A-ER and MiNR1 control vectors, and transduced cells were sorted into HSC and progenitor fractions. Sorted cells were stimulated with 100nM 4-OHT for 24 hours, and then RNA was isolated. Three independent transductions and stem cell/progenitor sorts were performed, and combined RNA fractions were used for hybridization with Illumina BeadChip arrays. Based on a > 2-fold gene expression change, 197 STAT5 probe sets were identified in the HSC compartment, 91 in the CMP fraction, 80 in the GMP fraction, and 323 in the MEP fraction (Figure 2A and supplemental Table 1). Remarkably, there was relatively little overlap between STAT5 target genes within the different stem cell and progenitor compartments, and the largest overlap in STAT5 target genes was observed between HSCs and MEPs (23%; Figure 2B). These data suggest that STAT5 fulfills distinct functions within these different compartments of the hematopoietic system. Several genes that were identified as STAT5 target genes within the HSC compartment were verified by Q-PCR analysis (Figure 2C). Next, we aimed to identify genes that would specifically associate with the long-term expansion phenotypes imposed on HSCs by STAT5. By down-modulating GATA1 in STAT5-transduced cord blood CD34+ cells, we have been able to dissect the erythroid differentiation phenotypes from HSC self-renewal phenotypes24 (Figure 2D). Lentiviral transduction of GATA1 RNAi vectors completely abrogated the STAT5-induced erythroid commitment, whereas long-term expansion, CAFC formation, and HSC self-renewal remained intact. Using this approach, we were able to identify 117 GATA1-independent STAT5 target genes.24 By comparing this list of GATA1-independent target genes with the STAT5 target genes in HSCs, we were able to identify 32 genes that would associate with long-term expansion and CAFC formation in the HSC compartment (Figure 2E and Table 1). Data for HIF2α, GPR171, PIM1, OSM, TUBB1, and SOX21 are shown in Figure 2F; also shown are potential STAT5-binding sites within the promoters of these genes, in which perfect palindromic STAT5-binding sites defined as TTC(n3)GAA are highlighted in green. Direct STAT5 binding to the HIF2α promoter was verified by ChIP experiments. UT7 cells were cytokine deprived overnight, stimulated with erythropoietin (10 U/mL) for 15 minutes to induce STAT5 tyrosine phosphorylation (Figure 3A), and real-time Q-PCR analysis was performed to confirm up-regulation of HIF2α expression (Figure 3B). As a positive control, the STAT5-induced up-regulation of CISH was also determined (Figure 3B). In parallel, lysates were prepared for ChIP. Immunoprecipitations were performed using antibodies against STAT5 or preimmune serum as a negative control. STAT5-bound DNA was analyzed by PCR using primer sets against the HIF2α promoter at a region without STAT5-binding sites (set 1), against the HIF2α promoter at regions that did contain perfect palindromic STAT5-binding sites (sets 2 and 3), and as a positive control, primers were used against the tandem STAT5-binding site within the CISH promoter (set 4; Figure 3C). As shown in Figure 3C, efficient STAT5 binding to the promoter region containing the STAT5 palindromic sequence located 344 bp upstream of the HIF2α start site, as well as to the CISH promoter, were observed. Region 1 was not bound by STAT5, and region 2, which did contain a perfect STAT5-binding site, was not efficiently coprecipitated. Thus, our data indicate that STAT5 can directly bind to the HIF2α promoter, and that the −344-bp site might be the STAT5 response element that regulates its expression.
Identification of STAT5 target genes in HSCs and progenitors. Cord blood cells were transduced with MiNR1 (control) and STAT5-wt ER retroviral vectors and sorted into HSC, CMP, GMP, and MEP fractions. Cells were stimulated with 100nM 4-OHT for 24 hours, and then RNA was extracted and used for Illumina BeadChip array analysis. Significantly expressed STAT5 target genes in HSC and progenitor compartments (> 2-fold change in gene expression) are shown. (B) Venn diagram showing STAT5 target genes in HSC, GMP, and MEP fractions. (C) Verification of STAT5 target genes by real-time Q-PCR. (D) Schematic representation of the identification of GATA1-independent STAT5 target genes. Cord blood CD34+ cells were transduced with MiNR1 or STAT5A-ER and luciferase RNAi or GATA1 RNAi vectors, as described previously.24 Cells were stimulated with 100nM 4-OHT for 24 hours, and then RNA was extracted and used for Illumina microarray analysis. Thus, significantly expressed STAT5 target genes could be identified in the absence or presence of GATA1 (> 2-fold change in gene expression). (E) Venn diagram showing STAT5 target genes in HSC and GATA1-independent target genes. (F) dataset from the Illumina BeadChip arrays showing expression of STAT5 target genes. Perfect palindromic STAT5-binding sites defined as TTC(n3)GAA are highlighted in green.
Identification of STAT5 target genes in HSCs and progenitors. Cord blood cells were transduced with MiNR1 (control) and STAT5-wt ER retroviral vectors and sorted into HSC, CMP, GMP, and MEP fractions. Cells were stimulated with 100nM 4-OHT for 24 hours, and then RNA was extracted and used for Illumina BeadChip array analysis. Significantly expressed STAT5 target genes in HSC and progenitor compartments (> 2-fold change in gene expression) are shown. (B) Venn diagram showing STAT5 target genes in HSC, GMP, and MEP fractions. (C) Verification of STAT5 target genes by real-time Q-PCR. (D) Schematic representation of the identification of GATA1-independent STAT5 target genes. Cord blood CD34+ cells were transduced with MiNR1 or STAT5A-ER and luciferase RNAi or GATA1 RNAi vectors, as described previously.24 Cells were stimulated with 100nM 4-OHT for 24 hours, and then RNA was extracted and used for Illumina microarray analysis. Thus, significantly expressed STAT5 target genes could be identified in the absence or presence of GATA1 (> 2-fold change in gene expression). (E) Venn diagram showing STAT5 target genes in HSC and GATA1-independent target genes. (F) dataset from the Illumina BeadChip arrays showing expression of STAT5 target genes. Perfect palindromic STAT5-binding sites defined as TTC(n3)GAA are highlighted in green.
GATA1-independent STAT5 target genes in human cord blood CD34+ HSCs
| Symbol . | Accession no. . | Fold change . | Definition . |
|---|---|---|---|
| C10orf128 | XM_931112.2 | 2.56 | chromosome 10 open reading frame 128 |
| C14orf139 | XR_017875.1 | 2.03 | chromosome 14 open reading frame 139 |
| CFH | NM_001014975.1 | 2.40 | complement factor H (CFH), transcript variant 2 |
| CISH | NM_145071.1 | 3.65 | cytokine inducible SH2-containing protein |
| CXCR4 | NM_003467.2 | 0.47 | chemokine (C-X-C motif) receptor 4 |
| EPAS1 | NM_001430.3 | 176.75 | endothelial PAS domain protein 1, HIF2α |
| FAM23B | NM_001013629.1 | 2.50 | family with sequence similarity 23, member B |
| FCGR2B | XM_938851.1 | 2.07 | Fc fragment of IgG, low affinity IIb, receptor |
| FILIP1L | NM_014890.2 | 3.40 | filamin A interacting protein 1-like |
| FILIP1L | NM_182909.2 | 2.87 | filamin A interacting protein 1-like |
| FRRS1 | NM_001013660.2 | 2.51 | ferric-chelate reductase 1 |
| GJA4 | NM_002060.2 | 0.47 | gap junction protein, alpha 4 |
| GPR171 | NM_013308.3 | 2.52 | G protein-coupled receptor 171 |
| GYPA | NM_002099.3 | 2.32 | glycophorin A |
| GYPB | NM_002100.3 | 3.70 | glycophorin B |
| GYPE | NM_198682.2 | 3.80 | glycophorin E |
| GYPE | NM_002102.3 | 3.62 | glycophorin E |
| HBE1 | NM_005330.3 | 4.69 | hemoglobin, epsilon 1 |
| HBZ | NM_005332.2 | 3.61 | hemoglobin, zeta |
| Hs.551847 | BE612775 | 2.00 | cDNA clone IMAGE:3856355 5 |
| HSPA6 | NM_002155.3 | 3.48 | heat-shock 70kDa protein 6 |
| IL18RAP | NM_003853.2 | 2.76 | interleukin 18 receptor accessory protein |
| LOC285016 | NM_001002919.2 | 3.57 | hypothetical protein LOC285016 |
| MGLL | NM_007283.5 | 0.49 | monoglyceride lipase |
| NRXN2 | NM_138734.1 | 0.43 | neurexin 2 |
| OSBP2 | NM_030758.3 | 3.17 | oxysterol binding protein 2 |
| OSM | NM_020530.3 | 22.21 | oncostatin M |
| PIM1 | NM_002648.2 | 4.05 | pim-1 oncogene |
| RAP1GAP | NM_002885.1 | 4.06 | RAP1 GTPase activating protein |
| SMAD6 | NM_005585.3 | 0.48 | SMAD family member 6 |
| SOD2 | NM_000636.2 | 2.19 | superoxide dismutase 2 |
| SOX21 | NM_007084.2 | 5.11 | SRY (sex determining region Y)-box 21 |
| TMEM158 | NM_015444.2 | 6.04 | transmembrane protein 158 |
| TUBB1 | NM_030773.2 | 2.96 | tubulin, beta 1 |
| Symbol . | Accession no. . | Fold change . | Definition . |
|---|---|---|---|
| C10orf128 | XM_931112.2 | 2.56 | chromosome 10 open reading frame 128 |
| C14orf139 | XR_017875.1 | 2.03 | chromosome 14 open reading frame 139 |
| CFH | NM_001014975.1 | 2.40 | complement factor H (CFH), transcript variant 2 |
| CISH | NM_145071.1 | 3.65 | cytokine inducible SH2-containing protein |
| CXCR4 | NM_003467.2 | 0.47 | chemokine (C-X-C motif) receptor 4 |
| EPAS1 | NM_001430.3 | 176.75 | endothelial PAS domain protein 1, HIF2α |
| FAM23B | NM_001013629.1 | 2.50 | family with sequence similarity 23, member B |
| FCGR2B | XM_938851.1 | 2.07 | Fc fragment of IgG, low affinity IIb, receptor |
| FILIP1L | NM_014890.2 | 3.40 | filamin A interacting protein 1-like |
| FILIP1L | NM_182909.2 | 2.87 | filamin A interacting protein 1-like |
| FRRS1 | NM_001013660.2 | 2.51 | ferric-chelate reductase 1 |
| GJA4 | NM_002060.2 | 0.47 | gap junction protein, alpha 4 |
| GPR171 | NM_013308.3 | 2.52 | G protein-coupled receptor 171 |
| GYPA | NM_002099.3 | 2.32 | glycophorin A |
| GYPB | NM_002100.3 | 3.70 | glycophorin B |
| GYPE | NM_198682.2 | 3.80 | glycophorin E |
| GYPE | NM_002102.3 | 3.62 | glycophorin E |
| HBE1 | NM_005330.3 | 4.69 | hemoglobin, epsilon 1 |
| HBZ | NM_005332.2 | 3.61 | hemoglobin, zeta |
| Hs.551847 | BE612775 | 2.00 | cDNA clone IMAGE:3856355 5 |
| HSPA6 | NM_002155.3 | 3.48 | heat-shock 70kDa protein 6 |
| IL18RAP | NM_003853.2 | 2.76 | interleukin 18 receptor accessory protein |
| LOC285016 | NM_001002919.2 | 3.57 | hypothetical protein LOC285016 |
| MGLL | NM_007283.5 | 0.49 | monoglyceride lipase |
| NRXN2 | NM_138734.1 | 0.43 | neurexin 2 |
| OSBP2 | NM_030758.3 | 3.17 | oxysterol binding protein 2 |
| OSM | NM_020530.3 | 22.21 | oncostatin M |
| PIM1 | NM_002648.2 | 4.05 | pim-1 oncogene |
| RAP1GAP | NM_002885.1 | 4.06 | RAP1 GTPase activating protein |
| SMAD6 | NM_005585.3 | 0.48 | SMAD family member 6 |
| SOD2 | NM_000636.2 | 2.19 | superoxide dismutase 2 |
| SOX21 | NM_007084.2 | 5.11 | SRY (sex determining region Y)-box 21 |
| TMEM158 | NM_015444.2 | 6.04 | transmembrane protein 158 |
| TUBB1 | NM_030773.2 | 2.96 | tubulin, beta 1 |
STAT5 binds directly to the −344-bp palindromic site within the HIF2α promoter. (A) UT7 cells were cytokine deprived overnight, after which time cells were either left unstimulated (unstim) or stimulated with erythropoietin (Epo; 10 ng/mL) for 15 minutes. Cell lysates were prepared and used for Western blotting. (B) Same experiment as in panel A, but mRNA was isolated from erythropoietin-stimulated cells at several time points, after which the expression of HIF2α and CISH was verified by real-time Q-PCR. (C) ChIP using antibodies against STAT5 or preimmune serum as a negative control. PCRs were performed using primer sets as indicated. Region 1 does not contain STAT5-binding sites, region 2 and 3 do contain STAT5-binding sites within the HIF2α promoter, and region 4 contains the tandem STAT5-binding site within the CISH promoter.
STAT5 binds directly to the −344-bp palindromic site within the HIF2α promoter. (A) UT7 cells were cytokine deprived overnight, after which time cells were either left unstimulated (unstim) or stimulated with erythropoietin (Epo; 10 ng/mL) for 15 minutes. Cell lysates were prepared and used for Western blotting. (B) Same experiment as in panel A, but mRNA was isolated from erythropoietin-stimulated cells at several time points, after which the expression of HIF2α and CISH was verified by real-time Q-PCR. (C) ChIP using antibodies against STAT5 or preimmune serum as a negative control. PCRs were performed using primer sets as indicated. Region 1 does not contain STAT5-binding sites, region 2 and 3 do contain STAT5-binding sites within the HIF2α promoter, and region 4 contains the tandem STAT5-binding site within the CISH promoter.
STAT5-induced long-term expansion, CFC numbers, and LTC-IC frequencies are impaired by down-modulation of HIF2α
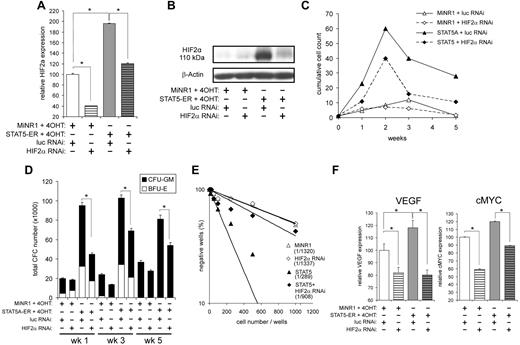
STAT5 significantly up-regulated HIF2α in HSCs in a GATA1-independent manner, which was confirmed both at the mRNA level (Figure 2C and 4A) and at the protein level (Figure 4B). Whereas HIF2α could also be up-regulated by STAT5 in CMPs, little up-regulation was observed in MEPs and no expression was observed in GMPs (Figure 2C and F). In contrast, no change in HIF1 mRNA expression was detected (data not shown). At normoxia (21% O2), very little HIF2α protein was observed in control cells (Figure 4B), possibly because of a combination of low levels of mRNA and degradation of hydroxylated HIF2α under these high-oxygen conditions. In contrast, STAT5 activation was sufficient to induce robust protein expression of HIF2α at normoxia, which was mediated at least in part by increased mRNA levels, but might also involve stabilization of the HIF2α protein (Figure 4B). To further study the involvement of HIF2α in the STAT5-induced phenotypes, we down-modulated HIF2α using a lentiviral RNAi approach. Cord blood CD34+ cells were cotransduced with MiNR1 control or STAT5-ER vectors together with pLVUT-luciferase-RNAi (a luciferase short-hairpin RNA control) or HIF2α-RNAi vectors. Double-transduced cells were sorted and, after 24 hours of stimulation with 100nM 4-OHT, total RNA was isolated from each sorted population. Down-modulation of HIF2α was confirmed by Q-PCR (Figure 4A) and by Western blotting (Figure 4B). Next, long-term cocultures on MS5 bone marrow stromal cells were initiated using double-transduced cord blood CD34+ cells. Down-modulation of HIF2α in STAT5-transduced cord blood cells resulted in a remarkable decrease in expansion throughout the 5-week culture period (Figure 4C). As expected, HIF2α down-modulation in control cord blood CD34+ cells did not significantly change the proliferation. Differentiation was also evaluated by FACS (CD11b, CD15, GPA, and CD71) and MGG staining of cytospins, but these experiments revealed that down-modulation of HIF2α did not change the differentiation pattern in either the STAT5-transduced or the MiNR1 control cells (data not shown). The decrease in expansion of STAT5-transduced cells upon down-modulation of HIF2α was also reflected by reduced CFC numbers at weeks 1, 3, and 5 (Figure 4D). There was no difference observed in the distribution of colony-forming unit granulocyte-macrophages or in burst-forming unit erythroid colonies. Annexin V staining was performed to determine whether decreased expansion was related to an increase in apoptosis, but no significant increase in Annexin V–positive cells was observed upon down-modulation of HIF2α in any of the groups (data not shown). Finally, stem-cell frequencies were determined in LTC-IC assays under limiting dilution conditions. Cells expressing activated STAT5 had an approximately 4-fold increase in LTC-IC frequency compared with controls (n = 3; MiNR1/Scr RNAi: 1/1322 ± 99; STAT5/Scr RNAi: 1/341 ± 43; an individual assay is shown in Figure 4E). When HIF2α was down-modulated in STAT5-transduced cells, LTC-IC frequencies were decreased approximately 2- to 3-fold compared with STAT5-transduced cells (n = 3; STAT5/Scr RNAi: 1/341 ± 43; STAT5/HIF2αRNAi: 1/836 ± 63; Figure 4E).
STAT5-induced long-term proliferation and increased CFC and LTC-IC frequencies are impaired by down-modulation of HIF2α. (A) Cord blood CD34+ cells were transduced with MiNR1 control or STAT5A-ER together with luciferase RNAi control or HIF2αRNAi constructs, and double-transduced cells were sorted. Sorted cells were stimulated with 100nM 4-OH for 24 hours, and then RNA was isolated. HIF2α mRNA expression levels were measured by Q-PCR. (B) Cord blood CD 34+ cells were transduced and sorted as described in panel A, but were used for Western blot analysis to determine HIF2α protein levels. (C) Double-transduced cord blood CD34+ cells as described in panel A were cultured in MS5 coculture. Cocultures were demi-depopulated weekly and cumulative cell counts are indicated. (D) Suspension cells from cocultures as described in panel C were analyzed in CFC assays. Data indicate total CFC numbers from a representative experiment of 3 independent experiments. (E) LTC-IC frequencies were determined in limiting dilution on MS5 stromal cells. Cultures were cultured for 5 weeks, and then methylcellulose was added. At week 7, LTC-IC frequencies were determined. Data shows the stem-cell frequencies of a representative experiment of 3 independent experiments. (F) RNA was isolated from double-transduced cells as described in panel A, RNA was isolated, and real-time Q-PCR was performed to identify HIF2α target genes. CFU-GM, colony-forming unit–granulocyte-macrophage; BFU-E, burst-forming unit–erythroid; *P < .05.
STAT5-induced long-term proliferation and increased CFC and LTC-IC frequencies are impaired by down-modulation of HIF2α. (A) Cord blood CD34+ cells were transduced with MiNR1 control or STAT5A-ER together with luciferase RNAi control or HIF2αRNAi constructs, and double-transduced cells were sorted. Sorted cells were stimulated with 100nM 4-OH for 24 hours, and then RNA was isolated. HIF2α mRNA expression levels were measured by Q-PCR. (B) Cord blood CD 34+ cells were transduced and sorted as described in panel A, but were used for Western blot analysis to determine HIF2α protein levels. (C) Double-transduced cord blood CD34+ cells as described in panel A were cultured in MS5 coculture. Cocultures were demi-depopulated weekly and cumulative cell counts are indicated. (D) Suspension cells from cocultures as described in panel C were analyzed in CFC assays. Data indicate total CFC numbers from a representative experiment of 3 independent experiments. (E) LTC-IC frequencies were determined in limiting dilution on MS5 stromal cells. Cultures were cultured for 5 weeks, and then methylcellulose was added. At week 7, LTC-IC frequencies were determined. Data shows the stem-cell frequencies of a representative experiment of 3 independent experiments. (F) RNA was isolated from double-transduced cells as described in panel A, RNA was isolated, and real-time Q-PCR was performed to identify HIF2α target genes. CFU-GM, colony-forming unit–granulocyte-macrophage; BFU-E, burst-forming unit–erythroid; *P < .05.
Several HIF target genes have been described that might potentially mediate the HIF2α-induced phenotypes, including VEGF and cMYC. We could indeed confirm that the expression of both VEGF and cMYC was down-modulated on transduction with HIF2α-RNAi vectors, both in MiNR1 control and in STAT5-transduced cord blood CD34+ cells (Figure 4F). No effects of STAT5 or HIF2α-RNAi were observed on the expression of HIF1α, HIF1β, or VHL (data not shown).
Glucose uptake is enhanced upon STAT5 activation
Because HIF signal transduction has been associated with alterations in glucose metabolism, particularly under hypoxic conditions, we next investigated whether STAT5 activation affected glucose uptake in human HSC and progenitor cells. As shown in Figure 5A and B, glucose uptake was significantly enhanced in cells expressing activated STAT5. The increase in glucose uptake was observed in both the HSC and the progenitor compartments. The expression of several glucose metabolism genes was then analyzed by real-time Q-PCR. A shown in Figure 5B, STAT5 activation resulted in the up-regulation of the glucose transporters GLUT1 and GLUT3, as well as in glycogen synthase 2 (GYS2) and UDP-glucose pyrophosphorylase 2 (UGP2). In GLUT1, GYS2, and UGP2, the STAT5-induced up-regulation was strongly mediated via HIF2α. Within the time frame of this experiment, no changes were observed in PDK2, PDK4, HK1, or HK2 (data not shown).
Glucose uptake is enhanced upon STAT5 activation. (A) Cord blood CD34+ cells were transduced with STAT5-ER and cells were stimulated with 100nM 4-OH for 24 hours as indicated. The glucose probe 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG) was added for 3 hours, and then glucose uptake was analyzed by FACS in the CD34+/CD38− HSC compartment and the CD34+/CD38+ multipotent progenitor (MPP) compartment. (B) Cord blood CD34+ cells were transduced with MiNR1 control or STAT5-ER together with luciferase RNAi control or HIF2α RNAi constructs, and double-transduced cells were sorted. Sorted cells were stimulated with 100nM 4-OH for 24 hours, and then RNA was isolated for real-time Q-PCR analysis. *P < .05.
Glucose uptake is enhanced upon STAT5 activation. (A) Cord blood CD34+ cells were transduced with STAT5-ER and cells were stimulated with 100nM 4-OH for 24 hours as indicated. The glucose probe 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG) was added for 3 hours, and then glucose uptake was analyzed by FACS in the CD34+/CD38− HSC compartment and the CD34+/CD38+ multipotent progenitor (MPP) compartment. (B) Cord blood CD34+ cells were transduced with MiNR1 control or STAT5-ER together with luciferase RNAi control or HIF2α RNAi constructs, and double-transduced cells were sorted. Sorted cells were stimulated with 100nM 4-OH for 24 hours, and then RNA was isolated for real-time Q-PCR analysis. *P < .05.
Discussion
Whereas various studies have highlighted the important role that STAT5 fulfills in normal hematopoiesis and in the development of leukemia, remarkably little has been revealed regarding the potential molecular mechanisms that are involved. By making use of an inducible model in which constitutive STAT5 activity can be introduced within specific compartments of the hematopoietic system, we have now shown that the STAT5-imposed long-term proliferative advantage and cobblestone formation are restricted to the HSC compartment. STAT5 was unable to induce long-term growth and cobblestone formation in myeloid or erythroid progenitor cells. Progenitor-initiated cultures showed only a transient STAT5-induced increase in cell numbers, and they could not be maintained for longer than 3 weeks regardless of constitutive STAT5 activity. To identify genes involved in the STAT5-induced phenotypes, we performed a genome-wide gene expression profiling on stem and progenitor cells, which resulted in the identification of 32 STAT5 target genes in the HSC compartment. One of the significantly altered genes was HIF2α. When HIF2α was down-modulated in STAT5-activated cells, we observed that the expansion of STAT5/HIF2α RNAi-transduced cells on MS5 bone marrow stromal cocultures was reduced, coinciding with reduced CFC and LTC-IC frequencies, whereas differentiation was not affected.
Recently, we demonstrated that the STAT5-induced long-term self-renewal phenotypes could be dissected from STAT5-induced erythropoiesis by lentiviral down-modulation of GATA1.24 This allowed the identification of STAT5-induced target genes associated with erythroid commitment, as well as target genes that did not associate with erythropoiesis. By comparing this last gene set with the list of target genes that were induced by STAT5 in HSCs, we were able to identify 32 genes that might potentially play a role in the long-term self-renewal phenotype induced by STAT5. HIF2α was studied in further detail because it has been shown to be overexpressed in various malignancies,28-30 because it plays a role in malignant transformation, and because it was overexpressed with the highest increase in the HSC compartment. Our data indicated that down-regulation of HIF2α reduced STAT5-induced cell proliferation, CFC numbers, and LTC-IC frequencies, whereas apoptosis and differentiation were not affected. Thus, the long-term phenotypes induced by STAT5 in HSCs are, at least in part, mediated via HIF2α. Under normoxic conditions, proline residues of HIF2 are hydroxylated, resulting in a reduction in protein levels via VHL-mediated proteasomal degradation. Under hypoxic conditions, such as in the presumed endosteal quiescent stem-cell niche, HIFs are stabilized and act as transcription factors.31 It is currently unknown whether and which HIF-induced target genes are essential to maintain the “stemness” of normal HSCs, but it was recently shown that in Hif1−/− mice HSC numbers decrease during stress, and this was associated with a loss of HSC quiescence.32 Another report indicated that HSCs in the quiescence niche use glycolysis for their energy demands, which depended on a Meis1-induced HIF1α-signaling network.33 Whether HIF1α and HIF2 α display similar or distinct functions in HSCs remains to be established.
Whereas HIF2α is normally only stabilized under hypoxic conditions, robust levels of HIF2α protein were observed in STAT5-transduced HSCs, even under normoxia. This was mediated via an up-regulation of HIF2α mRNA levels, but might also involve stabilization of the protein at normoxia, for example, via up-regulation of reactive oxygen species,34 because we have indeed observed increased reactive oxygen species levels in STAT5-transduced cells (our unpublished observations). HIF2α target genes might include those that provide adaptation to hypoxic stress and facilitate a transition from oxidative phosphorylation to glycolysis as a mode of ATP production. Glucose uptake was enhanced in STAT5-activated cells, and we observed that HIF2α is required for the STAT5-induced up-regulation of various genes associated with glucose metabolism, including GLU1, GYS2, and UGP2. These last 2 genes, GYS2 and UGP2, are involved in glycogen storage, which might ensure glucose availability for sustained ATP production. In addition, UGP2 is of particular interest because we identified this gene as one of the most differentially expressed genes in AML CD34+ cells compared with AML CD34− cells and normal bone marrow CD34+ cells (submitted manuscript). It has been demonstrated in lymphocytes that IL7 signaling can promote glucose uptake via a STAT5-dependent mechanism.35 Whereas we observed that STAT5 can directly bind to the HIF2 promoter and that STAT5-induced up-regulation of GLUT1, GYS2, and UGP2 depends on HIF2α, it is also possible that Akt activation is important, because it was shown that enhanced glucose uptake mediated via STAT5 transcriptional activity was impaired in Akt1-deficient T cells.35 Although not completely understood, enhanced glycolysis has been observed in many tumors, even under conditions in which there is enough oxygen available, and it is conceivable that HIF2α contributes to the proliferation of STAT5-transduced HSCs via a similar mechanism. In agreement with these data are the up-regulation of cMYC and VEGF in human cord blood CD34+ cells by STAT5 in an HIF2α-dependent manner. Although so far PIM1 has only been described as a HIF1α target gene,36,37 we do find that PIM1 is also up-regulated in STAT5-transduced HSCs. In addition to the growth-promoting activity of these genes, it has been suggested that HIF2α might contribute to the growth of tumor cells via activation of EGFR and IGFR1 tyrosine kinases.38 Future studies will reveal whether targeting of HIF2α will be sufficient to impair self-renewal of STAT5-transfomed leukemic stem cells as well.
The phenotype of the leukemic stem cell is under continuous debate, and it is not always clear whether leukemias arise from genetic mutations in HSCs or in progenitors. Our current data indicate that HSCs, but not myeloid or erythroid progenitors, are the target cells that are susceptible for long-term expansion imposed by STAT5. These data are in agreement with previous observations indicating that constitutive activation of STAT5 increased the number of self-renewing CAFCs that contained replating potential, particularly within the immature CD34+/CD38low compartment.17 In mice, a fatal myeloproliferative disease could be induced when activating STAT5 mutants were expressed in the primitive CD34−Lin−Sca1+c-Kit+ population, but not when expressed in committed progenitors.16 Apparently, STAT5 is not sufficient to reinstall a self-renewal program on progenitor cells that have already lost their self-renewal potential. This is in contrast to other oncogenes such as MLL-AF9 and MLL-ENL, which, particularly when expressed at high levels,39 can efficiently transform progenitor cells by reactivating an “embryonic stem cell–like” gene-expression program.40-42 This gene expression program includes, among others, Myb, Hmgb3, and Cbx5, which, next to the HoxA/Meis gene expression program, appear to be sufficient to impose self-renewal on progenitor cells and ultimately induce leukemic transformation. We indeed did not observe an up-regulation of these genes in STAT5-transduced HSCs or MPPs. In agreement with our observations, it has become clear that the BCR-ABL fusion protein also lacks the capability to reprogram more differentiated progenitor cells.43 More recently, using transgenic models in which BCR-ABL p210 was exclusively expressed in the immature Sca1+ compartment, it was shown that expression only in HSCs but not progenitors is sufficient to induce leukemic transformation.44
In our microarray analyses, we observed that STAT5 target genes did not overlap significantly among HSCs, CMPs, GMPs, and MEPs. Thus, our data show that STAT5 activates quite different sets of genes in HSCs and progenitors, which could explain why the STAT5-induced phenotypes are distinct in the different compartments. In our coculture assays, we observed that no growth advantage was imposed on GMPs. This is somewhat remarkable, because various myeloid growth factors such as IL3 and GM-CSF are quite efficient in activating STAT5 in myeloid cells. Initially, in STAT5−/− animals no defects were observed in myelopoiesis, suggesting that STAT5 is dispensable for myeloid development,10 although later it was shown that granulopoiesis under myelosuppressive conditions was impaired.45 Further studies are required to elucidate why constitutive activation of STAT5 appears insufficient to alter phenotypes in GMPs under steady-state conditions. Furthermore, it is noteworthy that whereas STAT5 activation efficiently induced erythroid commitment from HSC, CMP, and MEP compartments, which is in agreement with previously published data,17,25,46 no erythroid differentiation could be induced by STAT5 in GMPs. Apparently, GMPs have differentiated beyond a stage at which an erythroid-associated gene expression program can be induced by STAT5. Although it still needs to be elucidated why STAT5 target genes within HSC, CMP, GMP, and MEP compartments are so diverse, it is plausible that a tissue-specific expression of STAT5 cofactors, repressors, additional transcription regulators, or the epigenetic status of gene-regulatory elements are involved, which will certainly be a focus in future studies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We greatly appreciate the help of Dr J. J. Erwich and Dr A. van Loon and colleagues at the Obstetrics Department of University Medical Center in Groningen, Martini Hospital Groningen, for collecting cord blood. We also thank Annet Vos for help with cloning the HIF2α RNAi vector.
This work was supported by grants from the The Netherlands Organisation for Scientific Research (NWO) Innovational Research Incentives Scheme Veni 2004 and Vidi 2008 and from the Koningin Wilhelmina Fonds (KWF; Rijksuniversiteit Groningen [RUG] 2009-4275).
Authorship
Contribution: S.F. and A.T.J.W. performed experiments and analyzed data; S.M.G.J.D and E.V. analyzed and discussed data; and S.F. and J.J.S. designed the experiments, analyzed and discussed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr J. J. Schuringa, Division of Hematology, Department of Medicine, University Medical Center Groningen, Hanzeplein 1, 9713 GZ Groningen, The Netherlands; e-mail: j.schuringa@int.umcg.nl.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal